Abstract
Next generation sequencing (NGS) has been used to characterize the overall genomic landscape of melanomas. Here, we systematically examined mutations from recently published melanoma NGS data involving 241 paired tumor-normal samples to identify potentially clinically relevant mutations. Melanomas were characterized according to an in-house clinical assay that identifies well-known specific recurrent mutations in five driver genes: BRAF (affecting V600), NRAS (G12, G13, and Q61), KIT (W557, V559, L576, K642, and D816), GNAQ (Q209), and GNA11 (Q209). Tumors with none of these mutations are termed “pan-negative”. We then mined the driver mutation-positive and pan-negative melanoma NGS data for mutations in 632 cancer genes that could influence existing or emerging targeted therapies. First, we uncovered several genes whose mutations were more likely associated with BRAF- or NRAS-driven melanomas, including TP53 and COL1A1 with BRAF, and PPP6C, KALRN, PIK3R4, TRPM6, GUCY2C, and PRKAA2 with NRAS. Second, we found that the 69 “pan-negative” melanoma genomes harbored alternate infrequent mutations in the 5 known driver genes along with many mutations in genes encoding guanine nucleotide binding protein α-subunits. Third, we identified 12 significantly mutated genes in “pan-negative” samples (ALK, STK31, DGKI, RAC1, EPHA4, ADAMTS18, EPHA7, ERBB4, TAF1L, NF1, SYK, and KDR), including 5 genes (RAC1, ADAMTS18, EPHA7, TAF1L, and NF1) with a recurrent mutation in at least 2 “pan-negative” tumor samples. This meta-analysis provides a road map for the study of additional potentially actionable genes in both driver mutation-positive and pan-negative melanomas.
Keywords: Melanoma, Next-generation sequencing, Meta-analysis, Driver mutation, BRAF, NRAS, KIT, GNA11, GNAQ
Introduction
Melanoma is a malignant cancer of melanocytes. The disease accounts for 80% of deaths from skin cancer (1), with an estimated 76,690 new patients and 9,480 deaths expected in the United States in 2013 (1). The high mortality rate of advanced melanoma is largely due to its aggressive behavior and limited treatment options. As such, the 5-year survival rate for patients with advanced melanoma has historically been below 15% (2).
Melanoma has been traditionally classified based on clinical and histopathological characteristics, such as growth pattern and anatomic site of origin. More recently, driver mutations in genes encoding signaling proteins that activate the mitogen-activated protein kinase (MAPK) pathway (3) have been implicated in the pathogenesis of a majority of melanomas. Accordingly, we recently developed a melanoma-specific multiplex mutational profiling assay to detect 43 recurrent mutations in 6 genes relevant to targeted therapy for the disease (4). This SNaPshot assay, used routinely in our clinic, detects mutations in BRAF (at position V600), NRAS (G12/13, Q61), KIT (W557, V559, L576, K642, D816), GNAQ (Q209) and GNA11 (Q209) in tumors. We chose these specific mutations because 1) they occur in melanoma at a frequency of greater than 1% and 2) they have relevance to emerging or targeted therapies such as the BRAF inhibitors, vemurafenib and dabrafenib, the MEK inhibitor, trametinib, and the KIT inhibitor, imatinib. Using this assay, we have found that one-third of tumors lack any of these mutations and are herein referred to as “pan-negative” (4). Continued investigation for novel driver mutations in these cases is critical to improve therapeutic outcomes for patients. For example, we recently found that ~8% of “pan-negative” cases harbor non-V600 exon 15 BRAF mutations (5) and 4–8% contain activating BRAF fusions, both of which may be sensitive to MEK inhibition (6).
Rapid advances in next-generation sequencing (NGS) technologies have made comprehensive characterization of cancer genomes feasible (7–9). A large number of whole exome sequencing (WES) and whole genome sequencing (WGS) studies have already been performed in melanoma (10–18). Here, we analyzed 241 paired melanoma tumor/normal tissue samples from six recently published WES and WGS studies to identify mutation signatures associated with melanomas with the most common driver genes (i.e. BRAF and NRAS) and those without known recurrent BRAF, NRAS, KIT, GNAQ, or GNA11 mutations. This analysis provides important insight into the molecular events associated with melanomas and has identified potential therapeutic targets among “pan-negative” melanomas.
Materials and Methods
Melanoma NGS data collection and process
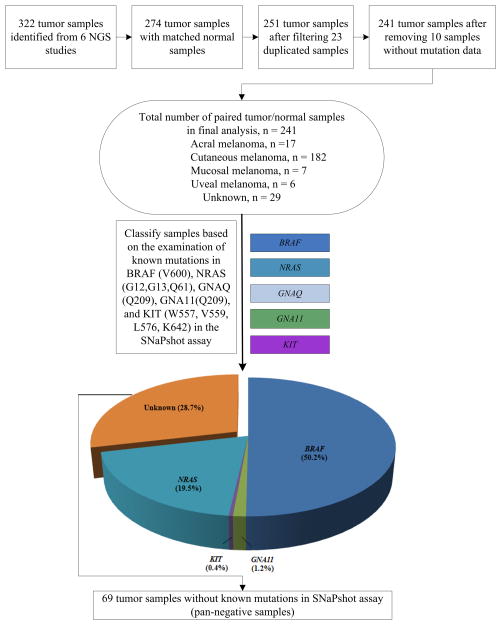
Similar to data collection procedures in our NGS Catalog database (8), we conducted a comprehensive literature search of melanoma NGS studies using the keywords “exome sequencing AND melanoma” and “whole genome sequencing AND melanoma” through PubMed (http://www.ncbi.nlm.nih.gov/pubmed). We performed a careful manual check of the search results. Our query revealed at least ten melanoma NGS studies published from 2010 to 2012 (as of September, 2012, right before we started the analysis) (8). Studies were excluded if only part of the NGS mutation data was available. The mutation data from (11) was not included in our study because only one tumor-normal pair was sequenced and it harbored the known driver mutation, BRAF V600E. Duplicate data were filtered by examining authors’ names and affiliations and tumor name/ID. As a result, 6 melanoma WGS or WES studies (12, 14–17, 19) were collected for our meta-analysis (Figure 1, Supplementary Table S1). The sequencing quality of these melanoma genomes/exomes was high, with the validation rate estimated to be > 95% in most of these studies.
Figure 1.
Flow diagram of the tumor sample selection and classification.
The number of sequenced tumor samples varied among the 6 studies, ranging from 7 to 121 samples. Here, we only used the NGS data from the tumors that had matched normal tissues in the same study. In addition, 23 of the 25 melanoma samples in (14) were sequenced in another study (19), so these 23 duplicated samples in (14) were removed in our study. The mutation rate is high in melanoma tumor genomes compared to other types of tumor genomes (9). Surprisingly, no somatic mutation data were identified in 10 melanoma samples in (15), most of which (6 out of 10) were mucosal or acral. Therefore, those samples were also excluded. In total, we analyzed NGS data from 241 tumor samples with mutation information, along with their matched normal samples (Figure 1, Table 1). Among them, 182 originated from cutaneous sites, 17 from acral sites, 7 from mucosal sites, 6 from uveal sites, and 29 from unknown primary sites (Supplementary Table S2).
Table 1.
Mutated genes associated with BRAF-mutated samples (frequency ≥ 10% and p-value < 0.05)
| Gene | # samples with BRAF mutation (N = 130) | Frequency (%) | # samples without BRAF mutation (N = 111) | Frequency (%) | p-value |
|---|---|---|---|---|---|
| TTN | 84 | 64.6 | 54 | 48.7 | 0.009 |
| TP53 | 28 | 21.5 | 11 | 9.9 | 0.011 |
| COL1A1 | 17 | 13.1 | 6 | 5.4 | 0.034 |
Genes were ordered by p-values.
Extraction of melanoma mutation data
The somatic mutation data were downloaded from the supplementary section of each of the 6 publications (12, 14–17, 19). For the current analysis, we primarily focused on somatic single nucleotide variants (SNVs) because they constitute the largest fraction of oncogenic drivers identified in melanoma. For these SNVs, we collected all non-silent parts, including missense, nonsense, nonstop, and splice site mutations, as well as accompanying information such as tumor name, tumor type, and gene name. We generally refer to these SNVs as mutations in this article.
Selection of candidate genes for analysis
NGS has enabled the identification of tens of thousands of somatic mutations in melanoma genomes (10). As a result, sifting through these mutations to pin down the few driver mutations present in melanoma is a challenge; therefore we generated a list of candidate genes to further analyze in the published data sets. We first compiled a list of genes that were potentially related to melanoma from literature reports (2, 20–23). Since mutations in protein kinase genes have been frequently reported in cancers (24), and they are often the targets in the design of antitumor therapies (25), we also included the 612 genes from Agilent’s SureSelect Human Kinome targeted NGS kit (http://www.genomics.agilent.com/en/home.jsp) in our candidate gene list. The resulting list contained 632 genes for further analysis (see Supplementary File S1). Unless specifically noted, the analysis below was conducted using these candidate genes.
Clinically relevant driver mutations and mutation analysis
Currently, Vanderbilt University Medical Center utilizes the SNaPshot genotyping assay to screen for 43 specific point mutations in 6 genes (BRAF, NRAS, KIT, GNAQ, GNA11, and CTNNB1) in melanoma (4) that are relevant to current and emerging targeted therapies in melanoma except for CTNNB1. Excluding CTNNB1 (CTNNB1 mutations commonly co-occur with mutations in the other 5 genes), we analyzed the melanoma NGS data against these drivers to determine mutations associated with these 5 driver genes, as well as to uncover potential novel drivers in “pan-negative” samples [i.e., samples which lack all the known, recurrent mutations in BRAF (V600), NRAS (G12, G13, and Q61), KIT (W557, V559, L576, K642, D816), GNAQ (Q209), and GNA11 (Q209)]. In-house Perl scripts were developed to analyze these data and a single-sided Fisher’s exact test was used to assess the significance of mutation association.
Results
Spectrum of known driver mutations in melanoma
To classify melanoma genomes according to our clinical SNaPshot-based assay, we queried WGS and WES data from 241 melanoma samples for known driver mutations in BRAF (V600), NRAS (G12/13, Q61), KIT (W557, V559, L576, K642, and D816), GNAQ (Q209) and GNA11 (Q209). Supplementary Table S2 summarizes the number of tumors, the tumor subtypes, and known driver mutation(s) that each tumor harbored. Briefly, 50.2% (121/241) tumors were found to harbor BRAF V600 mutations (Figure 1). Among them, 86.8% (105/121) had V600E missense mutations. Fifteen had V600K mutations (12.4%) and one had a V600R mutation (0.8%). Forty-seven samples (19.5%) had NRAS mutations, including Q61 mutations [44/47 (93.6%): Q61R (22/47, 46.8%), Q61K (12/47, 25.5%), Q61L (6/47, 12.8%), and Q61H (4/47, 8.5%)] and G12 mutations [3/47 (6.4%): G12V (2/47, 4.3%) and G12D (1/47, 2.1%)]. No G13 mutations were detected. Three uveal melanoma samples (3/241, 1.2%) had GNA11 Q209L mutations. Only one tumor (1/241, 0.4%) had a KIT mutation (V559A). No mutations were found in GNAQ. The remaining samples were wild type (WT) for these mutation hotspots. These data indicate that the 241 pooled genomes were generally representative of a typical melanoma population (4–5, 21).
Mutated genes associated with mutant BRAF in melanoma
We next sought to identify mutations that co-occurred with mutant BRAF in this dataset. From a biological standpoint, co-occurring mutations could contribute to disease progression and/or metastasis. From a clinical standpoint, such mutations could modify clinical responses to single-agent targeted therapies and/or contribute to drug resistance (26–27). Table 1 lists mutations that occurred in at least 10% of BRAF-mutated melanomas with a p-value < 0.05 (Fisher’s exact test). Genes were ranked according to their p-values for different frequencies between samples with and without BRAF mutations.
Mutations in the gene TTN were the most significantly associated with BRAF mutation, occurring in 64.6% of samples (p = 0.009). Encoding a functional component of striated muscle, TTN is the longest gene in the human genome (24). TTN mutations are often seen in cancer NGS studies and are likely passenger mutations due to gene length bias (9). The next most frequently mutated gene was TP53, found in 21.5% of BRAF-mutated tumors (p-value = 0.011). Although TP53 is a highly recognized tumor suppressor gene harboring numerous mutations, it is rarely mutated in melanoma relative to other cancers (28). The association between BRAF and TP53 mutations supports a previous report (29) based on Sanger resequencing of selected genes in melanoma cell lines. We used MutationAssessor (30) to predict the functional relevance of these TP53 mutations to melanoma. All missense mutations in TP53 were predicted to have a medium impact on the protein function. Since a nonsense mutation may lead to the loss of function of the encoding gene, we further examined the distribution of nonsense mutations in TP53 in the samples with versus without BRAF mutations. BRAF mutant samples harbored more nonsense mutations than BRAF wild-type samples (5.4% vs. 2.7%). The third most commonly mutated gene was COL1A1, which encodes a component of type I collagen (occurring in 13.1% of BRAF-mutated tumors, p = 0.034). Thus far, there is no evidence suggesting that COL1A1 is associated with melanoma tumorigenesis.
Mutated genes associated with mutant NRAS in melanoma
Table 2 shows the mutated genes associated with NRAS mutations (occurred in at least 10% of NRAS-mutated melanomas and p-value < 0.05, Fisher’s exact test). Aside from TTN, this list of mutated genes was surprisingly very different from the list associated with BRAF mutant tumors. For example, alterations in the cell cycle effector, PPP6C were frequently observed in NRAS-mutated melanomas, with a frequency of 17.7% and p = 0.011. This gene was recently found to be a novel candidate melanoma gene harboring putative somatic driver mutations (19). The genes encoding the Rho-GEF kinase, KALRN, and the phosphoinositide-3 kinase, PIK3R4, which ranked third and fourth in this association list based on p-values, were mutated in 27.5% and 11.8% of NRAS-mutated melanomas, respectively. Neither of these two genes has yet been reported to be involved in melanomagenesis. The fifth gene was TRPM6 (27.5%, p =0.020); its role in melanoma remains unknown, but other members of the Transient Receptor Potential ion channel subfamily M gene family (TRPM1, TRPM2, TRPM7 and TRPM8) have been proposed to play a crucial role in melanoma (31). The enterotoxin receptor, GUCY2C, which is an emerging prognostic molecular biomarker in colorectal cancer (32), was also found to co-occur with NRAS mutations (13.7%, p-value = 0.021) in melanoma. Finally, we observed that 13.7% of all NRAS mutations identified were coincident with mutations in PRKAA2 (p = 0.043), which encodes the catalytic subunit of AMP-activated protein kinase (AMPK). We further assessed the functional impact of the 8 PRKAA2 missense mutations that were found to be associated with the NRAS mutations. Four missense mutations in PRKAA2 were predicted to have a medium impact on the protein function by MutationAssessor. Shen et al. (33) reported that PRKAA2 (AMPK) is the long-sought kinase for BRAF Ser729. BRAF phosphorylation by PRKAA2 could attenuate BRAF signaling and inhibit cell proliferation. The mutation association we found here might suggest a new mechanism of tumorigenesis involving inhibitory mutations of PRKAA2 in BRAF wild-type, NRAS-mutant melanoma; however, future biological analyses outside the scope of this study are needed to confirm this concept. In addition, a recent study (34) suggested that PRKAA2 plays a potential role in cancer progression and may represent a possible therapeutic target for the treatment of early gastric cancer.
Table 2.
Mutated genes associated with NRAS-mutated samples (frequency ≥ 10% and p-value < 0.05)
| Gene | # samples with NRAS mutation (N = 51) | Frequency (%) | # samples without NRAS mutation (N = 190) | Frequency (%) | p-value |
|---|---|---|---|---|---|
| TTN | 37 | 72.6 | 101 | 53.2 | 0.009 |
| PPP6C | 9 | 17.7 | 11 | 5.8 | 0.011 |
| KALRN | 14 | 27.5 | 24 | 12.6 | 0.012 |
| PIK3R4 | 6 | 11.8 | 5 | 2.6 | 0.013 |
| TRPM6 | 14 | 27.5 | 26 | 13.7 | 0.020 |
| GUCY2C | 7 | 13.7 | 8 | 4.2 | 0.021 |
| PRKAA2 | 7 | 13.7 | 10 | 5.3 | 0.043 |
Genes were ordered by p-values.
For the above genes that were found to be significantly associated with BRAF or NRAS mutations, we manually reviewed their results in individual studies for reliability. Because most samples used in this study were from Hodis et al. (19) (121 paired tumor-normal samples) and Krauthammer et al. (15) (89 paired tumor-normal samples), we examined the mutations that co-occurred with mutant BRAF or NRAS only in these two studies (the details are available in Supplementary Tables S3–S10). We found some results in Table 2 were consistently observed, including PIK3R4, PPP6C, TRPM6, and KALRN using the Hodis et al. data (Supplementary Table S8) and TTN using the Krauthammer et al. data (Supplementary Table S10).
It is known that some driver mutations are associated with sun exposure while others are more commonly found in sun protected areas. We explored whether the observed mutation association was due to ultraviolet (UV) induction or a specific driver mutation in genes such as BRAF and NRAS. Specifically, we analyzed the mutation association patterns in each melanoma subtype (acral, mucosal, uveal, cutaneous, or unknown type). Three genes (TTN, TP53, and COL1A1) that were associated with BRAF-mutant melanomas in the whole cohort did not co-occur with mutant BRAF in any melanoma subtype (Supplementary Table S11). This lack of association might be due to small sample sizes in each subtype. However, we did not observe a statistically significant association in cutaneous melanoma either, which had 109 samples. This preliminary analysis suggested that the genes we reported are more likely due to the association with the driver gene BRAF, rather than sun exposure.
Interestingly, almost all genes associated with NRAS mutation (except PRKAA2) in melanomas (Table 2) were consistently observed in cutaneous melanoma (Supplementary Table S12). Since cutaneous melanoma is mainly caused by UV damage, we could not reject the hypothesis that the NRAS-associated genes (TTN, PPP6C, KALRN, PIK3R4, TRPM6, and GUCY2C) are due to sun exposure. Considering that UV specifically causes C>T transitions (i.e. C changes to T), we further compared C>T mutation frequency in NRAS mutant samples versus NRAS wild-type samples. The C>T frequency in NRAS mutant samples (80.7%) was slightly larger than that in NRAS wild-type samples (78.9%). For the purpose of comparison, the C>T frequency in BRAF mutant samples (78.3%) was slightly smaller than that in BRAF wild-type samples (80.5%). Further studies are needed to determine the influence of sun exposure on the association of mutations in NRAS-mutant melanomas.
Mutations enriched in “pan-negative” melanoma samples
Rare mutations in BRAF, NRAS, KIT, GNAQ, and GNA11
As stated above, our clinical SNaPshot-based assay examines tumors for mutations in BRAF (V600), NRAS (G12/13, Q61), KIT (W557, V559, L576, K642, D816), GNAQ (Q209) and GNA11 (Q209). A total of 69 tumors (28.6%) of the 241 tumor/normal pairs were “pan-negative” according to this mutation screening strategy (Figure 1). To identify other potential mutations in these genes, we interrogated the 69 tumor/normal pairs for non-hotspot mutations in BRAF, NRAS, KIT, GNAQ, and GNA11 (Table 3, Figure 2, and Supplementary Figures S1–S4). Two samples with distinct BRAF L597 mutations were identified. This frequency (2/69 = 2.9%) is consistent with our previous report, in which we identified two BRAF L597 mutations in 49 SNaPshot pan-negative samples (4.1%) upon screening of BRAF exon 15 (5). Six additional non-V600 BRAF mutations were found in these 69 pan-negative melanoma samples, including K601E (2 samples) (5), P75L (1 sample), G469R (1 sample) (35), N581T (1 sample), G593S (1 sample), and P731S (1 sample) (36). Some of these are already known to activate the MAPK pathway and confer sensitivity to MEK inhibition (5, 36). Thus, in this dataset, 10.1% (7 of 69) of pan-negative melanomas harbored non-V600 BRAF mutations, some of which are already suspected to benefit from treatment with targeted agents (37). This proportion (10.1%) was significantly higher than that in driver mutation-positive melanomas (3.5%, 6 of 172) (p = 0.039, Fisher’s exact test). In addition, 3.3% (4 of 121) and 4.3% (2 of 47) of melanomas with BRAF V600 and NRAS G12/G13/Q61 mutations, respectively, harbored non-V600 BRAF mutations. These two proportions were not significantly higher than that of their respective wild-type samples harboring non-V600 BRAF mutations: p = 0.960, Fisher’s exact test, BRAFV600 melanomas versus non-BRAFV600 melanomas (7.5%, 9 of 120); and p = 0.761, Fisher’s exact test, NRAS G12/G13/Q61 mutant melanomas versus non-NRAS G12/G13/Q61 melanomas (5.7%, 11 of 194). The rate of non-V600 BRAF mutations in the whole cohort is 5.4% (13 of 241).
Table 3.
Additional non-hot spot mutations detected in BRAF, NRAS, KIT, GNAQ, and GNA11 in the 69 pan-negative melanoma samples
| Gene | cDNA change | Amino acid change | Tumor name | Tumor subtype | Reference |
|---|---|---|---|---|---|
| BRAF | c.224C>T | p.P75L | YUKLAB | Cutaneous | Krauthammer et al. (15) |
| BRAF | c.1405G>A | p.G469R | MEL-Ma-Mel-48-Tumor | Cutaneous | Hodis et al. (19) |
| BRAF | c.1742A>C | p.N581T | YULAN | Cutaneous | Krauthammer et al. (15) |
| BRAF | c.1777G>A | p.G593S | LAU165 | Cutaneous | Nikolaev et al. (16) |
| BRAF | c.1789_1790CT>TC | p.L597S | MEL-JWCI-WGS-13-Tumor | Cutaneous | Hodis et al. (19) |
| BRAF | c.1790T>G | p.L597R | LAU165 | Cutaneous | Nikolaev et al. (16) |
| BRAF | c.1801A>G | p.K601E | ME017-Tumor | Cutaneous | Hodis et al. (19) |
| BRAF | c.1801A>G | p.K601E | LAU63 | Cutaneous | Nikolaev et al. (16) |
| BRAF | c.2191C>T | p.P731S | ME017-Tumor | Cutaneous | Hodis et al. (19) |
| NRAS | c.392A>G | p.H131R | MEL-Ma-Mel-119-Tumor | Cutaneous | Hodis et al. (19) |
| NRAS | c.64C>A | p.Q22K | MEL-Ma-Mel-103b-Tumor | Cutaneous | Hodis et al. (19) |
| KIT | c.1715T>C | p.L572P | YUBRUSE | Acral | Krauthammer et al. (15) |
| KIT | c.1912A>G | p.K638E | YUSAG | Acral | Krauthammer et al. (15) |
| KIT | c.2452A>T | p.N818Y | YURUB | Acral | Krauthammer et al. (15) |
| GNAQ | c.547C>T | p.R183X | MEL-JWCI-WGS-7-Tumor | Cutaneous | Hodis et al. (19) |
| GNAQ | c.553C>T | p.P185S | YUKLAB | Cutaneous | Krauthammer et al. (15) |
| GNAQ | c.632C>T | p.S211L | YUGADID | Uveal | Krauthammer et al. (15) |
| GNAQ | c.905C>G | p.A302G | YUKLAB | Cutaneous | Krauthammer et al. (15) |
| GNA11 | c.547C>T | p.R183C | YUBOO | Uveal | Krauthammer et al. (15) |
Figure 2. Location of the additional mutations detected in BRAF in the 69 pan-negative melanoma samples.
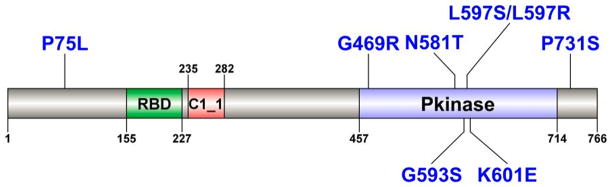
RBD: Raf-like Ras-binding domain (amino acids 155–227); C1_1: phorbol esters/diacylglycerol binding domain (amino acids 235–282); Pkinase: kinase domain (amino acids 457–714). Protein domain information was obtained from the Pfam database (http://pfam.sanger.ac.uk) and was visualized using DOG software (http://dog.biocuckoo.org). Mutations are indicated in blue.
Two somatic nonsynonymous mutations were identified in NRAS (Q22K and H131R), and three somatic nonsynonymous mutations were identified in KIT (L572P, K638E, and N818Y). The NRAS mutations were found in disease from sun damaged skin, and all KIT mutations were found in melanomas from acral sites. Additional GNAQ mutations included R183X, P185S, S211L, and A302G, of which P185S and A302G were present in a single patient. Interestingly, only one of these four GNAQ mutations originated from a uveal site; the other three GNAQ mutations were found in sun damaged skin. Only one GNA11 mutation (R183C) was identified in a uveal melanoma.
Somatic mutations in genes encoding other guanine nucleotide binding protein α-subunits (GNAs)
GNA11 and GNAQ encode GNAs, the α-subunits of G proteins (38), which play an important role in cell signaling as well as tumor initiation and progression (39). Recently, activating mutations in GNAS (another GNA) in breast cancer, colorectal cancer, and ovarian cancer were reported (40–41). Further interrogation of the “pan-negative” melanoma samples revealed 19 mutations in 9 genes encoding GNAs other than GNAQ and GNA11 (Supplementary Table S13). Interestingly, 17 of the 19 GNA mutations were found in cutaneous melanomas, unlike the known recurrent mutations in GNAQ and GNA11, which are primarily associated with uveal melanomas (42–43). Notably, 13 of these 19 GNA mutations are caused by C>T (G>A) mutations that are compatible with UV-mediated damage, providing evidence for UV exposure in melanoma pathogenesis. Among these 19 GNA-encoding genes, GNAI2 had 6 missense changes, of which 2 were present in a single sample. Taken together, these data indicate that GNA mutations potentially have a more significant role in cutaneous melanomas than previously thought. Future biological studies are necessary to determine whether these GNA mutations are drivers.
Other potential driver genes in pan-negative melanomas
To identify other potential novel driver mutations in pan-negative melanomas, we identified gene mutations that were enriched in the pan-negative samples (69) compared to the driver-positive samples (172). Twelve genes had statistically significant mutations in the pan-negative samples (mutated in ≥ 10% of pan-negative melanomas and p-value < 0.05, Fisher’s exact test) (Table 4, Supplementary Table S14) including ALK, STK31, DGKI, RAC1, EPHA4, ADAMTS18, EPHA7, ERBB4, TAF1L, NF1, SYK, and KDR (Supplementary Figures S5–S16). Here, the order of the genes was based on the p values. Among them, RAC1 (15, 19), ADAMTS18 (44), ERBB4 (45), and NF1 (19) have already been reported as potential contributors to melanoma tumor progression, providing validation of the effectiveness of this analysis strategy. The most significantly mutated gene was the tyrosine kinase, ALK (altered in 17.4% of pan-negative tumors, p = 0.001, Supplementary Figure S10). Among the 13 ALK missense mutations in 12 samples, one (E1197K) was predicted to have a high impact on the protein function by MutationAssessor and four were predicted to have a medium impact on the protein function. The remaining mutations had a low or no impact on protein function. All these missense mutations were C>T (G>A) transitions, which may be the result of UV light damage. Considering that activating ALK rearrangements are found in lung cancer (46) and more recently in colorectal cancer (47), and a subset of lung cancer patients with ALK fusion genes are sensitive to the kinase inhibitor crizotinib (48), the role of ALK in melanoma and the clinical efficacy of ALK inhibition in ALK mutation-positive melanoma warrants further investigation.
Table 4.
Summary of genes with mutations enriched in pan-negative melanoma samples (mutation frequency ≥ 10% and p-value < 0.05)
| Gene | # pan-negative samples (N = 69) | Frequency (%) | # driver mutation-positive samples (N = 172) | Frequency (%) | p-value |
|---|---|---|---|---|---|
| ALK | 12 | 17.4 | 6 | 3.5 | 0.001 |
| STK31 | 18 | 26.1 | 15 | 8.7 | 0.001 |
| DGKI | 11 | 15.9 | 8 | 4.7 | 0.005 |
| RAC1 | 8 | 11.6 | 5 | 2.9 | 0.011 |
| EPHA4 | 7 | 10.1 | 4 | 2.3 | 0.015 |
| ADAMTS18 | 16 | 23.2 | 19 | 11.1 | 0.015 |
| EPHA7 | 12 | 17.4 | 12 | 7.0 | 0.017 |
| ERBB4 | 16 | 23.2 | 20 | 11.6 | 0.021 |
| TAF1L | 11 | 15.9 | 11 | 6.4 | 0.022 |
| NF1 | 12 | 17.4 | 13 | 7.6 | 0.024 |
| SYK | 7 | 10.1 | 5 | 2.9 | 0.027 |
| KDR | 10 | 14.5 | 11 | 6.4 | 0.043 |
Genes were ordered by p-values.
Clustering of mutations in these 12 genes indicated that 5 genes (RAC1, ADAMTS18, EPHA7, TAF1L, and NF1) harbored at least one recurrent mutation present in at least 2 pan-negative tumor samples. Mutations in the GTPase RAC1 occurred in 8 (11.6%) of the 69 pan-negative tumors compared to 5 of the 172 (2.9%) driver-positive tumors (p = 0.011) (Supplementary Figure S5). Seven of these 8 RAC1 mutations were the alteration P29S, which was previously shown to activate RAC1 (19) and increase melanocyte proliferation (15); therefore, RAC1 P29S may be a potential therapeutic target in melanoma. Notably, the strong recurrent mutation pattern in RAC1 implies that it might act as an oncogene instead of tumor-suppressor gene in melanoma. ADAMTS18 (ADAM metallopeptidase with thrombospondin type I motif, 18), which typically acts as a tumor-suppressor, was recently found to be mutated in melanoma, and mutant ADAMTS18 could increase melanoma cell migration and metastasis (44). We found ADAMTS18 mutations in 23.2% of the 69 pan-negative melanomas (Supplementary Table S14), with a hotspot R172Q mutation in three pan-negative tumors (Supplementary Figure S6). The remaining three genes (EPHA7, TAF1L, and NF1) each contained one recurrent mutation in two pan-negative melanomas. EPHA7, which encodes ephrin receptor A7, has been shown to act as a tumor-suppressor in follicular lymphoma (49). We observed 14 EPHA7 mutations in 12 pan-negative tumors (17.4%, p = 0.017). Among them, a G642E mutation, present in the tyrosine kinase domain of EPHA7, occurred in two tumors (Supplementary Figure S7). This mutation may impact kinase activity of EPHA7, making it a potential clinical target. Furthermore, we found another significantly mutated gene in the ephrin family, EPHA4 (10.1%, p = 0.015, Supplementary Figure S13). Ephrin receptors have been proposed as potential biomarkers and therapeutic targets in prostate cancer (50). In TAF1L (TAF1 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 210kDa-like), we observed 15 mutations in 11 of the 69 pan-negative samples (15.9%, p = 0.022, Supplementary Figure S8). TAF1L was the fourth most frequently mutated gene by statistical ranking among the kinase genes in another study (24).
Many loss-of-function mutations were observed in the genes STK31 (serine/threonine kinase 31) and NF1 (neurofibromin 1). We found 22 STK31 mutations in 18 pan-negative tumors (26.1%, p = 0.001, Supplementary Figure S11), including 3 nonsense, 1 splice-site, and 18 missense mutations. STK31 is a highly conserved member of the serine/threonine kinase (STK) family. It has been shown that the expression of STK genes is frequently altered in human cancers (51). Considering that previous findings suggested a functional role for STK31 in regulating gastrointestinal (52) and colon cancer development (53), and considering that a variety of STK inhibitors are available, STK31 may represent a potent target in the treatment of melanoma. Moreover, 22 NF1 mutations, including 6 nonsense (include one recurrent mutation W784X), 4 splice-site, and 12 missense mutations, were observed in 12 pan-negative tumors (17.4%, p = 0.024, Supplementary Figure S9). Four of these 12 missense mutations in NF1 were predicted to have a medium impact on the protein function by MutationAssessor. For the splicing mutations, we did not find any of them in the SpliceDisease database, which is a comprehensive database linking splicing mutations with diseases (54). As a negative regulator of RAS signaling, NF1 inactivation was recently reported to promote BRAF inhibitor resistance in the context of BRAF-V600 mutant melanoma (36). In pan-negative melanoma, NF1 inactivating mutations (nonsense mutations) may also play a role in promoting MAPK pathway activation. Therefore, melanomas with NF1 inactivating mutations may respond to MEK1/2 inhibition.
It has been reported that mutation patterns are different among melanoma subtypes (28). Here, we further examined mutant genes in each melanoma subtype in the cohort of “pan-negative” tumor samples. No mutated genes were found in more than one melanoma samples originating from mucosal sites, uveal sites, or unknown primary sites. Two genes, KIT (mutated in 3 samples) and PREX2 (2 samples), were mutated in acral melanoma, however, this result did not reach statistical significance (KIT: p = 0.535; PREX2: p = 0.669). Results for sun-exposed melanomas (cutaneous sites) are summarized in Supplementary Table S15. Specifically, all 12 genes in Table 4 were consistently found in cutaneous melanoma. This is not unexpected because the majority of tumors in “pan-negative” samples were derived from cutaneous sites (44/69 = 63.8%). Besides the 12 genes, many other genes were also found to be enriched in pan-negative cutaneous melanoma samples, including MAPK1 (a.k.a. ERK2) and MARK1 (Supplementary Table S15).
Discussion
With the rapid advances in NGS technologies, the molecular profiles of many tumors have been extensively characterized (9). Knowledge gained from these mutation profiles can potentially be used to identify relevant cellular targets for the development of targeted therapeutics to implement a personalized medicine approach for patients whose tumors harbor driver mutations in these targets (55). In this study, we systematically examined mutations from recently published melanoma NGS data encompassing 241 paired tumor-normal samples, aiming to identify potential, clinically-relevant mutations in melanomas that did not present with any of the well-established and -characterized driver mutations in BRAF, NRAS, KIT, GNAQ, and GNA11, which we describe as “pan-negative” tumors. Notably, and consistent with other reports (2), we confirmed that approximately half of the melanoma tumors (50.2%) harbored BRAF V600 mutations. Through our statistical analyses, we identified several genes with non-silent mutations that were likely to co-occur with BRAF- and NRAS-driven melanomas. Genes that co-occurred with BRAF-mutant tumors included TP53 and COL1A1, while genes that co-occurred with NRAS-mutant tumors included PPP6C, KALRN, PIK3R4, TRPM6, GUCY2C, and PRKAA2. The co-occurring mutations could modify responsiveness to targeted therapies for BRAF- or NRAS-mutant melanoma. Subsequent analysis of the sequence data from 69 pan-negative samples revealed less common mutations in BRAF, NRAS, KIT, GNAQ, and GNA11 along with many mutations in other genes encoding guanine nucleotide binding protein α-subunits. Furthermore, our statistical analysis identified 12 significantly mutated genes in pan-negative samples, including ALK, STK31, DGKI, RAC1, EPHA4, ADAMTS18, EPHA7, ERBB4, TAF1L, NF1, SYK, and KDR. Among them, we discovered that 5 genes (RAC1, ADAMTS18, EPHA7, TAF1L, and NF1) harbored a recurrent mutation that presented in at least 2 pan-negative tumor samples. Interestingly, somatic nonsense mutations were frequently found in 2 genes: 6 nonsense mutations (including one recurrent mutation, W784X) in NF1 and 3 nonsense mutations in STK31 (for details, see Supplementary Table S14). These newly identified genes in pan-negative melanoma samples may serve as potential targets for therapeutic agents in the treatment of melanoma without known BRAF, NRAS, KIT, GNAQ, and GNA11 mutations.
If tumors containing these mutations are activated and inhibition of the mutation(s) leads to cell death or attenuated growth, then clinically targeting them through pharmacological approaches may be a useful strategy. For example, the kinase inhibitor crizotinib might be tested in tumors with ALK mutations; STK inhibitors could be tested in tumors with STK31 mutations; and MEK1/2 inhibitors may be tested in tumors with NF1 mutations. Moreover, the rare mutations in the 5 known driver genes (BRAF, NRAS, KIT, GNAQ, and GNA11) in this pan-negative melanoma panel clearly demonstrated that the commonly used SNaPshot screen could overlook a subset of melanoma patients that might also respond to the currently available, effective BRAF, NRAS, KIT, GNAQ, and GNA11 targeting inhibitors, or at least their effector pathways (MEK1/2 inhibitors). For example, non-V600E BRAF mutations, including L597R/S/Q/V and K601E, are actionable in melanoma with MEK1/2 inhibition (5).
The NGS analysis method in Hodis et al. (19) requires mutation data in both the exonic regions (both nonsynonymous and synonymous mutations) and intronic regions. Since some studies did not report mutations in the intronic regions or synonymous mutations in the exonic regions, we developed our method using only nonsynonymous mutations from all the available studies. However, Hodis et al. applied a filtering strategy to remove those exonic SNVs whose adjacent non-coding regions had high mutation rates. This would likely affect our pool-based association analyses for those mutations that occurred in the hypermutable regions. Because the raw sequence data from Hodis et al. is not immediately available, the results reported in this study are not definitive. Other factors, such as sequence coverage and variant calling pipelines, which affect SNV calling, might affect our mutant gene association analyses. Such factors, however, are currently the main challenges for NGS applications (56). Moreover, recent studies suggested that synonymous mutations might alter a protein’s expression, three-dimensional conformation and function (57). A recent WGS study has detected a recurrent functional synonymous mutation in melanoma (58). However, it is still too early to systematically explore the effects of synonymous mutations on melanoma. Additionally, we narrowed the candidate gene list based on their association with known melanoma driver genes. While this method seems effective, it also likely missed other potential candidate genes. In summary, different from each individual NGS study, which attempted to identify mutations and genes involved in melanoma as a whole, our study uniquely provides a roadmap of potentially druggable mutations and genes in specific melanoma subtypes, especially those lacking any known driver mutations (pan-negative samples).
Another limitation of this study is that only SNVs were examined. In future work, we will extend our analysis to other types of mutations, such as insertions and deletions (indels), copy number variations (CNVs), and structural variants (SVs), for a more comprehensive map of genetic alterations in melanoma. In addition, more work is needed to link the promising genomic alterations to clinical outcomes or drug activity. We will attempt to identify genetic variants as the predictors of drug sensitivity by mining datasets from the Cancer Cell Line Encyclopedia (CCLE, http://www.broadinstitute.org/ccle/home) project, the Genomics of Drug Sensitivity in Cancer project (GDSC, http://www.cancerrxgene.org), and other large-scale pharmacogenomic datasets in cancer cells.
To our knowledge, this study represents the largest effort to-date that characterizes potentially clinically-relevant mutations in melanomas without known BRAF, NRAS, KIT, GNAQ, and GNA11 driver mutations (i.e. pan-negative patients). Our comprehensive analyses of the mutation patterns in 69 pan-negative melanoma genomes, along with 172 melanoma genomes harboring known driver mutations, demonstrated the effectiveness of discovering underrepresented somatic mutations potentially relevant to targeted therapies in melanoma patients who otherwise could not receive targeted therapies. As clinical NGS rapidly emerges, the high capacity and low cost of these platforms will soon allow routine screening of more comprehensive sets of tumor mutations in CLIA-certified laboratories, and thus, will benefit numerous cancer patients.
In summary, we used existing published NGS data from 241 tumor-normal pairs to identify mutations that could influence existing and emerging targeted therapies in both driver mutation positive and pan-negative melanomas. Future preclinical studies will need to be conducted to determine whether these mutations are driver or passenger mutations and how they will affect response to targeted therapies specifically in melanoma. The strategies proposed in this study can also be applied to other types of cancer, like lung cancer, that have extensive NGS data available but still have many cases without well-defined, targetable driver mutations.
Supplementary Material
Acknowledgments
Financial support: This project was partially supported by National Institutes of Health grants (R01LM011177 and R03CA167695 to Z. Zhao, K12CA0906525 to D. Johnson, and K24CA097588 to J. Sosman), a Stand Up To Cancer Innovative Research Grant, a Program of the Entertainment Industry Foundation (SU2C-AACR-IRG0109 to W. Pao), and VICC Cancer Center Core Grant (P30CA068485 to J.A. Pietenpol). The authors also acknowledge support from the Joanna M. Nicolay Melanoma Foundation 2013 Research Scholar Award, the TJ Martell Foundation, the Robert J. Kleberg, Jr. and Helen C. Kleberg Foundation, the Ingram Professorship, the American Cancer Society Mary Hendrickson-Johnson Melanoma Professorship, and the Craig Chair Professorship Funds. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
We would like to thank the members of the Bioinformatics and Systems Medicine Laboratory for their valuable discussion in this project.
Footnotes
Conflicts of interest: All authors have no competing interest to declare.
References
- 1.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA-Cancer J Clin. 2013;63:11–30. doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- 2.Dutton-Regester K, Hayward N. Reviewing the somatic genetics of melanoma: from current to future analytical approaches. Pigm Cell & Melanoma Res. 2012;25:144–54. doi: 10.1111/j.1755-148X.2012.00975.x. [DOI] [PubMed] [Google Scholar]
- 3.Fecher LA, Amaravadi RK, Flaherty KT. The MAPK pathway in melanoma. Curr Opin Oncol. 2008;20:183–9. doi: 10.1097/CCO.0b013e3282f5271c. [DOI] [PubMed] [Google Scholar]
- 4.Lovly CM, Dahlman KB, Fohn LE, Su Z, Dias-Santagata D, Hicks DJ, et al. Routine multiplex mutational profiling of melanomas enables enrollment in genotype-driven therapeutic trials. PLoS ONE. 2012;7:e35309. doi: 10.1371/journal.pone.0035309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dahlman KB, Xia J, Hutchinson K, Ng C, Hucks D, Jia P, et al. BRAFL597 mutations in melanoma are associated with sensitivity to MEK inhibitors. Cancer Disc. 2012;2:791–7. doi: 10.1158/2159-8290.CD-12-0097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hutchinson KE, Lipson D, Stephens PJ, Otto G, Lehmann BD, Lyle PL, et al. BRAF fusions define a distinct molecular subset of melanomas with potential sensitivity to MEK inhibition. Clin Cancer Res. 2013;19:6696–702. doi: 10.1158/1078-0432.CCR-13-1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Meyerson M, Gabriel S, Getz G. Advances in understanding cancer genomes through second-generation sequencing. Nat Rev Genet. 2010;11:685–96. doi: 10.1038/nrg2841. [DOI] [PubMed] [Google Scholar]
- 8.Xia J, Wang Q, Jia P, Wang B, Pao W, Zhao Z. NGS Catalog: A database of next generation sequencing studies in humans. Human Mutat. 2012;33:E2341–E55. doi: 10.1002/humu.22096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lawrence MS, Stojanov P, Polak P, Kryukov GV, Cibulskis K, Sivachenko A, et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature. 2013 doi: 10.1038/nature12213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kunz M, Dannemann M, Kelso J. High-throughput sequencing of the melanoma genome. Exp Dermatol. 2013;22:10–7. doi: 10.1111/exd.12054. [DOI] [PubMed] [Google Scholar]
- 11.Pleasance ED, Cheetham RK, Stephens PJ, McBride DJ, Humphray SJ, Greenman CD, et al. A comprehensive catalogue of somatic mutations from a human cancer genome. Nature. 2010;463:191–6. doi: 10.1038/nature08658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wei X, Walia V, Lin JC, Teer JK, Prickett TD, Gartner J, et al. Exome sequencing identifies GRIN2A as frequently mutated in melanoma. Nat Genet. 2011;43:442–6. doi: 10.1038/ng.810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yokoyama S, Woods SL, Boyle GM, Aoude LG, MacGregor S, Zismann V, et al. A novel recurrent mutation in MITF predisposes to familial and sporadic melanoma. Nature. 2011;480:99–103. doi: 10.1038/nature10630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Berger MF, Hodis E, Heffernan TP, Deribe YL, Lawrence MS, Protopopov A, et al. Melanoma genome sequencing reveals frequent PREX2 mutations. Nature. 2012;485:502–6. doi: 10.1038/nature11071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Krauthammer M, Kong Y, Ha BH, Evans P, Bacchiocchi A, McCusker JP, et al. Exome sequencing identifies recurrent somatic RAC1 mutations in melanoma. Nat Genet. 2012;44:1006–14. doi: 10.1038/ng.2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nikolaev SI, Rimoldi D, Iseli C, Valsesia A, Robyr D, Gehrig C, et al. Exome sequencing identifies recurrent somatic MAP2K1 and MAP2K2 mutations in melanoma. Nat Genet. 2012;44:133–9. doi: 10.1038/ng.1026. [DOI] [PubMed] [Google Scholar]
- 17.Stark MS, Woods SL, Gartside MG, Bonazzi VF, Dutton-Regester K, Aoude LG, et al. Frequent somatic mutations in MAP3K5 and MAP3K9 in metastatic melanoma identified by exome sequencing. Nat Genet. 2012;44:165–9. doi: 10.1038/ng.1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Turajlic S, Furney SJ, Lambros MB, Mitsopoulos C, Kozarewa I, Geyer FC, et al. Whole genome sequencing of matched primary and metastatic acral melanomas. Genome Res. 2012;22:196–207. doi: 10.1101/gr.125591.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hodis E, Watson IR, Kryukov GV, Arold ST, Imielinski M, Theurillat J-P, et al. A landscape of driver mutations in melanoma. Cell. 2012;150:251–63. doi: 10.1016/j.cell.2012.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Walia V, Mu EW, Lin JC, Samuels Y. Delving into somatic variation in sporadic melanoma. Pigm Cell & Melanoma Res. 2012;25:155–70. doi: 10.1111/j.1755-148X.2012.00976.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wilson MA, Nathanson KL. Molecular testing in melanoma. Cancer J. 2012;18:117–23. doi: 10.1097/PPO.0b013e31824f11bf. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nathanson KL. Using genetics and genomics strategies to personalize therapy for cancer: focus on melanoma. Biochem Pharmacol. 2010;80:755–61. doi: 10.1016/j.bcp.2010.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Flaherty KT, Hodi FS, Fisher DE. From genes to drugs: targeted strategies for melanoma. Nat Rev Cancer. 2012;12:349–61. doi: 10.1038/nrc3218. [DOI] [PubMed] [Google Scholar]
- 24.Greenman C, Stephens P, Smith R, Dalgliesh GL, Hunter C, Bignell G, et al. Patterns of somatic mutation in human cancer genomes. Nature. 2007;446:153–8. doi: 10.1038/nature05610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Torkamani A, Verkhivker G, Schork NJ. Cancer driver mutations in protein kinase genes. Cancer Lett. 2009;281:117–27. doi: 10.1016/j.canlet.2008.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gibney GT, Smalley KS. An unholy alliance: cooperation between BRAF and NF1 in melanoma development and BRAFinhibitor resistance. Cancer Disc. 2013;3:260–3. doi: 10.1158/2159-8290.CD-13-0017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xing F, Persaud Y, Pratilas C, Taylor B, Janakiraman M, She Q, et al. Concurrent loss of the PTEN and RB1 tumor suppressors attenuates RAF dependence in melanomas harboring V600EBRAF. Oncogene. 2011;31:446–57. doi: 10.1038/onc.2011.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chin L, Garraway LA, Fisher DE. Malignant melanoma: genetics and therapeutics in the genomic era. Genes Dev. 2006;20:2149–82. doi: 10.1101/gad.1437206. [DOI] [PubMed] [Google Scholar]
- 29.Daniotti M, Oggionni M, Ranzani T, Vallacchi V, Campi V, Di Stasi D, et al. BRAF alterations are associated with complex mutational profiles in malignant melanoma. Oncogene. 2004;23:5968–77. doi: 10.1038/sj.onc.1207780. [DOI] [PubMed] [Google Scholar]
- 30.Reva B, Antipin Y, Sander C. Predicting the functional impact of protein mutations: application to cancer genomics. Nucleic Acids Res. 2011;39:e118. doi: 10.1093/nar/gkr407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guo H, Carlson JA, Slominski A. Role of TRPM in melanocytes and melanoma. Exp Dermatol. 2012;21:650–4. doi: 10.1111/j.1600-0625.2012.01565.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gong JP, Schulz S, Hyslop T, Waldman SA. GUCY2C molecular staging personalizes colorectal cancer patient management. Biomark Med. 2012;6:339–48. doi: 10.2217/bmm.12.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shen C-H, Yuan P, Perez-Lorenzo R, Zhang Y, Lee SX, Ou Y, et al. Phosphorylation of BRAF by AMPK impairs BRAF-KSR1 association and cell proliferation. Molecular cell. 2013;52:161–72. doi: 10.1016/j.molcel.2013.08.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim YH, Liang H, Liu X, Lee J-S, Cho JY, Cheong J-H, et al. AMPKα modulation in cancer progression: multilayer integrative analysis of the whole transcriptome in asian gastric cancer. Cancer Res. 2012;72:2512–21. doi: 10.1158/0008-5472.CAN-11-3870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Willmore-Payne C, Holden JA, Tripp S, Layfield LJ. Human malignant melanoma: detection of BRAF-and c-kit-activating mutations by high-resolution amplicon melting analysis. Human Pathol. 2005;36:486–93. doi: 10.1016/j.humpath.2005.03.015. [DOI] [PubMed] [Google Scholar]
- 36.Whittaker SR, Theurillat J-P, Van Allen E, Wagle N, Hsiao J, Cowley GS, et al. A genome-scale RNA interference screen implicates NF1 loss in resistance to RAF inhibition. Cancer Disc. 2013;3:350–62. doi: 10.1158/2159-8290.CD-12-0470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Heinzerling L, Kühnapfel S, Meckbach D, Baiter M, Kaempgen E, Keikavoussi P, et al. Rare BRAF mutations in melanoma patients: implications for molecular testing in clinical practice. Brit J Cancer. 2013 doi: 10.1038/bjc.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Blatt C, Eversole-Cire P, Cohn VH, Zollman S, Fournier R, Mohandas L, et al. Chromosomal localization of genes encoding guanine nucleotide-binding protein subunits in mouse and human. Proc Natl Acad Sci U S A. 1988;85:7642–6. doi: 10.1073/pnas.85.20.7642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hurst JH, Hooks SB. Regulator of G-protein signaling (RGS) proteins in cancer biology. Biochem Pharmacol. 2009;78:1289–97. doi: 10.1016/j.bcp.2009.06.028. [DOI] [PubMed] [Google Scholar]
- 40.Wood LD, Parsons DW, Jones S, Lin J, Sjoblom T, Leary RJ, et al. The genomic landscapes of human breast and colorectal cancers. Sci Sig. 2007;318:1108–13. doi: 10.1126/science.1145720. [DOI] [PubMed] [Google Scholar]
- 41.Kan Z, Jaiswal BS, Stinson J, Janakiraman V, Bhatt D, Stern HM, et al. Diverse somatic mutation patterns and pathway alterations in human cancers. Nature. 2010;466:869–73. doi: 10.1038/nature09208. [DOI] [PubMed] [Google Scholar]
- 42.Van Raamsdonk CD, Griewank KG, Crosby MB, Garrido MC, Vemula S, Wiesner T, et al. Mutations in GNA11 in uveal melanoma. N Engl J Med. 2010;363:2191–9. doi: 10.1056/NEJMoa1000584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Van Raamsdonk CD, Bezrookove V, Green G, Bauer J, Gaugler L, O’Brien JM, et al. Frequent somatic mutations of GNAQ in uveal melanoma and blue naevi. Nature. 2008;457:599–602. doi: 10.1038/nature07586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wei X, Prickett TD, Viloria CG, Molinolo A, Lin JC, Cardenas-Navia I, et al. Mutational and functional analysis reveals ADAMTS18 metalloproteinase as a novel driver in melanoma. Mol Cancer Res. 2010;8:1513–25. doi: 10.1158/1541-7786.MCR-10-0262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Prickett TD, Agrawal NS, Wei X, Yates KE, Lin JC, Wunderlich JR, et al. Analysis of the tyrosine kinome in melanoma reveals recurrent mutations in ERBB4. Nat Genet. 2009;41:1127–32. doi: 10.1038/ng.438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Soda M, Choi YL, Enomoto M, Takada S, Yamashita Y, Ishikawa S, et al. Identification of the transforming EML4–ALK fusion gene in non-small-cell lung cancer. Nature. 2007;448:561–6. doi: 10.1038/nature05945. [DOI] [PubMed] [Google Scholar]
- 47.Lipson D, Capelletti M, Yelensky R, Otto G, Parker A, Jarosz M, et al. Identification of new ALK and RET gene fusions from colorectal and lung cancer biopsies. Nat Med. 2012;18:382–4. doi: 10.1038/nm.2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kwak EL, Bang Y-J, Camidge DR, Shaw AT, Solomon B, Maki RG, et al. Anaplastic lymphoma kinase inhibition in non–small-cell lung cancer. N Engl J Med. 2010;363:1693–703. doi: 10.1056/NEJMoa1006448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Oricchio E, Nanjangud G, Wolfe AL, Schatz JH, Mavrakis KJ, Jiang M, et al. The Eph-receptor A7 is a soluble tumor suppressor for follicular lymphoma. Cell. 2011;147:554–64. doi: 10.1016/j.cell.2011.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lisle JE, Mertens-Walker I, Rutkowski R, Herington AC, Stephenson S-A. Eph receptors and their ligands: promising molecular biomarkers and therapeutic targets in prostate cancer. BBA-Rev Cancer. 2013;1835:243–57. doi: 10.1016/j.bbcan.2013.01.003. [DOI] [PubMed] [Google Scholar]
- 51.Capra M, Nuciforo PG, Confalonieri S, Quarto M, Bianchi M, Nebuloni M, et al. Frequent alterations in the expression of serine/threonine kinases in human cancers. Cancer Res. 2006;66:8147–54. doi: 10.1158/0008-5472.CAN-05-3489. [DOI] [PubMed] [Google Scholar]
- 52.Yokoe T, Tanaka F, Mimori K, Inoue H, Ohmachi T, Kusunoki M, et al. Efficient identification of a novel cancer/testis antigen for immunotherapy using three-step microarray analysis. Cancer Res. 2008;68:1074–82. doi: 10.1158/0008-5472.CAN-07-0964. [DOI] [PubMed] [Google Scholar]
- 53.Fok KL, Chung CM, Yi SQ, Jiang X, Sun X, Chen H, et al. STK31 maintains the undifferentiated state of colon cancer cells. Carcinogenesis. 2012;33:2044–53. doi: 10.1093/carcin/bgs246. [DOI] [PubMed] [Google Scholar]
- 54.Wang J, Zhang J, Li K, Zhao W, Cui Q. SpliceDisease database: linking RNA splicing and disease. Nucleic Acids Res. 2012;40:D1055–D9. doi: 10.1093/nar/gkr1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jia P, Zhao Z. VarWalker: Personalized mutation network analysis of putative cancer genes from next-generation sequencing data. PLoS Comp Biol. 2014;10:e1003460. doi: 10.1371/journal.pcbi.1003460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang Q, Jia P, Li F, Chen H, Ji H, Hucks D, et al. Detecting somatic point mutations in cancer genome sequencing data: a comparison of mutation callers. Genome Med. 2013;5:91. doi: 10.1186/gm495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sauna ZE, Kimchi-Sarfaty C. Understanding the contribution of synonymous mutations to human disease. Nat Rev Genet. 2011;12:683–91. doi: 10.1038/nrg3051. [DOI] [PubMed] [Google Scholar]
- 58.Gartner JJ, Parker SC, Prickett TD, Dutton-Regester K, Stitzel ML, Lin JC, et al. Whole-genome sequencing identifies a recurrent functional synonymous mutation in melanoma. Proc Natl Acad Sci U S A. 2013;110:13481–6. doi: 10.1073/pnas.1304227110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.