Abstract
Purpose
We examined expression profiles of 16 micro RNAs (miRNAs) in triple negative breast cancers to identify their potential as biomarkers for lymph node metastasis.
Methods
The expression profiles of miR-9, miR-21, miR-30a, miR-30d, miR-31, miR-34a, miR-34c, miR-100, miR-122, miR-125b, miR-146a, miR-146b, miR-155, miR-181a, miR-200c, and miR-205 were examined by using real-time quantitative reverse transcription polymerase chain reaction in tumor samples and corresponding benign breast tissues. Their associations with histopathological features and prognostic parameters were assessed.
Results
When compared with the expression in benign breast tissues, seven of the miRNAs (miR-31, miR-205, miR-34a, miR-146a, miR-125b, miR-34c, and miR-181a) were downregulated more than 1.5-fold in tumor tissues, whereas, only miR-21 was found to be upregulated more than 1.5-fold in tumor tissues. Although miR-200c levels were decreased only 1.12-fold in tumor tissues, the reduced expressions of miR-200c and miR-205 were significantly associated with lymph node metastasis (p=0.021 and p=0.016, respectively).
Conclusion
Our results demonstrate that miR-205 and miR-200c expression levels may be useful in predicting lymph node metastasis in triple negative breast cancer patients.
Keywords: Breast, Carcinoma, micro RNAs
INTRODUCTION
MicroRNAs (miRNAs) are small noncoding RNAs that regulate gene expression through base-pairing with their target messenger RNAs. Despite the fact that their functions are largely unknown, most miRNAs have the potential to regulate more than one target gene in the human genome. Studies have revealed that miRNAs may become deregulated in human cancers, acting as oncogenes or tumor suppressor genes [1,2]. miRNAs also regulate genes associated with prognostic parameters, which play a role in tumor progression, invasion, or metastasis. Oncogenic miRNAs (oncomirs) are usually overexpressed in cancer tissues, whereas tumor suppressor miRNAs usually demonstrate reduced expression [3].
Breast cancer may be divided into four therapeutic and prognostic subgroups according to expression of the estrogen receptor (ER) and/or progesterone receptor (PR) and overexpression of human epidermal growth factor receptor 2 (HER2) [4]. The ER and PR are expressed in the vast majority of tumors, and they are usually associated with a favorable prognosis. Overexpression of HER2 is associated with more aggressive behavior, but patients in this group do have the opportunity to benefit from treatment with trastuzumab. In contrast, patients in the triple negative breast cancer (TNBC) group have the poorest outcome, and these patients do not benefit from either endocrine treatment or trastuzumab. In our patient series, 50% of the TNBC patients were found to have lymph node metastasis at the time of surgery, a figure far higher than that of the other types, and one that reflects the aggressive behavior of this type of breast cancer. Although it represents a small fraction of breast cancer, TNBC is composed of a heterogeneous disease group with different histologic, biologic, and behavioral features [4,5,6].
In the present study, we aimed to investigate the expression profiles of 16 miRNAs for their utility as predictive markers of lymph node metastasis in TNBC. We also assessed their association with clinical and histopathological parameters.
METHODS
Patients
Formalin-fixed, paraffin-embedded pretreatment tissue samples from 32 TNBC patients who underwent radical mastectomy and axillary dissection between the 2004 and 2011 were included in the current study. The study was conducted in accordance with the Declaration of Helsinki and approved by our Institutional Ethics Committee (IRB approval number, 2013-107). None of the patients had a known history of familial breast cancer. Baseline characteristics of the patients are presented in Table 1. The ER, PR, and HER2 status of tumor samples that were determined by immunohistochemical methods in routine pathological examination and the histopathological features of tumor tissues, were re-evaluated by 1 pathologist in each case. In 12 of the cases, HER2 negativity was also confirmed by the fluorescence in situ hybridization (FISH) method.
Table 1.
Baseline characteristics of triple negative breast cancer (TNBC) patients
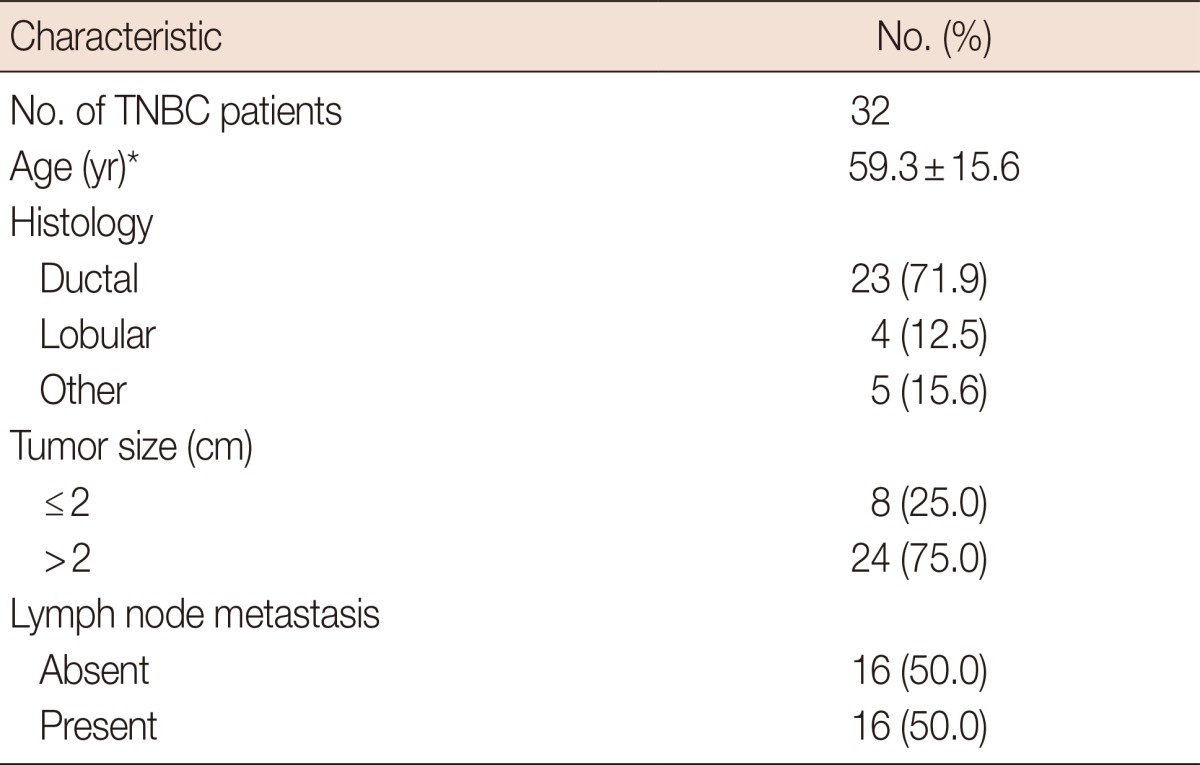
*Mean±SD.
Selection of miRNAs
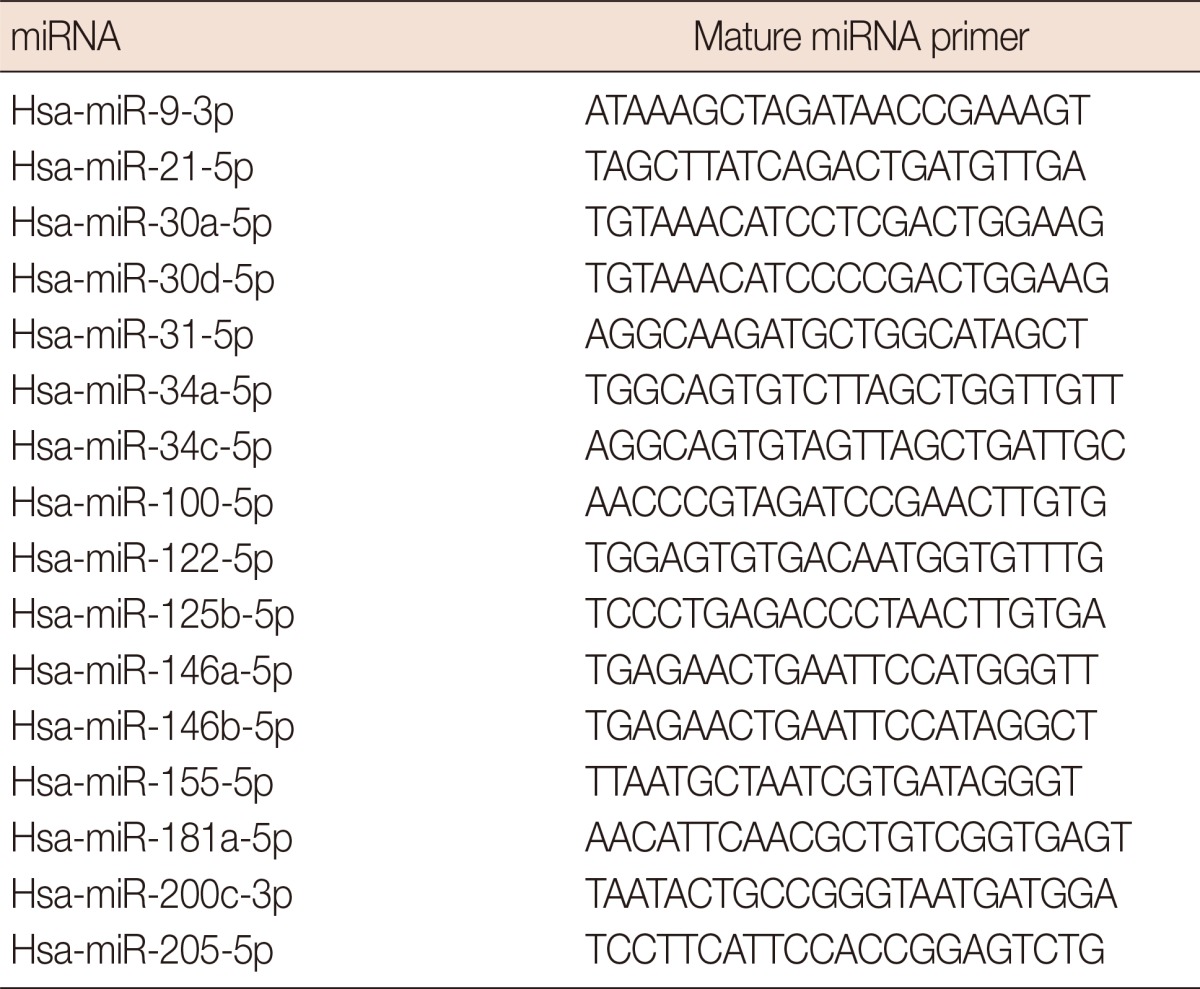
The miRNA selections were made based on a literature search for previous reports. To identify miRNAs with a predictive potential for lymph node metastasis, the search was focused on miRNAs which were found to 1) be related to the triple negative phenotype; 2) play critical roles in the cell cycle; 3) have prognostic or predictive values; 4) have a role in the invasion-metastasis cascade; or 5) be associated with aggressive behavior in several malignancies including breast cancer. Out of several candidates, further selection was made according to the target genes of the miRNAs for their involvement in the tumorigenic pathways of breast cancer. In an attempt to focus on relatively less-studied miRNAs, we excluded miRNAs such as miR-10b, miR-373, and miR-520c, the roles of which have been confirmed in the metastatic pathways of breast cancer [3]. As a result, out of hundreds of known human miRNAs, 16 candidates were considered to be suitable for the study. Selected miRNAs and their primers are listed in Table 2.
Table 2.
The list of micro RNA (miRNA) and mature miRNA primers used in the study
Isolation and relative quantification of miRNAs
For miRNA extraction from cancer tissues, previously prepared tissue macroarray blocks containing only tumor areas were used. In all cases, corresponding normal breast tissues were also selected for miRNA extraction.
Total RNA was isolated using a miRNeasy FFPE Kit (QIAGEN, Hilden, Germany). The quantity of the RNA was assessed by using a NanoDrop 1000 spectrophotometer (Thermo Scientific, Wilmington, USA). miRNAs were amplified using the SYBR Green method after reverse transcription and cDNA synthesis from isolated total RNA. In these steps, miScript technology kits were used according to the manufacturer's instructions (QIAGEN). RNU6B small nuclear RNA was used as a reference with serial dilutions of template cDNA. All reactions were performed in a final volume of 20 µL containing 2 µL of synthesized cDNA and carried out on RotorGene Q-5 Plex (QIAGEN) in duplicate by including no template and reverse transcriptase minus controls. After the amplification steps, melting curves were generated from melting data acquired from 50℃ to 85℃, at a ramping rate of 1℃/sec, to evaluate the polymerase chain reaction products. miRNA expression levels were calculated using the ΔΔCt method [7].
Statistical analysis
Results are presented as mean±standard deviation for continuous variables, and defined as percentages for categorical variables. We used the Kolmogorov-Smirnov test to assess the distributions of continuous variables. Mann-Whitney nonparametric tests were used to compare the differences in expression levels of miRNAs between different clinicopathologic disease groups such as metastatic and localized tumor groups. Unsupervised hierarchical cluster analysis was also performed to evaluate miRNA expression signatures in different tumor subtypes. All analyses were performed using SPSS for Windows software package, version 11.5 (SPSS Inc., Chicago, USA). All p-values were two-tailed, and p<0.05 was set as the level of statistical significance.
RESULTS
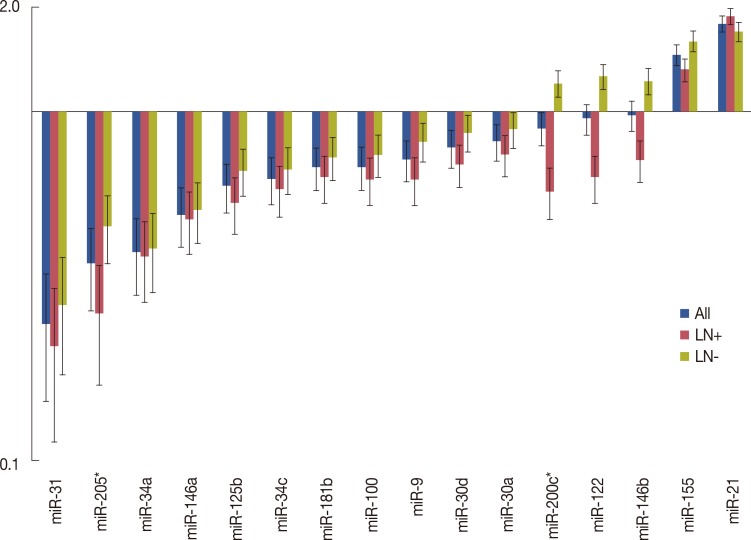
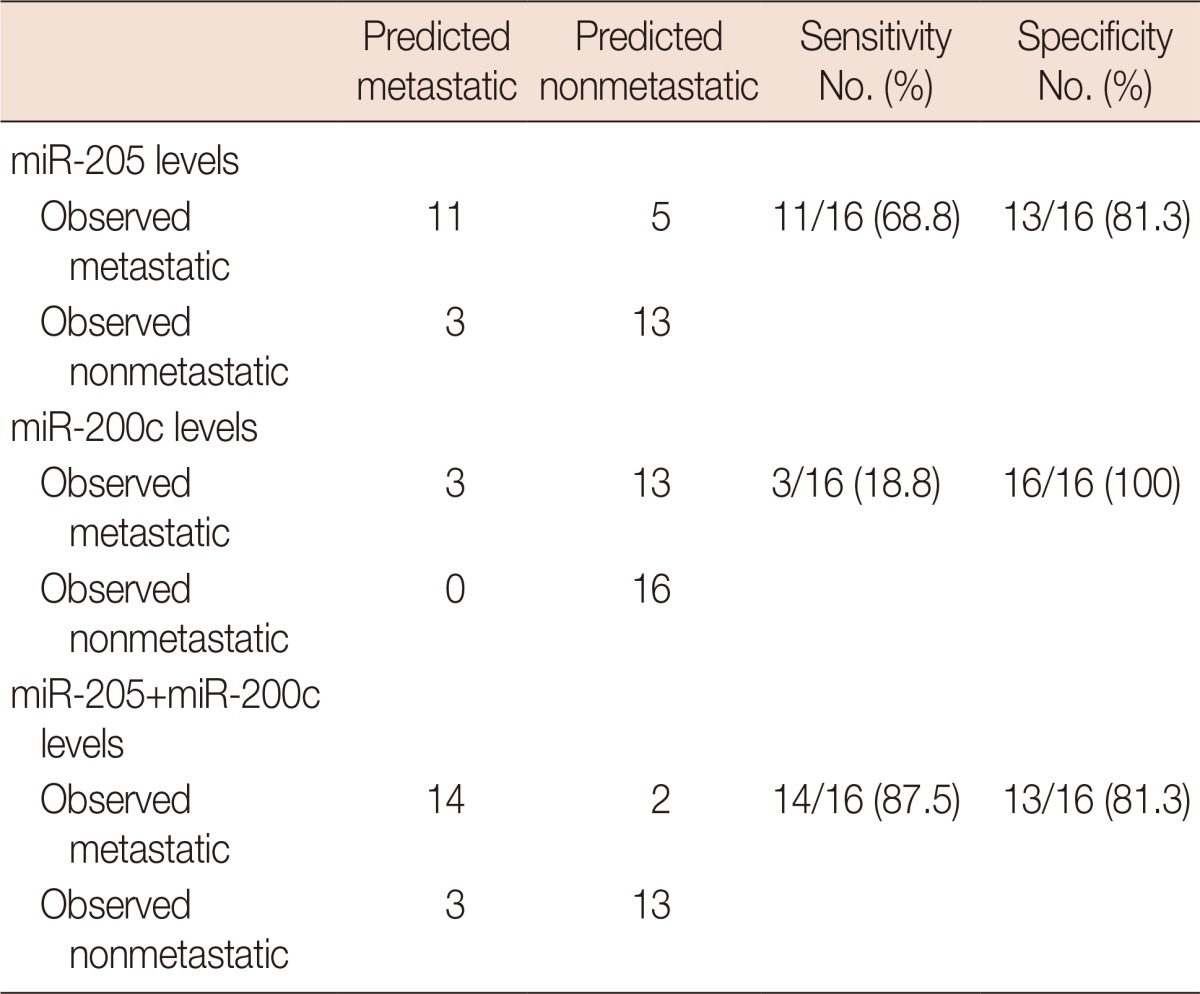
To identify their predictive potential for lymph node metastasis, we examined the expression profiles of 16 miRNAs in 32 TNBC samples and corresponding normal breast tissues. Seven of the miRNAs were downregulated in tumor tissues by 1.5-fold or more, and only miR-21 was found to be upregulated by more than 1.5-fold. The mean miRNA expression levels of cancer tissues normalized to normal breast tissues and the ratios in patients with and without metastasis are presented in Figure 1. miR-31 was the most strongly downregulated miRNA (mean, 0.247±0.299), followed by miR-205 (mean, 0.367±0.353) and miR-34a (mean, 0.396±0.303), respectively (Table 3). When the miRNA levels of the metastatic and nonmetastatic groups were compared, miR-205 and miR-200c levels were significantly decreased in the metastatic group (p=0.016 and p=0.021, respectively). In the metastatic group, miR-205 expression levels showed a more than 5-fold decrease in 11 (68.8%) cases, whereas this was observed in only three (18.8%) cases in the nonmetastatic group. miR-200c levels also exhibited a more than 5-fold decrease in four (25.0%) of the metastatic cases, whereas none of the cases in the nonmetastatic group demonstrated such a decrease. Collectively, a 5-fold decrease in either miR-205 or miR-200c expression levels was observed in 14 of 16 metastatic cases (87.5%). Using miR-205 and miR-200c expression levels, 22 of 32 patients (71.9%) could be correctly classified according to lymph node metastatic status. Using a 5-fold decrease as the threshold for either of these miRNAs, lymph node metastasis could be predicted with 87.5% sensitivity and 81.3% specificity (Table 4). Notably, miR-200c levels were decreased in the metastatic group (mean, 0.591±0.531), whereas these levels were unchanged in the nonmetastatic group (mean, 1.202±1.009). Similar results were also observed for miR-122 and miR-146b, which exhibited more than 1.5-fold higher levels in nonmetastatic tumors than in metastatic tumors; however, the differences between both groups did not reach the level of significance (p=0.051 and p=0.228, respectively). Except for miR-200c, miR-122, and miR-146b, all other miRNAs were either decreased or increased simultaneously in both the metastatic and nonmetastatic groups (Figure 1). miR-21 and miR-155 were the most highly upregulated miRNAs in both groups.
Figure 1.
Mean micro RNA (miRNA) expression levels of overall, metastatic and nonmetastatic triple negative breast cancer samples. Data were normalized to benign breast tissue levels and standardized by log2 transformation. Vertical lines indicate standard error.
LN+=lymph node metastasis; LN-=lymph node nonmetastasis.
*p<0.05.
Table 3.
Comparison of micro RNA (miRNA) expression levels in metastatic and nonmetastatic triple negative breast cancer
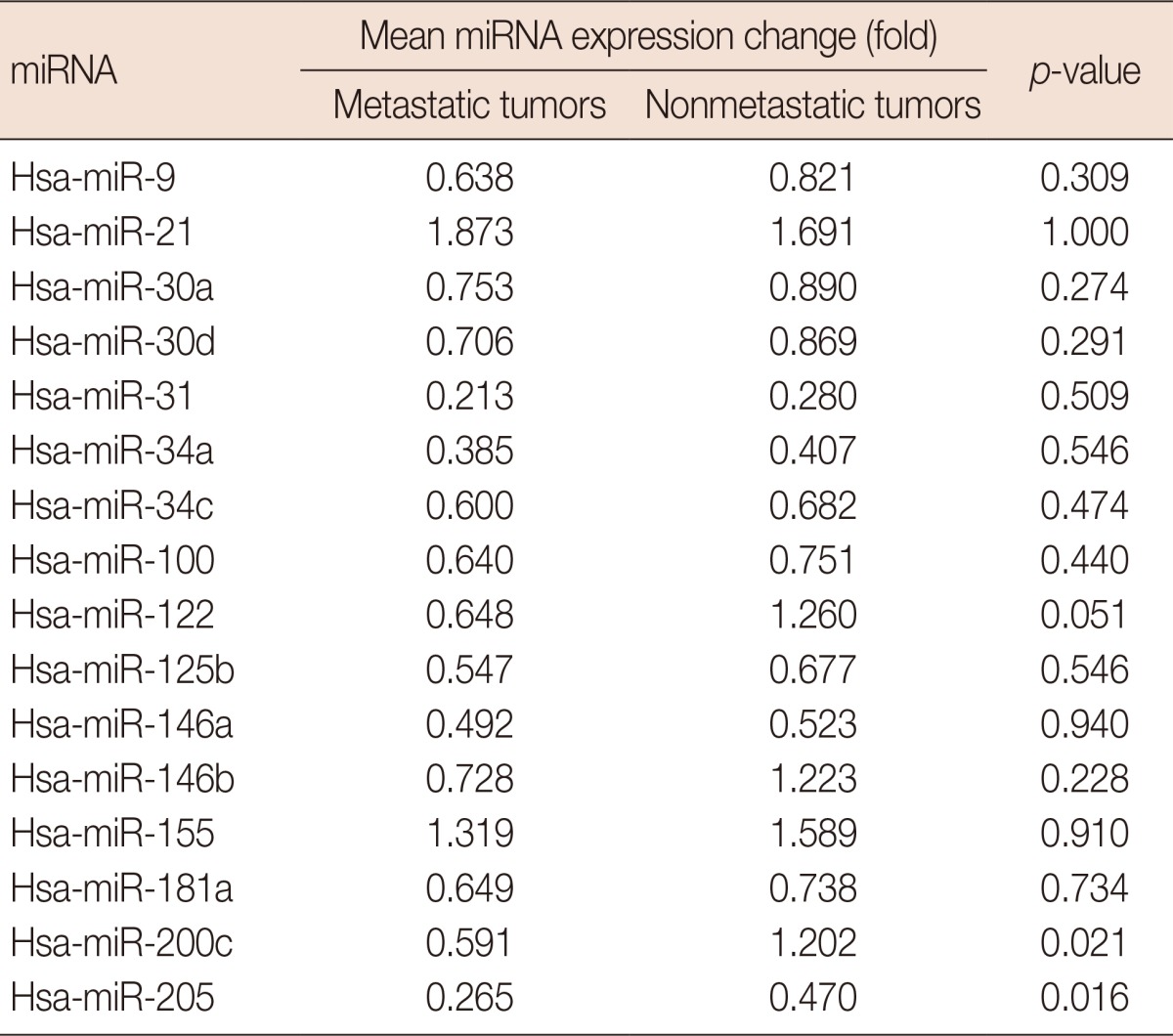
Table 4.
Predictive values of miR-205 and miR-200c expression changes using 5-fold decrease as treshold
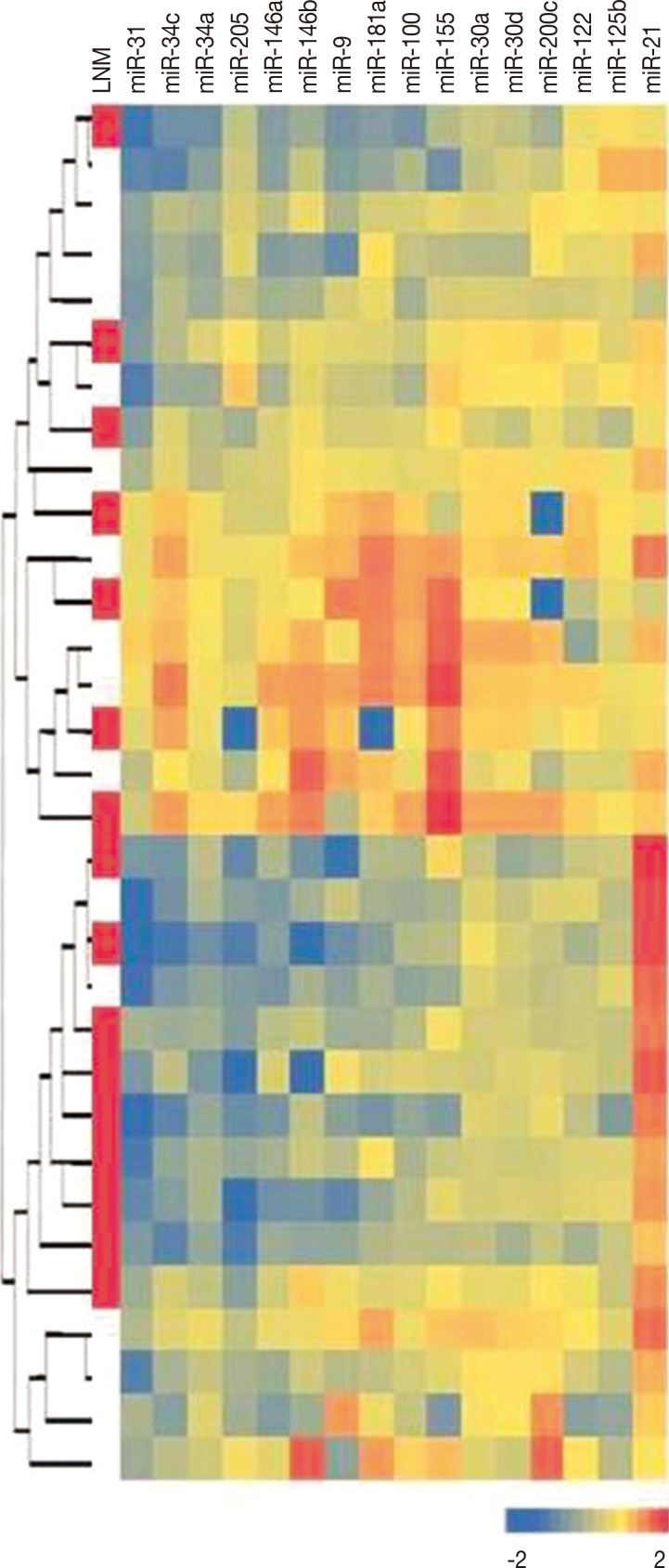
The normalized ratios of tumor/benign breast tissue miRNA expression levels in each case are shown by heatmap colors in Figure 2. We did not observe a separation of metastatic and nonmetastatic tumors in hierarchical clustering of tumor samples. However, miRNA expression levels revealed the presence of different tumor clusters, and most miRNA levels were highly correlated within these clusters.
Figure 2.
Unsupervised hierarchical clustering analysis result according to all micro RNA (miRNA) levels. Heatmap colors represent normalized miRNA levels in each case, and each line represents a case. Red boxes in the first column indicates lymph node metastasis (LNM). Data are standardized by log10 transformation.
We did not observe any significant association between miRNA expression levels and histological type, histological grade, nuclear grade, or tumor diameter (data not shown).
DISCUSSION
The accumulation of data from previous reports revealed that dysregulation of miRNA expression may play significant roles in the progression and dissemination of breast cancers. In order to identify their predictive potential for lymph node metastasis, we selected candidate miRNAs that play critical roles in the cell cycle or in tumor initiation or progression.
We found miR-31 to be the most strongly downregulated miRNA in TNBC tissues. miR-31 was previously shown to inhibit multiple steps of the invasion-metastasis cascade in breast cancer [8]. Its expression is reported to be highly decreased in metastatic breast cancer cell lines, and decreased levels have also been observed in nonmetastatic breast cancer cell lines [3]. The promoter-associated CpG island of the LOC554202 noncoding gene, the host gene of miR-31, was found to be heavily methylated in TNBC cell lines, which may lead to the loss of miR-31 expression and the activation of its target prometastatic genes [9]. Although the difference was not significant, metastatic tumors exhibited lower expression levels of miR-31 than did nonmetastatic tumors in our study. These findings were in accordance with previous reports and support the tumor suppressor activity and inhibitory effects of miR-31 on the invasion-metastasis cascade in TNBC.
miR-205 regulates the epithelial-mesenchymal transition (EMT), which is related to aggressive behavior, and it has been found to suppress the cell growth and invasive capacity of breast cancer cells [10,11]. miR-205 has been reported to be underexpressed in TNBC tissues in previous studies, a finding which supports its tumor suppressor function in breast cancer [12,13]. In our study, when compared with benign breast tissues, miR-205 expression levels were found to be decreased more than 2-fold in tumor tissues. Additionally, the decrease in the metastatic group was significantly higher than that in the nonmetastatic group in TNBCs.
In the present study, the second significantly decreased miRNA in the metastatic group was miR-200c. In contrast to miR-205, miR-200c levels were downregulated in metastatic tumors but the levels were not changed in nonmetastatic TNBCs. The MiR-200 family, including miR-200c, has been shown to induce the epithelial phenotype and to suppress EMT, as does miR-205. MiR-200c has been reported to suppress the tumorigenicity of breast cancer stem cells in vivo [14], and overexpression of miR-200s reduced lung metastases in an experimental model of pulmonary metastasis, findings that suggest the crucial role of these miRNAs in the metastasis of breast cancer [3]. Downregulation of miR-200c was also found to be associated with the triple negative phenotype in breast cancers [15].
Because miR-205 and miR-200c levels were significantly decreased in metastatic TNBCs, and their expression levels exhibited a trend with aggressive behavior, we stratified patients according to miR-205 and miR-200c expression levels using a 5-fold decrease as a threshold. In this manner, our data revealed that lymph node metastasis could be predicted with 87.5% sensitivity and 81.3% specificity in TNBCs. Although miR-205 was found to be more sensitive than miR-200c in predicting lymph node metastasis, it is worth noting that more decreased levels of miR-200c were only observed in metastatic tumors, and its levels were not changed in nonmetastatic tumors. Therefore, the results obtained with miR-200c in our study are promising for other members of the miR-200 family to be predictive markers of lymph node metastasis in TNBCs.
In our study, miR-34a, miR-34c, miR-146a, and miR-181a levels were found to be decreased more than 1.5-fold in TNBCs. However, we did not find any significant differences in their expression levels between metastatic and nonmetastatic tumors. On the other hand, miR-146b and miR-122 levels were decreased in the metastatic group, whereas they were unchanged in the nonmetastatic group. Although the differences in their expression levels between the metastatic and nonmetastatic groups did not reach the level of significance, the small number of patients limits the statistical power of our study, and thus the predictive values of miR-146b and miR-122 may be underestimated for this reason.
miR-21 and miR-155 were the only upregulated miRNAs in the present study. Circulating levels of miR-155 have been reported to be associated with overt metastases in breast cancer patients [16]. However, in another study, miR-155 levels did not correlate with metastatic relapse in stage II and III patients [15]. We did not observe a difference between the miR-155 levels of tumor tissues in metastatic and nonmetastatic patients. Moreover, the increase in levels in tumor tissues was only 1.45-fold when compared to that in benign breast tissues. MiR-21 has been found to be overexpressed in several human tumors, and its levels were also reported to be increased in TNBCs [12,17]. Our data revealed 1.8-fold overexpression in miR-21 levels in tumor tissues versus benign breast tissues. However, its levels were increased in both metastatic and nonmetastatic tumors, and we did not observe a difference in expression levels between the two groups.
We did not observe any change in expression levels of miR-9, miR-100, miR-30a, and miR-30d in tumor tissues. miR-30a and miR-30d belong to the miR-30 family and have similar functions. Although reduced expression of members of the miR-30 family has been reported to be associated with breast tumor progression, and downregulation of miR-30a was suggested as a novel plasma marker for breast cancer and especially for the triple negative phenotype, we did not observe a difference between tumor samples and corresponding benign tissues in our cases [3,18,19].
In conclusion, our data demonstrated that miR-31, miR-205, miR-34a, miR-146a, miR-125b, miR-34c, and miR-181a were downregulated and miR-21 was upregulated more than 1.5-fold in TNBC tissues. In addition, our data revealed that lower levels of miR-205 and miR-200c have the potential to predict lymph node metastasis in TNBCs with high sensitivity and specificity. Further research with well-designed prospective studies in larger patient groups may help to clarify the clinical utility of these two miRNAs as a predictive tool for lymph node metastasis in TNBCs.
Footnotes
This study was presented at 103rd United States and Canadian Academy of Pathology (USCAP) Annual Meeting 2014 held in San Diego, CA, USA on March 1st.
The authors declare that they have no competing interests.
References
- 1.Fabian MR, Sonenberg N. The mechanics of miRNA-mediated gene silencing: a look under the hood of miRISC. Nat Struct Mol Biol. 2012;19:586–593. doi: 10.1038/nsmb.2296. [DOI] [PubMed] [Google Scholar]
- 2.Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci U S A. 2006;103:2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tang J, Ahmad A, Sarkar FH. The role of microRNAs in breast cancer migration, invasion and metastasis. Int J Mol Sci. 2012;13:13414–13437. doi: 10.3390/ijms131013414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Harris L, Fritsche H, Mennel R, Norton L, Ravdin P, Taube S, et al. American Society of Clinical Oncology 2007 update of recommendations for the use of tumor markers in breast cancer. J Clin Oncol. 2007;25:5287–5312. doi: 10.1200/JCO.2007.14.2364. [DOI] [PubMed] [Google Scholar]
- 5.Dawson SJ, Provenzano E, Caldas C. Triple negative breast cancers: clinical and prognostic implications. Eur J Cancer. 2009;45(Suppl 1):27–40. doi: 10.1016/S0959-8049(09)70013-9. [DOI] [PubMed] [Google Scholar]
- 6.Rakha EA, Reis-Filho JS, Ellis IO. Basal-like breast cancer: a critical review. J Clin Oncol. 2008;26:2568–2581. doi: 10.1200/JCO.2007.13.1748. [DOI] [PubMed] [Google Scholar]
- 7.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 8.Valastyan S, Reinhardt F, Benaich N, Calogrias D, Szász AM, Wang ZC, et al. A pleiotropically acting microRNA, miR-31, inhibits breast cancer metastasis. Cell. 2009;137:1032–1046. doi: 10.1016/j.cell.2009.03.047. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 9.Augoff K, McCue B, Plow EF, Sossey-Alaoui K. miR-31 and its host gene lncRNA LOC554202 are regulated by promoter hypermethylation in triple-negative breast cancer. Mol Cancer. 2012;11:5. doi: 10.1186/1476-4598-11-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wu H, Zhu S, Mo YY. Suppression of cell growth and invasion by miR-205 in breast cancer. Cell Res. 2009;19:439–448. doi: 10.1038/cr.2009.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gregory PA, Bert AG, Paterson EL, Barry SC, Tsykin A, Farshid G, et al. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol. 2008;10:593–601. doi: 10.1038/ncb1722. [DOI] [PubMed] [Google Scholar]
- 12.Radojicic J, Zaravinos A, Vrekoussis T, Kafousi M, Spandidos DA, Stathopoulos EN. MicroRNA expression analysis in triple-negative (ER, PR and Her2/neu) breast cancer. Cell Cycle. 2011;10:507–517. doi: 10.4161/cc.10.3.14754. [DOI] [PubMed] [Google Scholar]
- 13.Savad S, Mehdipour P, Miryounesi M, Shirkoohi R, Fereidooni F, Mansouri F, et al. Expression analysis of MiR-21, MiR-205, and MiR-342 in breast cancer in Iran. Asian Pac J Cancer Prev. 2012;13:873–877. doi: 10.7314/apjcp.2012.13.3.873. [DOI] [PubMed] [Google Scholar]
- 14.Wellner U, Schubert J, Burk UC, Schmalhofer O, Zhu F, Sonntag A, et al. The EMT-activator ZEB1 promotes tumorigenicity by repressing stemness-inhibiting microRNAs. Nat Cell Biol. 2009;11:1487–1495. doi: 10.1038/ncb1998. [DOI] [PubMed] [Google Scholar]
- 15.Wu X, Somlo G, Yu Y, Palomares MR, Li AX, Zhou W, et al. De novo sequencing of circulating miRNAs identifies novel markers predicting clinical outcome of locally advanced breast cancer. J Transl Med. 2012;10:42. doi: 10.1186/1479-5876-10-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Roth C, Rack B, Müller V, Janni W, Pantel K, Schwarzenbach H. Circulating microRNAs as blood-based markers for patients with primary and metastatic breast cancer. Breast Cancer Res. 2010;12:R90. doi: 10.1186/bcr2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee JA, Lee HY, Lee ES, Kim I, Bae JW. Prognostic implications of microRNA-21 overexpression in invasive ductal carcinomas of the breast. J Breast Cancer. 2011;14:269–275. doi: 10.4048/jbc.2011.14.4.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ouzounova M, Vuong T, Ancey PB, Ferrand M, Durand G, Le-Calvez Kelm F, et al. MicroRNA miR-30 family regulates non-attachment growth of breast cancer cells. BMC Genomics. 2013;14:139. doi: 10.1186/1471-2164-14-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zeng RC, Zhang W, Yan XQ, Ye ZQ, Chen ED, Huang DP, et al. Down-regulation of miRNA-30a in human plasma is a novel marker for breast cancer. Med Oncol. 2013;30:477. doi: 10.1007/s12032-013-0477-z. [DOI] [PubMed] [Google Scholar]