Summary
Circadian clocks allow organisms to anticipate daily changes in the environment to enhance overall fitness. Transcription factors (TFs) play a prominent role in the molecular mechanism but are incompletely described possibly due to functional redundancy, gene family proliferation, and/or lack of context-specific assays. To overcome these, we performed a high-throughput yeast one-hybrid screen using the LUX ARRYHTHMO (LUX) gene promoter as bait against an Arabidopsis TF library. LUX is a unique gene because its mutation causes severe clock defects and transcript maintains high amplitude cycling in the cold. We report the well-characterized cold-inducible C-repeat (CRT)/drought-responsive element (DRE) binding factor CBF1/DREB1b is a transcriptional regulator of LUX. We show that CBF1 binds the CRT in the LUX promoter, and both genes overlap in temporal and spatial expression. CBF1 overexpression causes up-regulation of LUX and also alters other clock gene transcripts. LUX promoter regions including the CRT and Evening Element (EE) are sufficient for high amplitude transcriptional cycling in the cold, and cold-acclimated lux seedlings are sensitive to freezing stress. Our data show cold signaling is integrated into the clock by CBF-mediated regulation of LUX expression, thereby defining a new transcriptional mechanism for temperature input to the circadian clock.
Results and Discussion
CBF1 binds the LUX promoter
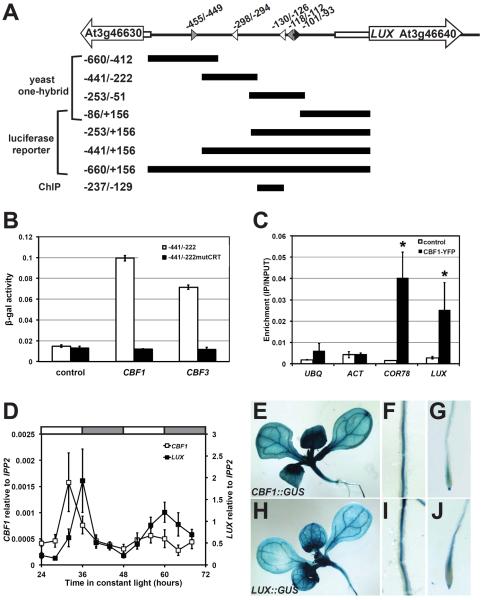
LUX is a key component of the circadian clock regulating growth and development in Arabidopsis [1–6]. The LUX transcript is circadian-regulated with peak expression in the evening [1–2]. It also maintains high amplitude oscillations under diurnal conditions in the cold [7]. The clock proteins CIRCADIAN CLOCK ASSOCIATED1 (CCA1) and REVEILLE8 (RVE8) regulate LUX promoter activity by binding the EE (Evening Element, AAAATATCT) motif [1,8], while the LBS (LUX binding site, GATA/TCG) mediates LUX auto-regulation [3]. TIMING OF CAB EXPRESSION1 (TOC1) also associates with the LUX promoter [9]. To identify other transcriptional regulators, we performed a yeast one-hybrid screen by challenging tiled LUX promoter fragments (Figure 1A) with an arrayed Arabidopsis transcription factor collection [10]. We found CBF1 (At4g25490) and CBF3 (At4g25480) strongly activated β-galactosidase (β-gal) reporter activity when the LUX promoter fragment −441/−222 was used as bait (Figure 1B). These factors are members of a small family of highly redundant AP2-domain transcription factors consisting of CBF1, CBF2 and CBF3 (also known as DREB1A-C) [11–15]. Although expressed under ambient conditions, cold treatment dramatically induces CBF expression and subsequent target genes to confer cold tolerance. The LUX promoter region −441/−222 possesses a single copy of the core C-repeat (CRT) element/dehydration responsive element (DRE) (CCGAC) at position −298/−294 [11,12,16,17]. This is specifically bound by CBF1 and CBF3 as mutation of 6bp including this core (−441/−222mutCRT) diminished β-gal activity (Figure 1B). An additional core CRT motif occurs at position −130/−126, however the CBFs were not recovered from our preliminary screen of the −253/−51 fragment nor in screens of the −660/−412 and −86/+156 regions that lack the CRT (data not shown). Although CBF2 was not recovered, the high functional redundancy between the CBFs suggests it might also bind the LUX promoter; additional studies are required to test this.
Figure 1.
CBF1 binds the LUX promoter
(A) Schematic of the LUX promoter. The transcriptional start occurs at +1. White arrows: gene bodies; white rectangles: 5'UTRs; arrowheads: DNA motifs with EELs in grey, CRTs in white, and EE in black; and black bars: promoter regions tested in different assays. Positions and sizes are roughly drawn to scale. (B) CBF1 binding to LUX promoter in yeast. Bars represent β-galactosidase activity where error bars represent standard error of three replicates. (C) CBF1 binding to the LUX promoter in Arabidopsis. ChIP assays were performed using control (LUX∷LUC −660/+156 line 139) or CBF1 overexpressing line 2–1 (CBF1ox2-1). Plants were grown under 12:12 and collected 16 hours after lights on for ChIP using an anti-GFP antibody. Results are normalized to input DNA. Bars represent average quantification from real-time PCR with error bars representing standard error of two independent experiments. Student's t-test was used to determine the significance of target enrichment relative to control (* p-value ≤0.05). UBQ, UBIQUITIN; and ACT, ACTIN. (D) Temporal expression of CBF1 and LUX. Wildtype plants were entrained under 12:12 before release to constant light at 22°C. mRNA levels were quantified by real-time reverse transcription PCR and normalized to IPP2. Data is the average of three biological replicates with error bars representing standard error. (E–J) Spatial expression of CBF1 and LUX. Seedlings carrying promoter∷GUS constructs of CBF1 (E–G) and LUX (H–J) were grown under 12:12 at 22°C. Expression detected throughout cotyledons, rosette leaves and hypocotyl (E and H); roots (F and I); and root tips (G and J).
To confirm CBF binding in vivo, we generated transgenic seedlings overexpressing yellow fluorescent protein (YFP)-tagged CBF1 and performed chromatin immunoprecipitations (ChIPs) using a representative line (CBF1ox2-1). This line exhibits phenotypes similar to those previously published [12,15,18,19], which include an enhanced resistance to freezing stress (Figure S1). Chromatin was isolated from control and CBF1ox2-1 seedlings grown under photocycles of 12-hours light/12-hours dark (abbreviated 12:12) at 22°C. In ChIP samples from control, specific binding was not detected at any tested regions including the negative control genes UBIQUITIN (UBQ) and ACTIN (ACT) (Figure 1C). In contrast, ChIP samples using CBF1ox2-1 showed specific enrichment at the COR78 promoter (amplicon −225/−144) that possesses three CRT motifs at positions −273/−268, −223/−218, −166/−161. Additionally enrichment was observed at the LUX promoter region −237/−129, which is flanked by CRTs at positions −298/−294 and −130/−126 (Figure 1A). Although the resolution of our ChIP experiments cannot distinguish between CRT sites, we confirmed CBF1 associates with the LUX promoter in vivo. Together with our yeast one-hybrid results, this indicates CBF1 occupies the LUX promoter by associating with the CRT motif.
CBF1 and LUX have overlapping expression patterns in vivo
To test whether CBF1 and CBF3 could regulate LUX expression in vivo, we compared expression patterns. Under ambient conditions, CBF expression is circadian with a peak phase in the afternoon [7, 20–22]. Using time-course array data from the Diurnal database [23], we compared expression under various diurnal and circadian conditions. CBF1, CBF3 and LUX all cycled in 7 diurnal and 1 circadian datasets (Figure S2). All three genes overlapped in their temporal expression, and in many datasets the CBFs had phases preceding LUX. For subsequent analyses, we focused on CBF1 as a representative of the CBF family. Using quantitative RT-PCR, we confirmed the sequential expression patterns of CBF1 and LUX under constant light at 22°C (Figure 1D).
To determine whether spatial expression patterns overlap in planta, we generated GUS reporter lines driven by the CBF1 and LUX promoters. The promoter fragments of CBF1 and LUX were sufficient to drive GUS activity throughout tissues (Figure 1E–J). GUS activity was detected in cotyledons, rosette leaves, hypocotyls, roots, and root tips. Similar CBF1 expression was reported previously [24]. The overlapping temporal and spatial expression patterns of CBF1 and LUX are consistent with a regulatory interaction.
Overexpression of CBF1 alters the levels of LUX and other clock gene transcripts
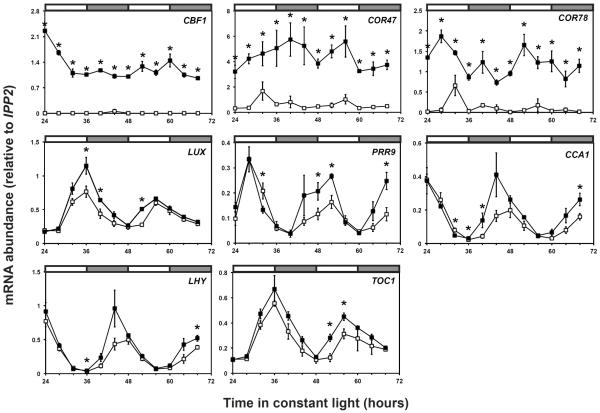
CBF regulation via the CRT motif causes activation of a suite of gene targets including the CORs [12,18]. To determine whether CBF1 regulates LUX expression, we analyzed the effect of CBF1 overexpression in seedlings. First we tested whether CBF1 overexpression alters the activity of a full-length LUX promoter-luciferase reporter (LUX∷LUC −660/+156) but saw no obvious difference relative to control (data not shown). Since CBF overexpression reduces plant size [15, 25] and a control luciferase reporter for normalization is not available for Arabidopsis, it is possible a subtle effect is undetectable by this assay. We then measured LUX transcript levels in a previously published CBF1 overexpression line [25]. This line expressed CBF1, COR47, and COR78 at considerably higher levels than wildtype at all time-points tested (Figure 2). In contrast, the LUX transcript exhibited a significant increase only at specific times of the day near the wildtype peak (Figure 2). Additionally PRR9, CCA1, LHY, and TOC1 showed significant alterations relative to the control. Unlike the CORs, the effects on LUX and the other clock genes are time specific. This may reflect their complex transcriptional regulation by multiple clock factors, and contribute to genetic robustness which is an inherent feature of circadian clocks that allows mutational perturbations to be accommodated without eliminating clock function [26]. As transcript patterns reflect the culmination of transcriptional and post-transcriptional events, other factors could also be involved. To confirm clock gene mis-regulation, we analyzed LUX transcripts in our CBF1 overexpression line used in ChIP experiments. We observed a significant up-regulation of the LUX transcript in the subjective night at CT36 (circadian time 36) and time-specific alterations in other clock genes (Figure S3). The changes in clock gene dynamics are not identical between overexpression lines and may be due to differences in CBF1 levels, ecotype backgrounds, and/or growth media. Nonetheless our observations support that CBF1 overexpression impacts LUX and other clock gene transcripts.
Figure 2.
CBF1 overexpression affects LUX and other clock gene transcripts
Transcript abundance was measured in control (open squares) and CBF1 overexpressor line (filled squares). Seedlings were entrained under 12:12 at 22°C for 10 days before transfer to constant light. Transcript levels were quantified by real-time reverse transcription PCR. Data is the average of three biological replicates with error bars representing standard error. Student's t-test was used to determine the significance of transcript levels relative to control (* p-value ≤0.05).
The LUX promoter confers high amplitude oscillations in the cold
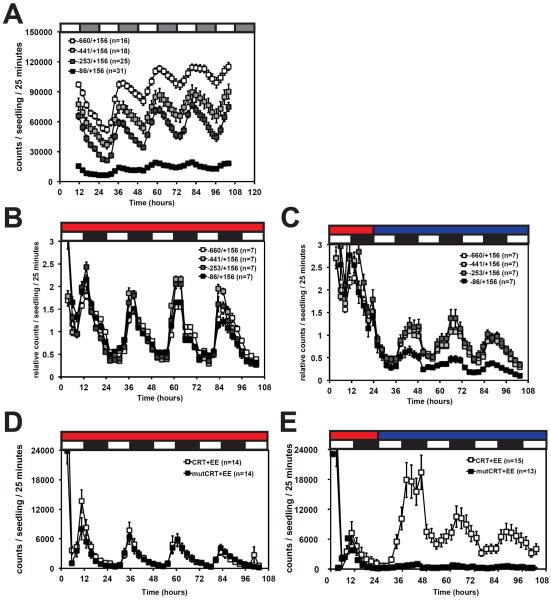
As the effects of CBF1 overexpression on the LUX transcript are subtle and attempts to produce CBF triple knockout lines have been unsuccessful [27], we characterized the role of the CBFs in regulating the LUX promoter by using various LUC reporters. We first generated LUX∷LUC lines that incorporated regions screened in our yeast one-hybrid assays (Figure 1A). Primary transgenic lines (T1) were imaged under constant light at 22°C. The full-length LUX promoter (−660/+156) was sufficient to drive circadian expression with a phase peak in the subjective evening similar to the endogenous LUX transcript (compare Figure 3A to Figure 2). Successive deletion of the 5' end caused small decreases in the overall expression as shown by comparing seedlings expressing fragment −660/+156 to those expressing −441/+156 and −253/+156. In contrast, further deletion caused a dramatic decrease in expression suggesting that the region spanning −253/−86 possesses regulatory elements important for transcriptional activation. Although other motifs may be involved, this decrease is consistent with the loss of EE and EE-like (EEL, AATATCT) motifs as the EE is sufficient for robust evening-phased expression [20,28].
Figure 3.
The CRT and EE in the LUX promoter confer evening-phased oscillations in the cold
Bioluminescence assays in Arabidopsis seedlings carrying luciferase reporters driven by LUX promoter fragments (A–C) or multimerized CRT and EE motifs (D and E). Ten-day old seedlings were entrained under 12:12 at 22°C. Seedlings remained under these conditions (B and D) or were transferred to constant light at 22°C (A) or 12:12 at 4°C (C and E). Red and blue bars represent 22°C and 4°C respectively. Seedlings in the T1 (A,D,E) or T2 (B,C) generation were assayed. Data shown is the averages of all plants imaged in a given experiment with error bars representing standard error. Values in B and C are relative to the average expression of that reporter line. Traces are representative of data from at least two independent experiments.
We next selected representative single insertion T2 transgenic lines to compare relative expression under diurnal conditions at 22°C and 4°C. This cold condition was previously reported to dampen the amplitudes of all tested clock transcripts except LUX [7]. At 22°C, all LUX promoter fragments produced high amplitude oscillations. Progressive truncation of the LUX promoter had little effect on amplitude at 22°C (Figure 3B). As seen in Figure 3C, the full-length LUX∷LUC construct recapitulated the reported transcript oscillations in the cold indicating the LUX promoter is sufficient to drive transcript cycling under these conditions. Interestingly we also noticed a small phase delay in expression after transfer to 4°C. This was reported previously for the transcript showing that the activity of the LUX promoter closely matches that of the endogenous gene [7]. After transfer to 4°C, deletion of the −600/−86 region caused a decrease in amplitude relative to the other fragments. Since this deletion removed both CRTs within the LUX promoter, this supports they may be required for the regulation of LUX expression in the cold.
To determine the minimal motifs sufficient for recapitulating the expression pattern of the full length LUX promoter in the cold, we generated reporters driven by short wildtype and mutant LUX sequences. The CRT at position −298/−294 is hypothesized to be important since CBF1 associates with the LUX promoter in yeast one-hybrid and ChIP assays (Figures 1B and 1C). As the LUX transcript oscillates and is evening-phased even though CBF transcripts are clamped high in the cold [7], the EE also appears important. CCA1 and RVE8 regulate LUX via the EE [1,8], and the EE and EEL are required for cold induction of COL1 and COR27 [29]. The LUX promoter possesses one EE and two EELs (Figure 1A). We cloned wildtype and mutant versions of the CRT (from position −298/−294) and EE in tandem to generate CRT+EE, mutant CRT+EE (mutCRT+EE), and CRT+mutant EE (CRT+mutEE) fragments. Primary transgenic lines were imaged at 22°C or 4°C. Because very few lines carrying the CRT+mutEE construct had detectable LUC levels at either temperature, they were omitted from analysis (data not shown). This lack of expression could be due to disrupted binding of circadian activators such as RVE8 [28] and cold regulators [29]. Transgenic plants carrying the CRT+EE construct displayed rhythmic oscillations at 22°C (Figure 3D). Peak expression occurred at the end of the light period similar to that reported previously for other EE lines [20,28]. Mutation of the CRT had no effect on this pattern at 22°C, confirming the EE is sufficient for rhythmic oscillations under ambient conditions. When seedlings carrying the CRT+EE were transferred to 4°C, high amplitude rhythms were observed, with the first peak after transfer showing higher amplitude than at 22°C (Figure 3E). This suggests the CRT+EE responds to the cold but this response is gated to the evening. The phase of expression in the cold was also slightly delayed relative to that at 22°C. This is reminiscent of the pattern observed with the full length LUX promoter (Figure 3C) as well as the endogenous transcript expression pattern reported previously [7]. High amplitude cycling at 4°C is specifically dependent on the CRT since its mutation greatly reduced evening-phased expression (Figure 3E). These results indicate that the CRT, in combination with the EE, recruits activators necessary for evening-phased expression at 4°C.
While the EE and CRT are sufficient to recapitulate the expression of the endogenous LUX promoter in the cold, the full complement of factors regulating LUX expression remains unknown. Based on our and other results, we speculate the CBFs (via the CRT) and CCA1/LHY/RVE8 (via the EE) are pivotal at cooler temperatures. Simplistically under ambient conditions, the clock proteins CCA1/LHY/RVE8 confer time-of-day information with CCA1/LHY repressing LUX expression during the early-morning and RVE8 activating evening-phased expression. Since the CBFs are also expressed under ambient conditions, they likely contribute to LUX expression but play a lesser role as suggested by the subtle effects of CBF1 overexpression (Figure 2). As temperatures decrease however, the expression of CCA1/LHY (and other clock genes) loses rhythmicity while LUX maintains high amplitude oscillations. Parallel to this, the expression of the CBFs increases with cold. We hypothesize as temperatures decrease and the oscillations of clock genes like CCA/LHY dampen, the CBFs play an increasingly important role to ensure LUX oscillations are maintained over a broad temperature range. However as CBF rhythms also dampen in the cold, post-transcriptional regulation and/or additional factors are also likely involved.
Freezing tolerance is disrupted in lux mutants
The cold response is a well-characterized output of the clock. CCA1 and LHY regulate freezing tolerance by directly associating with CBF promoter regions to gate their expression [30,31]. Loss of CCA1/LHY impairs freezing tolerance [30–32] while CCA1 overexpression promotes freezing tolerance [33]. In addition, mutation of PRR9/7/5 and TOC1 enhances freezing tolerance, and PRR5, PRR7, and TOC1 associate with CBF promoters [9,34–37]. While the mechanism is unknown, loss of GIGANTEA (GI) also causes freezing sensitivity but independently of changes in CBF expression [38].
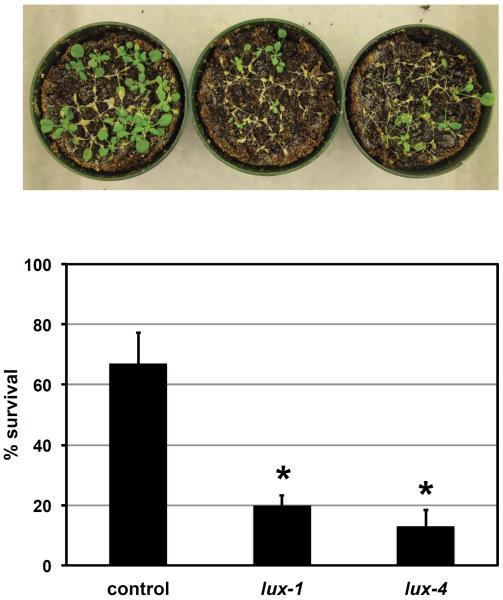
To test the functional relevance of LUX in the cold, we assessed the response of lux seedlings to freezing stress after cold acclimation (Figure 4). Plants were cold acclimatized before exposure to −5°C for 5 hours. Strikingly most lux-1 and lux-4 seedlings died while the majority of control seedlings survived (Figure 4). This demonstrates that LUX contributes to freezing tolerance. Recently the lux-2 mutant was reported to have similar sensitivity as wildtype to freezing stress in the absence of cold-acclimation [37]. Since the lux-1, lux-2, and lux-4 mutations all cause premature stop codons and affect clock processes similarly [1], this suggests LUX may have a very specific role during the acclimation process.
Figure 4.
LUX is required for survival to freezing stress after cold acclimation
Freezing tolerance is expressed as percent (%) survival. Control (CAB2∷LUC), lux-1, and lux-4 seedlings were grown under 12:12 at 22°C for approximately 4 weeks. Plants were then transferred at 4 hours after lights on into 4°C darkness for 3 days cold acclimation before freezing at −5°C for 5 hours. After 24 hours recovery at 4°C, plants were returned to 12:12 at 22°C for 7 days. Data is the average of four independent experiments with error bars representing standard error. Student's t-test was used to determine the significance of survival relative to control (* p-value ≤0.05).
How LUX contributes to freezing tolerance remains to be determined. Under ambient conditions, lux has low and high expression of CCA/LHY and TOC1, respectively [1,2]. Since CCA1/LHY activates and TOC1 represses CBF expression, it is possible that the freezing sensitivity of lux is indirectly due to their regulation of CBF and target gene expression [31,37]. The lux mutant also exhibits high GI expression [2] but as the mechanism of GI involvement is unknown, this observation is not simple to resolve. Because LUX is a DNA-binding transcription factor, we speculate it may also directly regulate genes involved in cold tolerance. LUX functions in a transcriptional complex with other evening-phased proteins including ELF3 [4,6]. ELF3 was recently reported to associate with the CBF3 promoter [37] so it is possible LUX also directly regulates CBF3. It was previously reported that the lux mutant under ambient conditions exhibits high CBF1/3 expression [37]. However, we observed lux-1 and lux-4 are sensitive to freezing after cold-acclimation (Figure 4). Together this suggests LUX may regulate cold response genes in parallel to the CBF pathway. Determining the direct target genes of LUX may help clarify the mechanisms involved.
Like light sensing, we anticipate cold input to the Arabidopsis clock occurs through multiple molecular mechanisms. We have shown that the well-characterized cold-inducible CBF1 transcription factor plays an important role in maintaining LUX oscillations in the cold and that LUX plays an essential role in freezing tolerance after cold-acclimation. In addition to LUX, the CBFs could regulate other clock genes; for example the LHY and TOC1 promoters contain CRT motifs and their transcripts are altered in CBF1 overexpressing lines (Figures 2 and S3). Another mode of cold sensing in Arabidopsis occurs post-transcriptionally via alternative splicing of CCA1 [33]. As the alternative CCA1 transcript presumably encodes a protein that interferes with the full-length protein, its suppression is necessary for freezing tolerance. In other organisms, post-transcriptional mechanisms are common for cold input to the clock. Thermosensitive splicing of the Drosophila clock gene period affects clock phasing and circadian-regulated locomotor activity [39,40]. The Neurospora clock gene FREQUENCY (FRQ) also undergoes alternative splicing in a temperature-dependent manner to affect free-running rhythms and temperature compensation [41]. In addition, FRQ translation biases towards noncoding upstream open reading frames in the 5'UTR in order to restrict translation under decreasing temperatures [42,43]. Non-optimal codon usage for FRQ was also reported, and in cyanobacteria this mechanism limits translation of the central clock KaiB and KaiC proteins under cooler temperatures [44,45]. Finally in mouse, the cold-inducible RNA-binding protein (CIRP) is required for circadian rhythms in fibroblasts and directly regulates mRNA stability of the circadian oscillator CLOCK gene [46]. Our discovery that LUX is transcriptionally regulated by CBF1 adds a new layer of regulation to cold input to the circadian clock. Uncovering additional mechanisms of connectivity between temperature and the circadian clock will undoubtedly further our understanding of growth and development.
Experimental Procedures
For detailed protocols, see Supplemental Information.
Supplementary Material
Highlights.
-
-
The LUX clock gene promoter is bound by the cold-induced CBF1 transcription factor
-
-
CBF1 overexpression up-regulates LUX and alters other clock gene transcripts
-
-
The CRT and EE motifs are sufficient to confer evening-phased expression in the cold
-
-
lux mutants are sensitive to freezing after cold-acclimation
Acknowledgments
We thank J. Gendron, E. Kolmos, and M.-A. Nohales-Zafra for discussion and reviewing of the manuscript; A. Kwon, J. King, and M. Tien for technical assistance; and A. Lestelle for administrative support. Research reported in this publication was supported by the National Institute of General Medical Sciences of the National Institutes of Health under award numbers R01GM056006, R01GM067837, and RC2GM092412 to S.A.K.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hazen SP, Schultz TF, Pruneda-Paz JL, Borevitz JO, Ecker JR, Kay SA. LUX ARRHYTHMO encodes a Myb domain protein essential for circadian rhythms. Proc Natl Acad Sci U S A. 2005;102:10387–10392. doi: 10.1073/pnas.0503029102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Onai K, Ishiura M. PHYTOCLOCK 1 encoding a novel GARP protein essential for the Arabidopsis circadian clock. Genes Cells. 2005;10:963–972. doi: 10.1111/j.1365-2443.2005.00892.x. [DOI] [PubMed] [Google Scholar]
- 3.Helfer A, Nusinow DA, Chow BY, Gehrke AR, Bulyk ML, Kay SA. LUX ARRHYTHMO encodes a nighttime repressor of circadian gene expression in the Arabidopsis core clock. Curr Biol. 2011;21:126–133. doi: 10.1016/j.cub.2010.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nusinow DA, Helfer A, Hamilton EE, King JJ, Imaizumi T, Schultz TF, Farre EM, Kay SA. The ELF4-ELF3-LUX complex links the circadian clock to diurnal control of hypocotyl growth. Nature. 2011;475:398–402. doi: 10.1038/nature10182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chow BY, Helfer A, Nusinow DA, Kay SA. ELF3 recruitment to the PRR9 promoter requires other Evening Complex members in the Arabidopsis circadian clock. Plant Signal Behav. 2012;7:170–173. doi: 10.4161/psb.18766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Herrero E, Kolmos E, Bujdoso N, Yuan Y, Wang M, Berns MC, Uhlworm H, Coupland G, Saini R, Jaskolski M, Webb A, Goncalves J, Davis SJ. EARLY FLOWERING4 recruitment of EARLY FLOWERING3 in the nucleus sustains the Arabidopsis circadian clock. Plant Cell. 2012;24:428–443. doi: 10.1105/tpc.111.093807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bieniawska Z, Espinoza C, Schlereth A, Sulpice R, Hincha DK, Hannah MA. Disruption of the Arabidopsis circadian clock is responsible for extensive variation in the cold-responsive transcriptome. Plant Physiol. 2008;147:263–279. doi: 10.1104/pp.108.118059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hsu PY, Devisetty UK, Harmer SL. Accurate timekeeping is controlled by a cycling activator in Arabidopsis. Elife. 2013;2:e00473. doi: 10.7554/eLife.00473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang W, Perez-Garcia P, Pokhilko A, Millar AJ, Antoshechkin I, Riechmann JL, Mas P. Mapping the core of the Arabidopsis circadian clock defines the network structure of the oscillator. Science. 2012;336:75–79. doi: 10.1126/science.1219075. [DOI] [PubMed] [Google Scholar]
- 10.Pruneda-Paz JL, Breton G, Para A, Kay SA. A functional genomics approach reveals CHE as a component of the Arabidopsis circadian clock. Science. 2009;323:1481–1485. doi: 10.1126/science.1167206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stockinger EJ, Gilmour SJ, Thomashow MF. Arabidopsis thaliana CBF1 encodes an AP2 domain-containing transcriptional activator that binds to the C-repeat/DRE, a cis-acting DNA regulatory element that stimulates transcription in response to low temperature and water deficit. Proc Natl Acad Sci U S A. 1997;94:1035–1040. doi: 10.1073/pnas.94.3.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu Q, Kasuga M, Sakuma Y, Abe H, Miura S, Yamaguchi-Shinozaki K, Shinozaki K. Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low-temperature-responsive gene expression, respectively, in Arabidopsis. Plant Cell. 1998;10:1391–1406. doi: 10.1105/tpc.10.8.1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gilmour SJ, Zarka DG, Stockinger EJ, Salazar MP, Houghton JM, Thomashow MF. Low temperature regulation of the Arabidopsis CBF family of AP2 transcriptional activators as an early step in cold-induced COR gene expression. Plant J. 1998;16:433–442. doi: 10.1046/j.1365-313x.1998.00310.x. [DOI] [PubMed] [Google Scholar]
- 14.Medina J, Bargues M, Terol J, Perez-Alonso M, Salinas J. The Arabidopsis CBF gene family is composed of three genes encoding AP2 domain-containing proteins whose expression Is regulated by low temperature but not by abscisic acid or dehydration. Plant Physiol. 1999;119:463–470. doi: 10.1104/pp.119.2.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gilmour SJ, Fowler SG, Thomashow MF. Arabidopsis transcriptional activators CBF1, CBF2, and CBF3 have matching functional activities. Plant Mol Biol. 2004;54:767–781. doi: 10.1023/B:PLAN.0000040902.06881.d4. [DOI] [PubMed] [Google Scholar]
- 16.Baker SS, Wilhelm KS, Thomashow MF. The 5'-region of Arabidopsis thaliana cor15a has cis-acting elements that confer cold-, drought- and ABA-regulated gene expression. Plant Mol Biol. 1994;24:701–713. doi: 10.1007/BF00029852. [DOI] [PubMed] [Google Scholar]
- 17.Yamaguchi-Shinozaki K, Shinozaki K. A novel cis-acting element in an Arabidopsis gene is involved in responsiveness to drought, low-temperature, or high-salt stress. Plant Cell. 1994;6:251–264. doi: 10.1105/tpc.6.2.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jaglo-Ottosen KR, Gilmour SJ, Zarka DG, Schabenberger O, Thomashow MF. Arabidopsis CBF1 overexpression induces COR genes and enhances freezing tolerance. Science. 1998;280:104–106. doi: 10.1126/science.280.5360.104. [DOI] [PubMed] [Google Scholar]
- 19.Gilmour SJ, Sebolt AM, Salazar MP, Everard JD, Thomashow MF. Overexpression of the Arabidopsis CBF3 transcriptional activator mimics multiple biochemical changes associated with cold acclimation. Plant Physiol. 2000;124:1854–1865. doi: 10.1104/pp.124.4.1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harmer SL, Hogenesch JB, Straume M, Chang HS, Han B, Zhu T, Wang X, Kreps JA, Kay SA. Orchestrated transcription of key pathways in Arabidopsis by the circadian clock. Science. 2000;290:2110–2113. doi: 10.1126/science.290.5499.2110. [DOI] [PubMed] [Google Scholar]
- 21.Edwards KD, Anderson PE, Hall A, Salathia NS, Locke JC, Lynn JR, Straume M, Smith JQ, Millar AJ. FLOWERING LOCUS C mediates natural variation in the high-temperature response of the Arabidopsis circadian clock. Plant Cell. 2006;18:639–650. doi: 10.1105/tpc.105.038315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kidokoro S, Maruyama K, Nakashima K, Imura Y, Narusaka Y, Shinwari ZK, Osakabe Y, Fujita Y, Mizoi J, Shinozaki K, Yamaguchi-Shinozaki K. The phytochrome-interacting factor PIF7 negatively regulates DREB1 expression under circadian control in Arabidopsis. Plant Physiol. 2009;151:2046–2057. doi: 10.1104/pp.109.147033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mockler TC, Michael TP, Priest HD, Shen R, Sullivan CM, Givan SA, McEntee C, Kay SA, Chory J. The DIURNAL project: DIURNAL and circadian expression profiling, model-based pattern matching, and promoter analysis. Cold Spring Harb Symp Quant Biol. 2007;72:353–363. doi: 10.1101/sqb.2007.72.006. [DOI] [PubMed] [Google Scholar]
- 24.Novillo F, Medina J, Salinas J. Arabidopsis CBF1 and CBF3 have a different function than CBF2 in cold acclimation and define different gene classes in the CBF regulon. Proc Natl Acad Sci U S A. 2007;104:21002–21007. doi: 10.1073/pnas.0705639105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Achard P, Gong F, Cheminant S, Alioua M, Hedden P, Genschik P. The cold-inducible CBF1 factor-dependent signaling pathway modulates the accumulation of the growth-repressing DELLA proteins via its effect on gibberellin metabolism. Plant Cell. 2008;20:2117–2129. doi: 10.1105/tpc.108.058941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hogenesch JB, Ueda HR. Understanding systems-level properties: timely stories from the study of clocks. Nat Rev Genet. 2011;12:407–416. doi: 10.1038/nrg2972. [DOI] [PubMed] [Google Scholar]
- 27.Gery C, Zuther E, Schulz E, Legoupi J, Chauveau A, McKhann H, Hincha DK, Teoule E. Natural variation in the freezing tolerance of Arabidopsis thaliana: effects of RNAi-induced CBF depletion and QTL localisation vary among accessions. Plant Sci. 2011;180:12–23. doi: 10.1016/j.plantsci.2010.07.010. [DOI] [PubMed] [Google Scholar]
- 28.Harmer SL, Kay SA. Positive and negative factors confer phase-specific circadian regulation of transcription in Arabidopsis. Plant Cell. 2005;17:1926–1940. doi: 10.1105/tpc.105.033035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mikkelsen MD, Thomashow MF. A role for circadian evening elements in cold-regulated gene expression in Arabidopsis. Plant J. 2009;60:328–339. doi: 10.1111/j.1365-313X.2009.03957.x. [DOI] [PubMed] [Google Scholar]
- 30.Fowler SG, Cook D, Thomashow MF. Low temperature induction of Arabidopsis CBF1, 2, and 3 is gated by the circadian clock. Plant Physiol. 2005;137:961–968. doi: 10.1104/pp.104.058354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dong MA, Farre EM, Thomashow MF. Circadian clock-associated 1 and late elongated hypocotyl regulate expression of the C-repeat binding factor (CBF) pathway in Arabidopsis. Proc Natl Acad Sci U S A. 2011;108:7241–7246. doi: 10.1073/pnas.1103741108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Espinoza C, Degenkolbe T, Caldana C, Zuther E, Leisse A, Willmitzer L, Hincha DK, Hannah MA. Interaction with diurnal and circadian regulation results in dynamic metabolic and transcriptional changes during cold acclimation in Arabidopsis. PLoS One. 2010;5:e14101. doi: 10.1371/journal.pone.0014101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Seo PJ, Park MJ, Lim MH, Kim SG, Lee M, Baldwin IT, Park CM. A self-regulatory circuit of CIRCADIAN CLOCK-ASSOCIATED1 underlies the circadian clock regulation of temperature responses in Arabidopsis. Plant Cell. 2012;24:2427–2442. doi: 10.1105/tpc.112.098723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nakamichi N, Kusano M, Fukushima A, Kita M, Ito S, Yamashino T, Saito K, Sakakibara H, Mizuno T. Transcript profiling of an Arabidopsis PSEUDO RESPONSE REGULATOR arrhythmic triple mutant reveals a role for the circadian clock in cold stress response. Plant Cell Physiol. 2009;50:447–462. doi: 10.1093/pcp/pcp004. [DOI] [PubMed] [Google Scholar]
- 35.Nakamichi N, Kiba T, Kamioka M, Suzuki T, Yamashino T, Higashiyama T, Sakakibara H, Mizuno T. Transcriptional repressor PRR5 directly regulates clock-output pathways. Proc Natl Acad Sci U S A. 2012;109:17123–17128. doi: 10.1073/pnas.1205156109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu T, Carlsson J, Takeuchi T, Newton L, Farre EM. Direct regulation of abiotic responses by the Arabidopsis circadian clock component PRR7. Plant J. 2013;76:101–114. doi: 10.1111/tpj.12276. [DOI] [PubMed] [Google Scholar]
- 37.Keily J, Macgregor DR, Smith RW, Millar AJ, Halliday KJ, Penfield S. Model selection reveals control of cold signalling by evening-phased components of the plant circadian clock. Plant J. 2013;76:247–257. doi: 10.1111/tpj.12303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cao S, Ye M, Jiang S. Involvement of GIGANTEA gene in the regulation of the cold stress response in Arabidopsis. Plant Cell Rep. 2005;24:683–690. doi: 10.1007/s00299-005-0061-x. [DOI] [PubMed] [Google Scholar]
- 39.Cheng Y, Gvakharia B, Hardin PE. Two alternatively spliced transcripts from the Drosophila period gene rescue rhythms having different molecular and behavioral characteristics. Mol Cell Biol. 1998;18:6505–6514. doi: 10.1128/mcb.18.11.6505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Majercak J, Sidote D, Hardin PE, Edery I. How a circadian clock adapts to seasonal decreases in temperature and day length. Neuron. 1999;24:219–230. doi: 10.1016/s0896-6273(00)80834-x. [DOI] [PubMed] [Google Scholar]
- 41.Garceau NY, Liu Y, Loros JJ, Dunlap JC. Alternative initiation of translation and time-specific phosphorylation yield multiple forms of the essential clock protein FREQUENCY. Cell. 1997;89:469–476. doi: 10.1016/s0092-8674(00)80227-5. [DOI] [PubMed] [Google Scholar]
- 42.Liu Y, Garceau NY, Loros JJ, Dunlap JC. Thermally regulated translational control of FRQ mediates aspects of temperature responses in the Neurospora circadian clock. Cell. 1997;89:477–486. doi: 10.1016/s0092-8674(00)80228-7. [DOI] [PubMed] [Google Scholar]
- 43.Diernfellner AC, Schafmeier T, Merrow MW, Brunner M. Molecular mechanism of temperature sensing by the circadian clock of Neurospora crassa. Genes Dev. 2005;19:1968–1973. doi: 10.1101/gad.345905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhou M, Guo J, Cha J, Chae M, Chen S, Barral JM, Sachs MS, Liu Y. Non-optimal codon usage affects expression, structure and function of clock protein FRQ. Nature. 2013;495:111–115. doi: 10.1038/nature11833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xu Y, Ma P, Shah P, Rokas A, Liu Y, Johnson CH. Non-optimal codon usage is a mechanism to achieve circadian clock conditionality. Nature. 2013;495:116–120. doi: 10.1038/nature11942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Morf J, Rey G, Schneider K, Stratmann M, Fujita J, Naef F, Schibler U. Cold-inducible RNA-binding protein modulates circadian gene expression posttranscriptionally. Science. 2012;338:379–383. doi: 10.1126/science.1217726. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.