Abstract
The polymorphic hepatic enzyme CYP2C19 catalyzes the metabolism of clinically important drugs such as clopidogrel, proton-pump inhibitors, and others and clinical pharmacogenetic testing for clopidogrel is increasingly common. The CYP2C19*10 SNP is located 1 bp upstream the CYP2C19*2 SNP. Despite the low frequency of the CYP2C19*10 allele, its impact on metabolism of CYP2C19 substrates and CYP2C19*2 genotyping makes it an important SNP to consider for pharmacogenetic testing of CYP2C19. However, the effect of the CYP2C19*10 allele on clopidogrel metabolism has not been explored to date. We measured the enzymatic activity of the CYP2C19.10 protein against clopidogrel. The catalytic activity of CYP2C19.10 in the biotransformation of clopidogrel and 2-oxo-clopidgorel was significantly decreased relative to wild type CYP2C19.1B. We also report that the CYP2C19*10 SNP interferes with the CYP2C19*2 TaqMan® genotyping assay, resulting in miscalling of CYP2C19*10/*2 as CYP2C19*2/*2. Our data provide evidence of CYP2C19.10’s reduced metabolism of clopidogrel and 2-oxo-clopidogrel.
Keywords: CYP2C19*10, Clopidogrel, Pharmacokinetic, Pharmacogenetic, genotyping
INTRODUCTION
CYP2C19 contains the largest frequency of null alleles among the CYP2C gene family as well as various reduced function alleles, and it catalyzes the metabolism of many clinically important drugs. The contribution of CYP2C19 catalytic activity varies between different substrates such as the anticonvulsant mephenytoin, the proton-pump inhibitors omeprazole and lansoprazole, and the bioactivation of antiplatelet drug clopidogrel [1–3]. There are more than 30 variant alleles of the CYP2C19 gene (http://www.cypalleles.ki.se/cyp2c19.htm). The CYP2C19*2 and CYP2C19*3 null alleles and the CYP2C19*17 gain of function allele are the most common CYP2C19 alleles. The frequency of CYP2C19*2 allele varies from ~15% for Caucasians and Africans to ~29–35% in Asians [4].
The non-synonymous CYP2C19*10 SNP rs6413438 (C680T, Pro227Leu) is located 1 bp upstream of the null CYP2C19*2 SNP rs4244285 (G681A, a cryptic splice site). The CYP2C19*10 SNP codes for a reduced function allelic protein that was first reported by Blaisdell et al. 2002. The lower in vitro metabolic activity of recombinant CYP2C19.10 protein against the two commonly used probe substrates of CYP2C19, S-mephenytoin and omeprazole, has been previously reported [1, 2]. The effect of the CYP2C19.10 allelic protein on clopidogrel metabolism has not been evaluated. In this study, we measured the enzymatic activity of recombinant CYP2C19.10 protein on clopidogrel, S-mephenytoin and omeprazole in vitro.
It has also been reported that the presence of CYP2C19*10 SNP before CYP2C19*2 SNP on exon 5 of CYP2C19 gene interferes with CYP2C19*2 genotyping in a PCR-RFLP assay [5] and the INFINITI CYP2C19*2 genotyping assay (www.autogenomics.com/pdf/INFINITICYP2C19PackageInsertUSA) resulting in misclassification of the CYP2C19*10 allele as the CY2C19*2 allele.
Clopidogrel is an antiplatelet prodrug that undergoes a complex multi-step oxidative metabolism and bioactivation to its active metabolite in the liver by CYP2C19 and a few other CYP enzymes. The CYP2C19*2 and CYP2C19*3 null polymorphisms in CYP2C19 gene has been associated with reduced clinical response to clopidogrel [4]. Clopidogrel is a major focus of our Personalized Medicine Program at the University of Florida, and in other clinical phamacogenetic programs. As a part of our clinical pharmacogenetic program, we noted a potential problem with genotyping CYP2C19*2 when CYP2C19*10 is present. We then recognized the absence of data on CYP2C19.10 on clopidogrel metabolism. Hence we describe the impact of CYP2C19.10 on metabolism of clopidogrel and 2-oxo-clopidogrel and also the effect of the CYP2C19*10 SNP on the CYP2C19*2 TaqMan® genotyping assay.
MATERIALS AND METHODS
Chemicals and Materials
Clopidogrel, 2-oxo-clopidogrel, d4-clopidogrel, and the 2-bromo-3’-methoxy acetophenone (MPB) derivative of racemic cis-clopidogrel thiol active metabolite (clopidogrel-AM) were obtained from Toronto Research Chemicals (catalogue number C587256, Ontario, Canada). Omeprazole, S-mephenytoin, 5-hydroxyomeprazole, 4-hydroxymephenytoin, recombinant human NADPH-P450 oxidoreductase, human cytochorome b5, and other regents were purchased from Sigma-Aldrich (St. Louis, MO). Partially purified recombinant bacterially expressed CYP2C19.1B (wild type) and CYP2C19.10 proteins were provided by J. Goldstein’s laboratory [1].
CYP2C19.10 kinetic study
Recombinant CYP2C19.1 and CYP2C19.10 were modified at the N-terminus for optimal expression in bacteria by replacing the first 8 amino acids with those of bovine cytochrome P450 17-α. These amino acids are not involved in substrate specificity. The allelic proteins were expressed in E coli DH5α and purified as described extensively in [1, 2, 12].
The in vitro incubation study was conducted following a previously published method with some minor modifications [1]. In brief, the partially purified recombinant CYP2C19.1B (wild type) and CYP2C19.10 proteins were pre-incubated with recombinant human NADPH-P450 oxidoreductase (4 pmol/pmol P450) and human cytochrome b5 (2 pmol/ pmol P450) at 37°C for 5 min, followed by adding various concentrations of the tested substrates, including clopidogrel (300, 400, 500 µM), 2-oxo-clopidogrel (10, 20, 50, 100, 200, 386 µM), omeprazole (2, 5, 10, 20, 50, 100, 200, 500 µM), or S-mephenytoin (50, 100, 200, 400, 600, 1000 µM) in a reaction buffer containing 20 mM HEPES, 0.1 mM EDTA and 1.25 mM MgCl2 (pH 7.4) for 3 min. The reaction was initiated by adding NADPH-generating reagents (0.1 mg/ml of yeast glucose-6-phosphate dehydrogenase, 3 mg/ml of NADP+, and 0.07 M glucose-6-phosphate). The final concentrations of the CYP2C19 enzymes were 2.24 µM, 1.12 µM, 0.046 µM, and 0.11 µM for the reactions of clopidogrel, 2-oxo-clopidogrel, omeprazole, and S-mephenytoin, respectively. The incubation times were 2 h for clopidogrel, 20 min for 2-oxo-clopidogrel, and 10 min for omeprazole and S-mephenytoin. The amounts of the CYP2C19 enzymes and the incubation times were evaluated in preliminary studies to ensure that the formation of the metabolites was linear with respect to the concentrations of enzymes and incubation time.
The metabolic reactions of clopidogrel and 2-oxo-clopidogrel were terminated by adding a two-fold volume of acetonitrile containing the internal standard d4-clopidogrel (10 ng/ml) and the derivatizing reagent MPB, (5 mM). MPB was utilized to convert the unstable clopidogrel-AM to its stable MPB derivative to facilitate analysis. The samples were vortexed and left at room temperature for 10 min to allow the completion of derivatization. The reactions for the hydroxylation of omeprazole and mephenyoin were terminated by adding two-fold volume of acetonitrile containing the internal standards d3-hydroxyomeprazole (10 ng/ml). All samples were then vortexed and centrifuged at 16,000 xg for 20 min at 4°C. The supernatants, containing parent compounds and formed metabolites (clopidogrel-AM, 5-hydroxyomeprazole, and 4-hydroxymephenytoin) were collected and subjected to liquid chromatograph tandem mass spectrometry (LC-MS/MS) analysis utilizing assays described below.
LC-MS/MS assays
The LC-MS/MS system consisted of a Shimadzu HPLC system coupled with an AB Sciex API 3000 triple quadrupole mass spectrometer. The concentrations of clopidogrel, 2-oxo-clopidogrel, and clopidogrel-AM were determined using an LC-MS/MS assay described in our recent publication [13]. The assays for the quantification of omeprazole, mephenytoin and their respective hydroxylation metabolites were developed based on the published methods with some modifications [14, 15]. Briefly, the analytes were separated on a Phenomenex (Torrance, CA) 3-µm C18 reverse phase column (2.0×150 mm) at a flow rate of 0.2 mL/min. The mobile phase was 90% methanol with 0.1% formic acid for omeprazole and 80% acetonitrile with 0.2% formic acid for S-mephenytoin. Ionization was achieved via ESI in the positive mode and ions were monitored by multiple reaction monitoring. Omeprazole, 5-hydroxyomeprazole, S-mephenytoin, 4-hydroxymephenytoin and d3-hydroxyomeprazole (internal standard for omeprazole and S-mephenytoin) were monitored via the m/z transition 346.1/198.1, 362.3/213.9, 219.1/134.1, 235.1/150.2 and 365.3/213.9, respectively. The collision energy was 32V and 25V for omeprazole and S-mephenytoin, respectively. Ionspray voltage, source temperature, and curtain gas was 5000 V, 400 °C and 12 L/h, respectively, for both methods. The assays were validated with precision (CV% < 10%) and accuracy (88% – 114%) for all analytes.
CYP2C19*10 allele genotyping
Genomic DNA from the INternatioinal Verapamil SR Trandolapil Study (INVEST), and Pharmacogenomic Evaluation of Antihypertensive Responses (PEAR) were isolated from mouthwash or blood by using commercially available kits (PureGene, Gentra Systems Inc., Minneapolis, MN, and QIAGEN DNA Blood Isolation Kit, QIAGEN, Valencia, CA). DNA samples from a total of 181 African-American and 202 Hispanic-Latinos DNA samples from PEAR [9] and INVEST [10] clinical studies were genotyped for CYP2C19*2 and *10 by pyrosequencing [11]. The pyrosequencing genotyping assay for CYP2C19*2 and CYP2C19*10 were designed by Pyrosequencing assay design software. The PCR and sequencing primers, annealing temperature and sequence to analyze for pyrosequencing assays are as follow, the CYP2C19*10 and CYP2C19*2 forward Bio-5’-TTACAACCAGAGCTTGGCATAT-3’, Reverse-5’-CGCAAGCAGTCACATAACTAAGC-3’PCR primers, reverse sequencing primer 5’- AAGTAATTTGTTATGGGTTC-3’, annealing temperature 58° C, and sequence to analyze, CC/TG/AGGAAATAATCAATGA. Both CYP2C19*2 and CYP2C19*10 SNPs are genotyped by single PCR and pyrosequencing reaction. The predicted histogram and pyrogram along with genotype calls for a sample are shown in figure 1.
Figure 1.
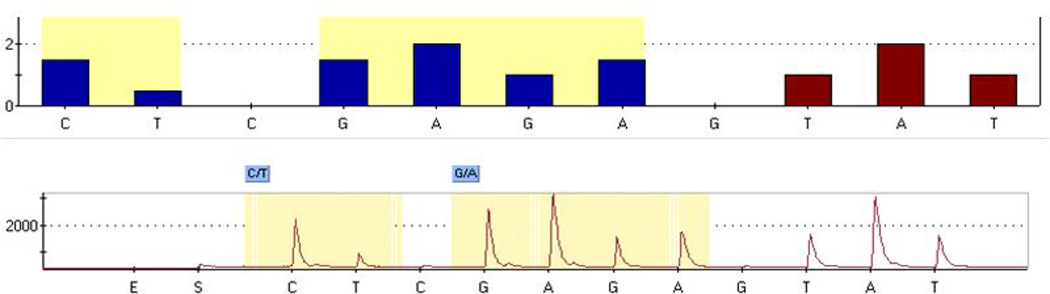
Predicted histogram and pyrogram along with genotype call for an individual with CYP2C19*10 (C/T) and CYP2C19*2 (G/A) genotype are shown. The order of nucleotide dispensation is located at the bottom of histogram and pyrogram.
Statistical Analysis
Data are presented as mean ± SD of duplicated independent experiments. Data were fit to the Michaelis-Menten equation, and the kinetic parameters Km and Vmax were calculated via nonlinear regression analysis (Graphpad Prism software version 4.0; Graphpad Software, Inc., San Diego, CA). Apparent intrinsic clearance (Clint) was calculated as the ratio of Vmax to Km. The differences in kinetic parameters between CYP2C19.1B and CYP2C19.10 were analyzed using the student’s t-test and P<0.05 is considered statistically significant. Hardy-Weinberg Equilibrium (HWE) was tested using a χ2 (chi square) goodness-of-fit test with one degree of freedom.
RESULTS
CYP2C19.10 kinetic analysis
The in vitro incubation studies demonstrated that CYP2C19.1B can efficiently catalyze the hydroxylation of the known CYP2C19 substrates omeprazole and S-mephenytoin (Fig. 2A and Fig. 2B). The Vmax values of CYP2C19.1B were determined to be 2.77 ± 0.04 nmole/min/nmole and 1.17 ± 0.13 nmole/min/nmole for omeprazole and S-mephenytoin, respectively, under the experimental conditions. The Km values for omeprazole and S-mephenytoin, were 32 ± 2 µM and 119 ± 21 µM, respectively. Consistent with previous reports, CYP2C19.10 behaved as a loss-of-function variant in the metabolism of S-mephenytoin, while exhibiting markedly decreased enzymatic activity towards omeprazole hydroxylation. The observed Clint values of CYP2C19.10 (2.2 ± 0.04) for omeprazole 5’-hydroxylation were less than 3% of that of CYP2C19.1B (87 ± 2.6).
Figure 2.
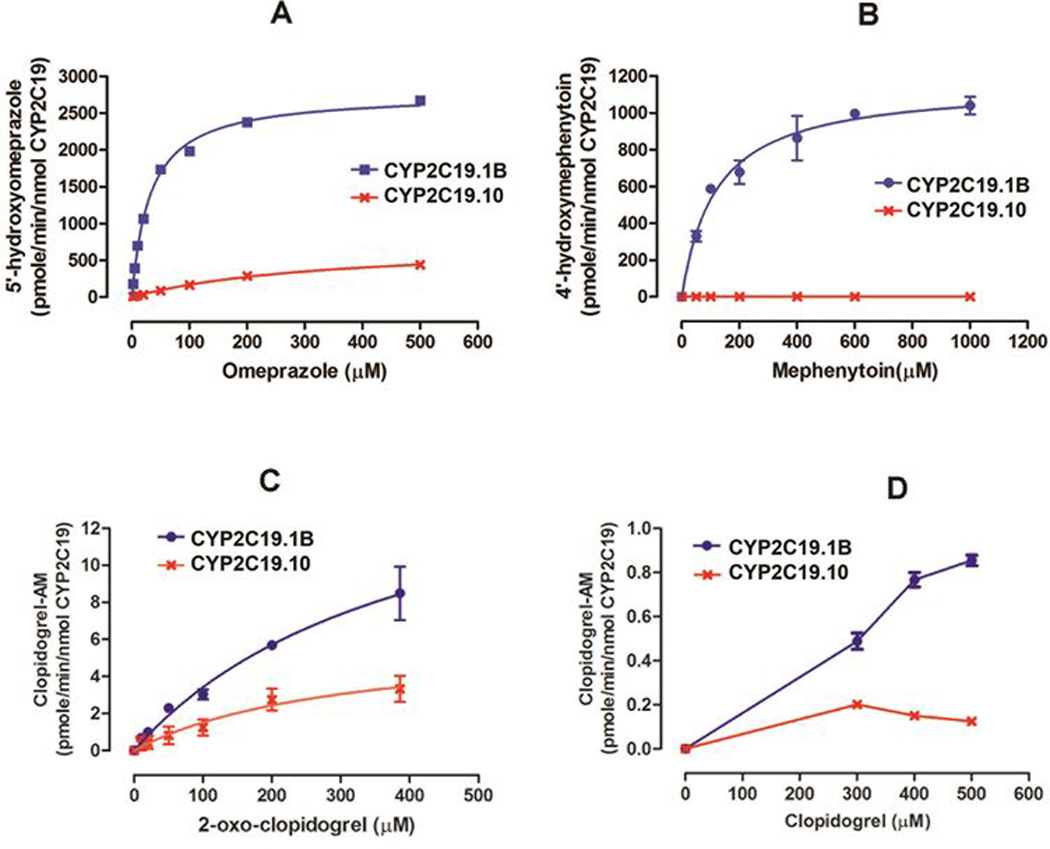
Kinetic analysis of catalytic activity of CYP2C19.1B (wild type) and CYP2C19.10 proteins on the metabolism of omeprazole (A), S-mephenytoin (B), 2-oxo-clopidogrel (C), and clopidogrel (D). Enzymatic activity was determined by the measurements of respective metabolites as indicated in the Y axis title of each figure. Values are means ± SD of two independent experiments.
The clopidogrel intermediate metabolite 2-oxo-clopidogrel was efficiently metabolized to clopidogrel-AM by CYP2C19.1B (Fig. 2C). However, the catalytic activity of CYP2C19.10 on the activation of 2-oxo-clopidgorel was significantly impaired relative to the wild type enzyme. The calculated Clint value of CYP2C19.10 on 2-oxo-clopdigrel activation was approximately 25% that of CYP2C19.1B (Table 1). Significantly decreased enzymatic activity of CYP2C19.10 was also observed for the activation of clopidogrel (Fig. 2D). The observed velocities of clopidogrel-AM formation from 2-oxo-clopidogrel and clopidogrel (Fig. 2C and Fig. 2D) indicate that the activation of clopidogrel mediated by CYP2C19 was less efficient relative to 2-oxo-clopidogrel activation. We were only able to measure the activation of clopidogrel at three relatively high substrate concentrations (i.e. 300, 400, 500 µM). The kinetic parameters Vmax and Km of clopidogrel activation were not determined due to the limited range of substrate concentrations available for analysis. The kinetic parameters of other tested substrates are summarized in Table 1.
Table 1.
Enzyme kinetic parameters for CYP2C19.1B and CYP2C19.10
| 2-oxo-clopidogrel | Omeprazole | S-mephenytoin | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Vmax | Km (µM) | Clint | Vmax | Km (µM) | Clint | Vmax | Km (µM) | Clint | |
| 2C19.1B | 17.4 ± 4.1 | 409.2 ± 159.5 | 0.052 ± 0.008 | 2.77 ± 0.04 | 32 ± 2 | 87.2 ± 2.6 | 1.17 ± 0.13 | 119 ± 21 | 9.7 ± 2.7 |
| 2C19.10 | 6.1 ± 2.3 | 302.3 ± 202.4 | 0.013 ± 0.001** | 0.74 ± 0.02** | 336 ± 18** | 2.2 ± 0.04** | N/A | N/A | N/A |
Vmax pmole/min/nmole CYP2C19 for 2-oxo-clopidogrel; nmole/min/nmole CYP2C19 for omeprazole and S-mephenytoin
Km µM
Clint nl/min/nmole CYP2C19
N/A: Not available due to undetectable metabolite
P<0.01 versus CYP 2C19.1B
CYP2C19*10 genotyping and frequency
The presence of the CYP2C19*10 SNP in close proximity to CYP2C19*2 interferes with the CYP2C19*2 TaqMan® genotyping assay and results in miscalling of CYP2C19*10/*2 as CYP2C19*2/*2. This only has been observed with *2 allele and does not affect other alleles such as *1 (*1/*10) or *17 (*10/*17). The frequency of the CYP2C19*10 allele is 0.21% for African Americans and Mexicans and 0.04% for Caucasians (http://www.ncbi.nlm.nih.gov/snp). The presence of CYP2C19*10 SNP disrupted the CYP2C19*2 TaqMan® assay genotyping process and heterozygous samples with CYP2C19*10/*2 genotypes were called as homozygous CYP2C19*2/*2. The sample was then genotyped by a pyrosequencing method and the actual genotype was confirmed as CYP2C19*10/*2. The genotype calls for an individual sample with CYP2C19*10 (C/T) and CYP2C19*2 (G/A) genotype are shown in figure 1. The CYP2C19*10 allele frequency for African-Americans, Hispanics and Caucasians was 0.8%, 0.25%, and 0%, respectively.
DISCUSSION
The non-synonymous CYP2C19*10 SNP results in substitution of proline 227, a highly conserved amino acid among the CYP2C family, to leucine. Proline 227 is located within the F-G loop, forming a part of the substrate access channel in CYP2C19 that plays an important role in substrate specificity [6, 7]. Although CYP2C19.10 is not a null variant, it was shown to have significantly decreased enzymatic activity toward menphenytoin and omeperazole relative to the wild type enzyme, behaving functionally like a loss of function allele [1, 8, 2]. The potential impact of the CYP2C19.10 variant on metabolism of other CYP2C19 substrates, CYP2C19*2 genotyping and the absence of kinetic data on clopidogrel bioactivation were the reasons we performed this study. To our knowledge, this is the first study to measure the enzymatic activity of CYP2C19.10 protein toward clopidogrel and 2-oxo-clopidogrel. S-mephenytoin and omeprazole were used as positive controls in this study and consistent with previous reports (1, 8), the CYP2C19.10 variant behaved as a loss of function allele for the metabolism of S-mephenytoin, and exhibited a significantly decreased enzymatic activity toward omeprazole hydroxylation. The activation of 2-oxo-clopidogrel by CYP2C19.10 variant was significantly impaired and its calculated Clint value was approximately 25% that of the wild type CYP2C19.1B enzyme. Thus while the CYP2C19.10 allelic protein shows dramatic loss of function for S-mephenytoin and omeperazole, it clearly exhibits substrate specificity and would be characterized as a reduced function allele towards clopidogrel and 2-oxo-clopidogrel. Thus the impact of CYP2C19.10 on metabolism of various substrates cannot be precisely defined and will require further study with each substrate of interest.
The CYP2C19*1B-mediated activation of clopidogrel was less efficient relative to 2-oxo-clopidogrel activation. We speculate that this may be due to the complex two-step metabolism of clopidogrel compared to one step metabolism of 2-oxo-clopidogrel. Additionally, clopidogrel is a mechanism-based CYP2C19 inhibitor as determined in an in vitro study [16]. Thus, the presence of the parent compound clopidogrel may have attenuated the second step activation (i.e. 2-oxo-clopidogrel > clopidogrel-AM) catalyzed by CYP2C19. We were also not able to determine the kinetic parameters Vmax and Km of clopidogrel metabolism due to the limited range of substrate concentrations.
Since the presence of CYP2C19*10 allele affects the CY2C19*2 SNP genotyping and results in misclassification of CYP2C19*10/*2 as CYP2C19*2/*2 in some reported genotyping methods, we suggest that samples with CYP2C19*2/*2 genotypes be checked for the presence of the CYP2C19*10 SNP. Particularly since CYP2C19.10 does not behave as null allelic protein for clopidogrel. For CYP2C19 substrates such as S-mephenytoin in which CYP2C19*10 is essentially a loss of function allele, the metabolism status wouldn’t differ from that of carriers of CYP2C19*2; however, for substrates such as clopidogrel that are partially metabolized by CYP2C19.10, it is important to genotype for both CYP2C19*10 and CYP2C19*2 SNPs. Although the CYP2C19*10 allele frequency for African-Americans (0.8%), Hispanics (0.25), and Caucasians (0%), was relatively low, the important impact of this allele on CYP2C19 substrate metabolism and CY2C19*2 SNP genotyping justifies its inclusion in CYP2C19 pharmacogenetic testing panels. For CYP2C19 genotyping, we suggest to use assays that can distinguish CYP2C19*10 from CYP2C19*2.
In conclusion, our data provide evidence that CYP2C19.10 variant partially metabolizes clopidogrel and 2-oxo-clopidogrel, and the presence of CYP2C19*10 allele affects the CY2C19*2 TaqMan® genotyping assay and results in misclassification of CYP2C19*10/*2 as CYP2C19*2/*2.
Acknowledgments
This work was partially supported by the Intramural Research Program of the National Institute of Environmental Health Sciences (Project ES021024-32) (J.A.G.) and partially by Grants NIAID 1R21AI096345 (J.M and H.J.Z.), UL1RR029890-03S3 (UL1TR000064) (J.A.J.), U01 GM074492 (J.A.J and T.L.), and U01 GM074492 (J.A.J. and T.L.).
Footnotes
Conflict of Interest
There are no conflicts of interest declared.
References
- 1.Blaisdell J, Mohrenweiser H, Jackson J, Ferguson S, Coulter S, Chanas B, et al. Identification and functional characterization of new potentially defective alleles of human CYP2C19. Pharmacogenetics. 2002;12:703–711. doi: 10.1097/00008571-200212000-00004. [DOI] [PubMed] [Google Scholar]
- 2.Wang H, Kim RA, Sun D, Gao Y, Wang H, Zhu J, et al. Evaluation of the effects of 18 non-synonymous single-nucleotide polymorphisms of CYP450 2C19 on in vitro drug inhibition potential by a fluorescence-based high-throughput assay. Xenobiotica. 2011;41(9):826–835. doi: 10.3109/00498254.2011.582893. [DOI] [PubMed] [Google Scholar]
- 3.Ancrenaz V, Desmeules J, James R, Fontana P, Reny JL, Dayer P, et al. The paraoxonase-1 pathway is not a major bioactivation pathway of clopidogrel in vitro. Br J Pharmacol. 2012;166(8):2362–2370. doi: 10.1111/j.1476-5381.2012.01946.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Scott SA, Sangkuhl K, Gardner EE, Stein CM, Hulot JS, Johnson JA, et al. Clinical Pharmacogenetics Implementation Consortium. Clinical Pharmacogenetics Implementation Consortium guidelines for cytochrome P450-2C19 (CYP2C19) genotype and clopidogrel therapy. Clin Pharmacol Ther. 2011;90(2):328–332. doi: 10.1038/clpt.2011.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rasmussen H, Werge T. Misclassification of allele CYP2C19*10 as CY2C19*2 by a commonly used PCR-RFLP procedure. GenetTest. 2008;12(1):57–58. doi: 10.1089/gte.2007.0055. [DOI] [PubMed] [Google Scholar]
- 6.Tsao CC, Wester MR, Ghanayem B, Coulter SJ, Chanas B, Johnson EF, et al. Identification of human CYP2C19 residues that confer S-mephenytoin 4'-hydroxylation activity to CYP2C9. Biochemistry. 2001;40(7):1937–1944. doi: 10.1021/bi001678u. [DOI] [PubMed] [Google Scholar]
- 7.Wada Y, Mitsuda M, Ishihara Y, Watanabe M, Iwasaki M, Asahi S. Important amino acid residues that confer CYP2C19 selective activity to CYP2C9. J Biochem. 2008;144(3):323–333. doi: 10.1093/jb/mvn070. [DOI] [PubMed] [Google Scholar]
- 8.Lee SJ, Kim WY, Kim H, Shon JH, Lee SS, Shin JG. Identification of new CYP2C19 variants exhibiting decreased enzyme activity in the metabolism of S-mephenytoin and omeprazole. Drug Metab Dispos. 2009;37(11):2262–2269. doi: 10.1124/dmd.109.028175. [DOI] [PubMed] [Google Scholar]
- 9.Johnson JA, Boerwinkle E, Zineh I, Chapman AB, Bailey K, Cooper-DeHoff RM, et al. Pharmacogenomics of antihypertensive drugs: rationale and design of the Pharmacogenomic Evaluation of Antihypertensive Responses (PEAR) study. Am. Heart. J. 2009;157:442–449. doi: 10.1016/j.ahj.2008.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pepine CJ, Handberg-Thurmond E, Marks RG, Conlon M, Cooper-DeHoff R, Volkers P, et al. Rationale and design of the international verapamil SR/trandolapril study (INVEST): an internet-based randomized trial in coronary artery disease patients with hypertension. J. Am Coll. Cardiol. 1998;32:1228–1237. doi: 10.1016/s0735-1097(98)00423-9. [DOI] [PubMed] [Google Scholar]
- 11.Langaee TY, Ronaghi M. Genetic Variation Analyses by Pyrosequencing. Mutation Research. 2005;573(1–2):96–102. doi: 10.1016/j.mrfmmm.2004.07.023. [DOI] [PubMed] [Google Scholar]
- 12.Ibeanu GC, Blaisdell J, Ghanayem BI, Beyeler C, Benhamou S, Bouchardy C, et al. An additional defective allele, CYP2C19*5, contributes to the S-mephenytoin poor metabolizer phenotype in Caucasians. Pharmacogenetics. 1998;8(2):129–135. doi: 10.1097/00008571-199804000-00006. [DOI] [PubMed] [Google Scholar]
- 13.Zhu HJ, Wang X, Gawronski BE, Brinda BJ, Angiolillo DJ, Markowitz JS. Carboxylesterase 1 as a determinant of clopidogrel metabolism and activation. J Pharmacol Exp Ther. 2013;344(3):665–672. doi: 10.1124/jpet.112.201640. [DOI] [PubMed] [Google Scholar]
- 14.Kim MJ, Kim H, Cha IJ, Park JS, Shon JH, Liu KH, Shin JG. High-throughput screening of inhibitory potential of nine cytochrome P450 enzymes in vitro using liquid chromatography/tandem mass spectrometry. Rapid communications in mass spectrometry : RCM. 2005;19:2651–2658. doi: 10.1002/rcm.2110. [DOI] [PubMed] [Google Scholar]
- 15.Zhang W, Han F, Guo P, Zhao H, Lin ZJ, Huang MQ, Bertelsen K, Weng N. Simultaneous determination of tolbutamide, omeprazole, midazolam and dextromethorphan in human plasma by LC-MS/MS--a high throughput approach to evaluate drug-drug interactions. Journal of chromatography B, Analytical technologies in the biomedical and life sciences. 2010;878:1169–1177. doi: 10.1016/j.jchromb.2010.03.026. [DOI] [PubMed] [Google Scholar]
- 16.Nishiya Y, Hagihara K, Kurihara A, Okudaira N, Farid NA, Okazaki O, Ikeda T. Comparison of mechanism-based inhibition of human cytochrome P450 2C19 by ticlopidine, clopidogrel, and prasugrel. Xenobiotica. 2009;39:836–843. doi: 10.3109/00498250903191427. [DOI] [PubMed] [Google Scholar]


