Abstract
Single-walled carbon nanotubes (SWNTs) have been used in a wide range of fields, and the surface modification via carboxyl functionalization can further improve their physicochemical properties. However, whether carboxyl-modified SWNT poses potential risks to microbial denitrification after its release into the environment remains unknown. Here we present the possible effects of carboxyl-modified SWNT on the growth and denitrification activity of Paracoccus denitrificans (a model denitrifying bacterium). It was found that carboxyl-modified SWNT were present both outside and inside the bacteria, and thus induced bacterial growth inhibition at the concentrations of 10 and 50 mg/L. After 24 h of exposure, the final nitrate concentration in the presence of 50 mg/L carboxyl-modified SWNT was 21-fold higher than that in its absence, indicating that nitrate reduction was substantially suppressed by carboxyl-modified SWNT. The transcriptional profiling revealed that carboxyl-modified SWNT led to the transcriptional activation of the genes encoding ribonucleotide reductase in response to DNA damage and also decreased the gene expressions involved in glucose metabolism and energy production, which was an important reason for bacterial growth inhibition. Moreover, carboxyl-modified SWNT caused the significant down-regulation and lower activity of nitrate reductase, which was consistent with the decreased efficiency of nitrate reduction.
Due to the extraordinary structural, electrical, and mechanical properties, single-walled carbon nanotubes (SWNTs) have been used in a wide variety of fields, including medicinal chemistry, electronics, optics, and materials science1,2. Nevertheless, previous studies pointed out that the very low solubility is still a major stumbling block in the applications of SWNTs2,3. Recently, chemical functionalization has attracted enormous interest, because it has the ability to improve the solubility of nanomaterials4. It was reported in the literature that SWNTs could be chemically derivatized with hydrophilic substituents (such as carboxylic acid groups) for medical and biological applications, and these functional groups were able to provide sites for covalent integration of SWNTs into organic or inorganic polymer structures to produce excellent composite materials5,6. However, the chemical modification can change the cell-SWNT interactions7, which suggests the necessity to consider the potential risks of the surface modified SWNT.
Recent studies have indicated that some emerging pollutants such as engineered nanomaterials cause negative effects on biological denitrification, a unique pathway by which fixed nitrogen such as nitrate is converted into dinitrogen gas (N2)8. For example, the presence of quantum dots was observed to affect the denitrification process of Pseudomonas stutzeri9, and the exposure to oxide nanoparticles inhibited the nitrate reduction of mixed culture in activated sludge10,11,12. However, it is still unclear whether carboxyl-modified SWNT can pose the possible risks to microbial denitrification (especially bacterial growth and nitrate reduction) once it has been intentionally or unintentionally released into the environment.
Denitrifying bacteria usually use nitrate as an electron acceptor and sequentially reduce it to N2. At the same time, to achieve successful denitrification, these denitrifiers need to use organic matter as an electron donor8. It has been reported that the denitrifying enzymes, such as nitrate reductase (NAR), nitrite reductase (NIR), nitric oxide reductase (NOR), and nitrous oxide reductase (N2OR), perform the vital functions in these reduction steps8. Therefore, it can be concluded that the denitrification capacity of the denitrifier is closely relevant to the gene expressions and catalytic activities of these key enzymes involved in denitrification13,14.
Here we present the potential effects of carboxyl-modified SWNT on the bacterial growth and denitrification process. Firstly, we adopted scanning and transmission electron microscopes to show the interactions between SWNTs and bacterial cells. Then, lactate dehydrogenase (LDH) release assays were used to indicate the bacterial membrane integrity after exposure to SWNTs. Also, the growth curves and transformations of nitrogen (including nitrate, nitrite, and nitrous oxide) were determined to assess the influences of SWNTs on microbial denitrification. Finally, we conducted the RNA sequencing (RNA-Seq) and reverse transcriptase quantitative PCR (RT-qPCR), and measured the catalytic activities of key denitrifying enzymes in order to reveal the underlying mechanisms of SWNTs affecting bacterial growth and denitrification activity.
Results
Effects of carboxyl-modified SWNT on bacterial cell surface
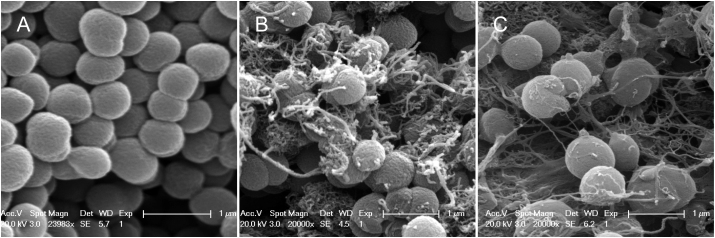
In this study, scanning electron microscope (SEM) images were used to directly display the cell morphology of P. denitrificans exposed to non-modified SWNT and carboxyl-modified SWNT. Figure 1A shows that the bacterial cells of P. denitrificans had smooth membrane surfaces in the culture medium without the presence of non-modified SWNT or carboxyl-modified SWNT. When the culture medium contained non-modified SWNT (Figure 1B) and carboxyl-modified SWNT (Figure 1C), these bacterial cells were trapped in the aggregated nanotubes. The membrane integrity measurements further indicated that there was no significant difference in the relative LDH releases between the control and the non-modified SWNT or carboxyl-modified SWNT exposure (p > 0.05) (Figure S1, Supporting Information), which was consistent with the SEM observations (Figure 1).
Figure 1. SEM images of P. denitrificans cells in the absence (A) and presence of 50 mg/L non-modified SWNT (B) and carboxyl-modified SWNT (C) after 24 h of exposure.
Effects of carboxyl-modified SWNT on bacterial growth
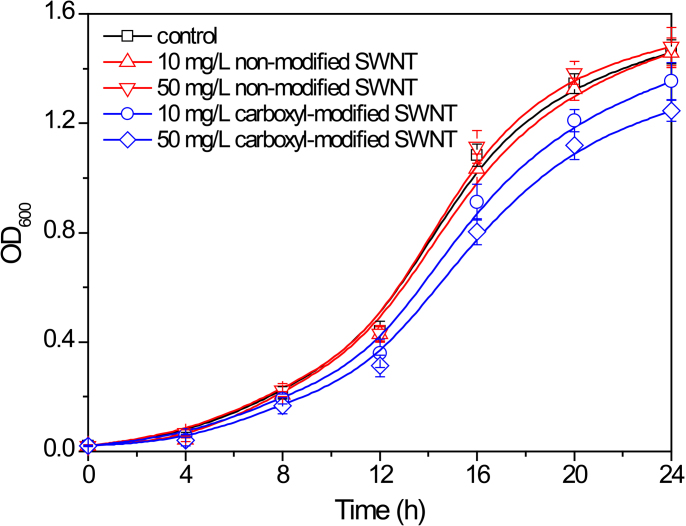
Figure 2 shows the growth curves of P. denitrificans in the absence and presence of non-modified SWNT and carboxyl-modified SWNT during 24 h of exposure. We found that there were no significant differences in bacterial density (optical density at 600 nm, OD600) during the adaptation period (0–8 h). Nevertheless, in the exponential growth phase (8–20 h), the growth rates and bacterial densities in the presence of 10 and 50 mg/L carboxyl-modified SWNT became remarkably lower than those in the control. After 24 h of exposure, the average OD600 value of P. denitrificans in the control was 1.46, which was significantly higher than those exposed to 10 and 50 mg/L carboxyl-modified SWNT (1.35 and 1.24) (Figure 2). It should be noted that the presence of 10 or 50 mg/L of non-modified SWNT had no measurable influences on the bacterial growth during 24 h of exposure.
Figure 2. Growth curves of P. denitrificans in the absence (control) and presence of non-modified SWNT and carboxyl-modified SWNT during 24 h of exposure.
Error bars represent standard deviations of triplicate measurements.
Effects of carboxyl-modified SWNT on bacterial denitrification
In addition to the surface integrity and bacterial growth, the variations of NO3−-N, NO2−-N, and N2O in the presence of non-modified SWNT and carboxyl-modified SWNT were further investigated. Figure 3 illustrates that compared with the control, the exposure to non-modified SWNT (10 or 50 mg/L) had no negative influences on the transformations of NO3−-N, NO2−-N, and N2O (p > 0.05). When P. denitrificans was exposed to 10 and 50 mg/L carboxyl-modified SWNT, the nitrate residue concentrations were 24.3 and 44.5 mg/L, respectively, greatly higher than that in the control (2.1 mg/L) (Figure 3A). However, carboxyl-modified SWNT showed no measurable effects on the variations of NO2−-N, and N2O (p > 0.05), indicating that carboxyl-modified SWNT caused the significant inhibitory effect on the nitrate reduction process of P. denitrificans.
Figure 3. Transformations of NO3−-N (A, solid line), NO2−-N (A, dashed line) (A) and N2O in the gas (B, solid line) and liquid phases (B, dashed line) in the absence (control) and presence of non-modified SWNT and carboxyl-modified SWNT.
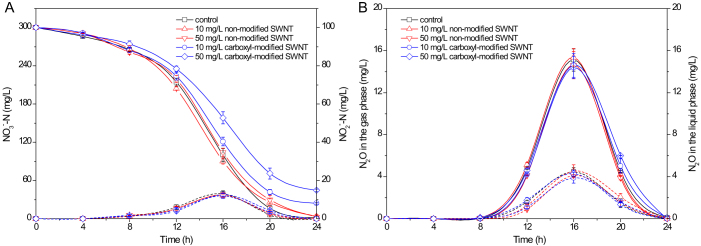
Error bars represent standard deviations of triplicate measurements.
Effects of carboxyl-modified SWNT on the transcriptional profiling and denitrifying enzyme activity
Because carboxyl-modified SWNT obviously affect the bacterial growth and denitrification activity, we further explored the transcriptional profiles of P. denitrificans in the absence (the control) and presence of 50 mg/L carboxyl-modified SWNT in order to reveal potential mechanisms underlying these negative effects. It can be seen in Table 1 that a total number of 44912380 reads were obtained in this study after raw sequences processing and filtering. Among these clean reads, 92.8% and 87.7% of the total reads matched to the reference genome in the absence and presence of carboxy-modified SWNT, respectively, suggesting that the sequencing reads could well reflect the transcriptional expressions of P. denitrificans.
Table 1. Summary of the reads mapped to the reference genomes and genes of P. denitrificans in the absence (Control) and presence of 50 mg/L carboxyl-modified SWNT.
| Control | Carboxyl-modified SWNT | ||||
|---|---|---|---|---|---|
| Number of reads | Percentage | Number of reads | Percentage | ||
| Total number of high-quality reads | 23080806 | 100.0% | 21831574 | 100.0% | |
| Total basepairs of high-quality reads | 1881207366 | 100.0% | 1666282416 | 100.0% | |
| Map to Genome | Total mapped reads | 21414916 | 92.8% | 19136498 | 87.7% |
| Unique matches | 18935555 | 82.1% | 14784420 | 67.8% | |
| Multi-position matches | 2479361 | 10.7% | 4352078 | 19.9% | |
| Total unmapped reads | 1665890 | 7.2% | 2695076 | 12.3% | |
| Map to Gene | Total mapped reads | 15015022 | 65.1% | 13585102 | 62.2% |
| Unique matches | 12168362 | 52.7% | 9259014 | 42.4% | |
| Multi-position matches | 2846660 | 12.4% | 4326088 | 19.8% | |
| Total unmapped reads | 8065784 | 34.9% | 8246472 | 37.8% | |
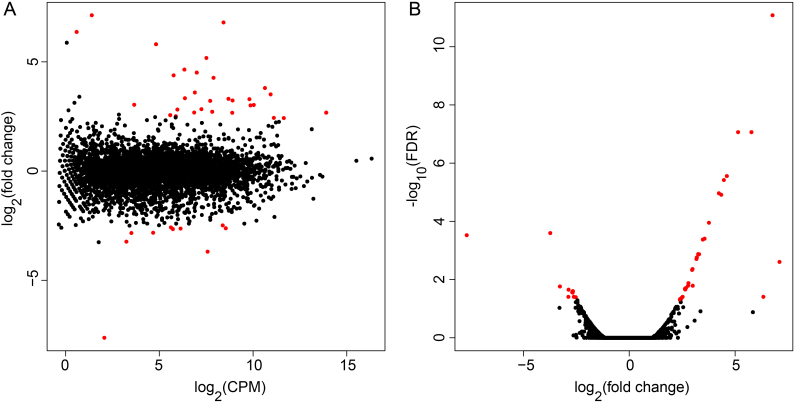
Gene Ontology (GO) annotation has become a major tool for analysis of genome-scale experiments. Figure 4 illustrates the functional annotation of the clean reads at the GO category of biological process. We observed that the gene expressions were mainly involved in the metabolic processes of organic compound, nitrogen compound, macromolecule, small molecule, and cellular biosynthesis. Meanwhile, there were no significant differences between the control and carboxyl-modified SWNT exposure experiments. Figure 5 further gives the MA plot and volcano plot of genome-wide expression changes. Using the criteria of >2-fold change and FDR < 0.05, 37 significant differentially expressed genes (DEGs) with 26 up-regulations and 11 down-regulations were identified in the presence of 50 mg/L carboxyl-modified SWNT.
Figure 4. GO analysis of the sequencing reads from the exposure experiments with (A) and without (B) the presence of 50 mg/L carboxyl-modified SWNT.
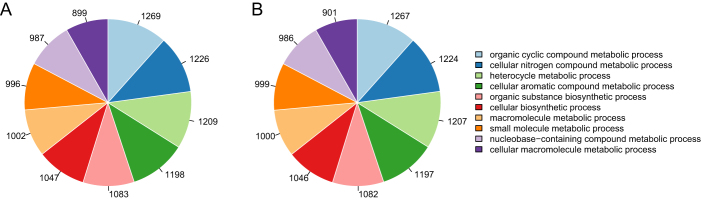
Figure 5. Distribution of differentially expressed genes (red dots) caused by 50 mg/L carboxyl-modified SWNT in MA plot (A) and volcano plot (B).
Red and black dots represent the genes with significantly differential expression (>2-fold change and FDR < 0.05) and with no significant difference, respectively.
The bacterial growth and denitrification of P. denitrificans require the uptake and degradation of organic molecules to obtain energy and reducing power8. For example, in this study, glucose was used as the sole carbon and energy source. Previous studies indicated that some enzymes, such as phosphoglucomutase and pyruvate kinase, play important roles in the metabolism of glucose15. The energy production is observed to be closely related to the key enzymes responsible for the transformation of ATP such as ATPase16. Figure 6 shows that the gene expressions of phosphoglucomutase, pyruvate kinase, and ATPase were significantly inhibited by carboxyl-modified SWNT. These results suggested that the presence of carboxyl-modified SWNT caused the negative effects on glucose utilization and energy production processes, which might be an important reason for the growth inhibition.
Figure 6. Gene expressions of key DNA sequences in the absence (control) and presence of 50 mg/L carboxyl-modified SWNT.
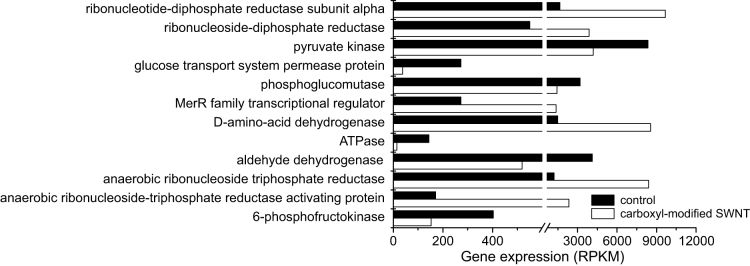
The gene expression level was calculated using the RPKM (reads per kilobase of exon region per million mapped reads) method.
It is well-know that NAR, NIR, NOR, and N2OR are the key enzymes responsible for microbial denitrification. Many studies have used narG, nirS, norB, and nosZ genes as molecular markers to examine the expressions of NAR, NIR, NOR, and N2OR, respectively17. Thus, we quantified the expressions of narG, nirS, norB, and nosZ genes in the absence and presence of 50 mg/L carboxyl-modified SWNT in order to understand their possible influences on microbial denitrification. Figure 7A illustrates that carboxyl-modified SWNT caused the down-regulation of narG gene, indicating that the expression of NAR was inhibited by carboxyl-modified SWNT. Moreover, Figure 7B reveals that the presence of carboxyl-modified SWNT significantly decreased the catalytic activity of NAR (p < 0.05), which was consistent with the inhibition to nitrate reduction caused by carboxyl-modified SWNT.
Figure 7. Gene expressions (A) and relative activities (B) of nitrate reductase (NAR), nitrite reductase (NIR), nitric oxide reductase (NOR), and nitrous oxide reductase (N2OR) in the absence (control) and presence of 50 mg/L carboxyl-modified SWNT.
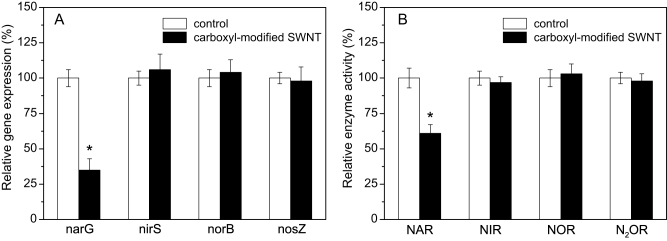
Error bars represent standard deviations of triplicate measurements. Asterisks indicate statistical differences (p < 0.05) from the control test.
Discussion
Engineered nanomaterials usually lead to cell leakage after contact with cell membranes, because their small sizes and high reactivities can easily cause the production of reactive oxygen species (ROS)18,19. It is well-known that the ROS formation is mainly resulted from the reduction of oxygen, but microbial denitrification occurs under the anoxic condition. Our results indicated that no matter whether SWNT was modified with carboxyl groups or not, its direct contact with P. denitrificans did not disrupt cell membranes (Figure 1), and thus had no measurable effects on the cell membrane integrity. Although the cell membrane integrity of P. denitrificans was unaffected by the presence of non-modified SWNT and carboxyl-modified SWNT, the increased concentrations of carboxyl-modified SWNT could induce the growth inhibition of P. denitrificans (Figure 2). On the other hand, carboxyl-modified SWNT also decreased the rate of nitrate reduction, and thus led to the higher nitrate residue concentration after 24 h of exposure (Figure 3).
It was reported that the dispersion status of nanomaterials plays an important role in their negative effects on living organisms18,20. SWNTs can form the aggregation easily in aqueous solution owing to the strong π-π interaction, but the surface functionalization has the ability to enhance the dispersion efficiency of nanomaterials4. The zeta potential data showed that the carboxyl modification highly improved the dispersion of SWNTs (Figure S2, Supporting Information), which supported the previous results observed in the literature4,5. The highly dispersed SWNTs can attach bacterial surfaces easily, and has the ability to cause the negative influences on bacterial cells21, which was consistent with the inhibitory effects of carboxyl-modified SWNT on bacterial growth and denitrification activity of P. denitrificans.
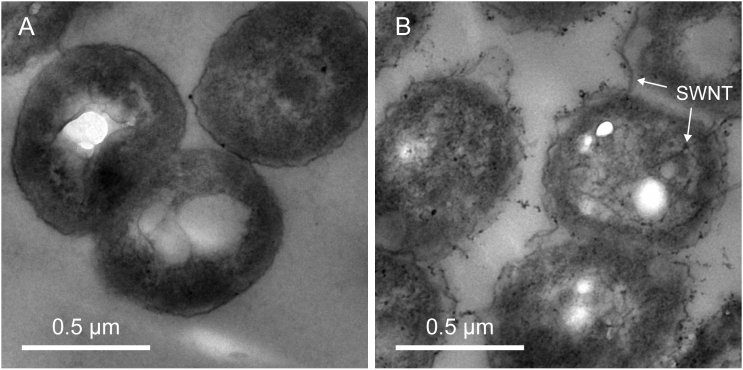
Previous publications pointed out that bacterial growth and metabolism are closely related to intracellular complex regulations, such as the transcriptional regulation22,23. TEM images showed that some carboxyl-modified SWNTs were indeed present inside the bacterial cells (Figure 8). RNA-Seq analysis further revealed that carboxyl-modified SWNT resulted in the significant changes of gene expressions in P. denitrificans, indicating that the direct interference with cytoplasmic proteins or with specific promoters affected gene expression. It was reported in the literature that cells could respond to DNA damage by arresting the cell cycle and activating the genes involved in DNA repair24. Ribonucleotide reductase is an essential enzyme for DNA synthesis in all prokaryotic and eukaryotic cells, because this key enzyme catalyzes the rate-limiting step in the production of deoxyribonucleoside triphosphates, the precursors for DNA synthesis25. It was found that the ribonucleotide reductase genes were subject to transcriptional induction after DNA damage26. Our results showed that carboxyl-modified SWNT greatly increased the gene expressions of ribonucleotide diphosphate reductase and ribonucleoside triphosphate reductase (Figure 6), indicating that the presence of carboxyl-modified SWNT led to the DNA damage of P. denitrificans.
Figure 8. TEM images of P. denitrificans cells in the absence (A) and presence of 50 mg/L carboxyl-modified SWNT (B) after 24 h of exposure.
Biological denitrification is carried out by large numbers of denitrifying bacteria8. Among these denitrifiers, only several species of bacteria such as P. denitrificans are able to achieve the complete reduction of nitrate to N227. It has been reported that P. denitrificans can use ATP-binding cassette (ABC) transporters to uptake glucose in the medium, and then consume this sugar to grow and reduce nitrate8. However, the gene encoding a glucose transport system permease protein was down-regulated in the presence of 50 mg/L carboxyl-modified SWNT (Figure 6). Concurrently, the gene expressions of the key enzymes involved in glucose metabolism, such as phosphoglucomutase, pyruvate kinase, and ATPase, were also significantly decreased, which was consistent with the observation of glucose utilization being significantly suppressed by 50 mg/L carboxyl-modified SWNT (Figure S3, Supporting Information). These results indicated that the presence of carboxyl-modified SWNT caused the negative effects on glucose utilization and energy production, which was mainly responsible for the growth inhibition. On the other hand, NAR is able to catalyze the reduction of nitrate to nitrite, and we found that carboxyl-modified SWNT inhibited the expression and activity of NAR. Therefore, the decreases in the gene expression and catalytic activity of NAR could lead to the inhibition to nitrate reduction caused by carboxyl-modified SWNT.
In summary, the exposure to carboxyl-modified SWNT had no measurable impacts on bacterial membrane integrity after direct contact with cell membranes. However, the presence of carboxyl-modified SWNT led to the growth inhibition of P. denitrificans. TEM images showed that some carboxyl-modified SWNT were present inside the bacterial cells. Transcriptional profiles further indicated that the growth inhibition caused by carboxyl-modified SWNT was due to the bacterial DNA damage and inhibition of gene expressions involved in glucose and ATP metabolism. Also, the presence of carboxyl-modified SWNT resulted in the negative influences on the gene expression and catalytic activity of NAR, which finally caused the decrease in the nitrate reduction of P. denitrificans.
Methods
SWNTs and their suspensions
The commercial non-modified SWNT and carboxyl-modified SWNT were purchased from Nanjing Xianfeng Nano Co. (Nanjing, China). The average diameters of these SWNTs were in the ranges of 1–2 nm, whereas their lengths were 0.5–2 μm. To remove residual carbon and metal catalyst, both SWNTs were first heated at 350°C for 3 h and cooled to room temperature. Then, SWNTs were washed using 12 M HCl, sonicated for 1 h, filtered through a 5 μm polytetrafluoroethylene (PTFE) membrane, resuspended in deionized (DI) water repeatedly, and adjusted to neutral pH. The final solution was dried in an oven at 60°C overnight to get SWNT powders28. To prepare the stock suspensions, 1 g SWNTs were added into 1 L of Milli-Q water, followed by ultrasonication (25°C, 250 W, 40 KHz) for 1 h to enhance the dispersion.
Cell culture
Paracoccus denitrificans (ATCC 19367) was purchased from American Type Culture Collection, and grown in Difco nutrient broth at 30°C and 200 rpm to an optical density at 600 nm (OD600) of 0.8–1.0. Then, the culture of P. denitrificans was used as the inoculum.
Effects of SWNTs on bacterial growth and denitrification
To investigate the potential effects of SWNTs, P. denitrificans was exposed to 0, 10, and 50 mg/L non-modified SWNT and carboxyl-modified SWNT in a mineral medium modified from the literature27. The mineral medium contained (per liter) 7.0 g K2HPO4, 3.0 g KH2PO4, 0.5 g MgSO4, 1.0 g (NH4)2SO4, 2.16 g KNO3, 5.0 g glucose, and 50 μL trace element feed. In particular, the addition of ammonium sulfate was used to preclude assimilatory nitrate reduction, which ensured that nitrate consumption was due to respiration9. The trace element feed contained (per liter) 7.4 g CaCl2·2H2O, 1.0 g MnSO4·H2O, 3.6 g ZnSO4·7H2O, 0.4 g CoCl2·6H2O, 0.96 g FeCl3·6H2O, 0.027 g CuCl2·2H2O, 0.3 g H3BO3, 0.01 g NiCl2·6H2O, and 3.7 g EDTA10. The cultures of P. denitrificans were inoculated in the medium at an initial OD600 of 0.05. Then, these serum bottles were bubbled with argon gas, sealed with rubber stoppers, and placed into a shaker at 30°C and 200 rpm. During 24 h of exposure, the cultures were withdrawn from the serum bottles at intervals of 4 h, and their OD600 values were measured via a spectrophotometer (Shimadzu UV-1800, Japan). It should be noted that the OD600 values of SWNTs were also determined and subtracted from those of the cultures in order to obtain the OD600 values of bacterial cells according to the literature29. Meanwhile, the transformations of NO3−-N, NO2−-N, and N2O were examined in order to show the possible effects of SWNTs on microbial denitrification processes.
RNA-Seq analysis
Bacterial cells were harvested in the exponential growth phase (16 h) by centrifugation at 10000 g for 10 min at 4°C, and then lysed in TRIzol reagent (Invitrogen) for extraction of total RNA. To avoid DNA contamination, the extracted RNA was treated with DNase I (Ambion) according to the manufacturer's protocol. Thereafter, mRNA was isolated from the DNA-free total RNA using the MICROBExpress Bacterial mRNA Enrichment Kit (Ambion), and prepared for Illumina sequencing using the mRNA-Seq Sample Preparation Kit (Illumina) according to manufacturer's instructions. The RNA-Seq libraries were finally sequenced using an Illumina HiSeq 2000. All sequences have been deposited in NCBI SRA database under accession numbers SRX400081 and SRX400082. Raw reads were filtered by removing the reads with (1) sequence adapters, (2) more than 5% ‘N' bases, and (3) more than 50% QA ≤ 15 bases. Then, the clean reads were aligned to the reference genome using SOAP230, and no more than 3 mismatches were allowed in the alignment for each read. The gene expression level was calculated using the RPKM (reads per kilobase of exon region per million mapped reads) method31, and the differentially expressed genes (DEGs) were identified based on the criteria of absolute fold change > 2 and FDR < 0.05. Gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses were performed using Blast2GO using default annotation parameters32.
Quantification of gene expressions of denitrifying enzymes
The gene expressions of key denitrifying enzymes, such as NAR, NIR, NOR, and N2OR, were determined by the quantification of narG, nirS, norB, and nosZ genes via reverse transcriptase quantitative PCR (RT-qPCR). The extracted total RNA described in the “RNA-Seq analysis” section was used to synthesize cDNA at 42°C according to the literature9. Thereafter, cDNA was purified using a QIAquick PCR Purification Kit (Qiagen) according to the manufacturer's instruction. The qPCR was performed via a StepOne Real-Time PCR System (Applied Biosystems, Foster City, USA) in a total volume of 20 μL containing 1 × SYBR Green PCR Master Mix, 0.5 μM each primer, and 1 μL of cDNA. The primers and amplification conditions were documented in Table S1 (Supporting Information). All qPCR assays were performed using three replicates per sample, and contained the control reactions without cDNA.
Enzyme activity assays
After 24 h of exposure, bacterial cells were harvested by centrifugation at 10000 g for 10 min, washed thrice with 0.1 M phosphate-buffered saline (PBS buffer) (pH 7.4), and resuspended in the same buffer. Then, the resuspended pellets were disrupted by ultrasonication at 4°C for 5 min, and then the cell debris was removed by centrifugation at 12000 g for 10 min at 4°C. The crude cell extracts were immediately used for determination of the activities of NAR, NIR, NOR, and N2OR. The protein concentrations of cell extracts were measured using Protein Assay Kit (Bio-Rad) with bovine serum albumin (BSA) as a standard. The reaction mixture (2 mL) contained 10 mM potassium phosphate buffer (pH 7.1), 10 mM methyl viologen, 5 mM Na2S2O4, 1 mM electron acceptor (NO3−, NO2−, NO, or N2O), and 100 μL of cell extract33. The reaction mixture was incubated at 30°C, and the consumption of electron acceptor were measured every 10 min according to our previous publication34.
Scanning electron microscopy
SEM images were used to show the surface morphology of bacterial cells exposed to non-modified SWNT and carboxyl-modified SWNT10. Briefly, bacterial cells were centrifuged at 1000 g for 5 min after 24 h of exposure. The pellets were washed thrice with 0.1 M PBS buffer (pH 7.4), and fixed in 0.1 M PBS buffer (pH 7.4) containing 2.5% glutaraldehyde at 4°C for 4 h. After rinsing twice with 0.1 M PBS buffer (pH 7.4), the pellets were dehydrated in ethanol serials (50%, 70%, 80%, 90%, and 100%, 15 min per step), and then dried in air. Finally, the images were obtained using the FEI Quanta 200 SEM at 20 kV.
Transmission electron microscopy
After exposure to carboxyl-modified SWNT, cells were harvested by centrifugation, washed thrice with 0.1 M PBS (pH 7.4), and fixed in the perfluorocarbon containing osmium tetroxide (1%) for 1 h. After three rinses in pure perfluorocarbon, the samples were dehydrated, embedded, sectioned, stained, and imaged with a JEM-1230 transmission electron microscope (JEOL, Tokyo, Japan).
Analytical methods
The zeta potentials of SWNTs were measured via a Zetasizer Nano ZS (Malvern Instruments, UK). The LDH release was examined using the Cytotoxicity Detection Kit (Roche Applied Science) according to the manufacturer's instruction. The measurements of NO3−-N and NO2−-N were conducted according to the Standard Methods35. The detailed procedures for determining N2O in the gas and liquid phases were documented in the literature36,37. Briefly, the gaseous N2O was analyzed using a gas chromatograph (GC, Agilent 7820A) equipped with an electron capture detector (ECD). The dissolved N2O was measured by GC using a headspace method37, and the equilibrium temperature and time were 25°C and 3 h, respectively. This GC contained a pre-column (Porapak Q 80/100 mesh, 1 m × 2 mm) and a main column (Porapak Q 80/100 mesh, 3 m × 2 mm). The temperatures of the columns and ECD were 100 and 300°C, respectively, and a mixture of 91% Ar + 9% CH4 was used as the carrier gas.
Statistical analysis
All tests were performed in triplicate and the results were expressed as mean ± standard deviation. An analysis of variance (ANOVA) was used to test the significance of results and p < 0.05 was considered to be statistically significant.
Author Contributions
X.Z. and Y.C. conceived and designed the experiments. X.Z., Y.S., R.W. and M.L. carried out the experiments. X.Z., Y.S., Y.W. and H.H. collected data and performed the statistical analysis. X.Z., Y.S. and Y.C. wrote the paper. All authors discussed the results and reviewed the manuscript.
Supplementary Material
Supporting Information
Acknowledgments
This work was supported by the National Hi-Tech Research and Development Program of China (Grant no. 2011AA060903), National Natural Science Foundation of China (Grant no. 41301558 and 51278354), and China Postdoctoral Science Foundation (Grant no. 2013M530210).
References
- De Volder M. F. L., Tawfick S. H., Baughman R. H. & Hart A. J. Carbon nanotubes: Present and future commercial applications. Science 339, 535–539 (2013). [DOI] [PubMed] [Google Scholar]
- Tasis D., Tagmatarchis N., Bianco A. & Prato M. Chemistry of carbon nanotubes. Chem. Rev. 106, 1105–1136 (2006). [DOI] [PubMed] [Google Scholar]
- Chen J. et al. Solution properties of single-walled carbon nanotubes. Science 282, 95–98 (1998). [DOI] [PubMed] [Google Scholar]
- Hirsch A. Functionalization of single-walled carbon nanotubes. Angew. Chem. Int. Edit. 41, 1853–1859 (2002). [DOI] [PubMed] [Google Scholar]
- Peng H., Alemany L. B., Margrave J. L. & Khabashesku V. N. Sidewall carboxylic acid functionalization of single-walled carbon nanotubes. J. Am. Chem. Soc. 125, 15174–15182 (2003). [DOI] [PubMed] [Google Scholar]
- Du J. et al. The interaction of serum proteins with carbon nanotubes depend on the physicochemical properties of nanotubes. J. Nanosci. Nanotechnol. 11, 10102–10110 (2011). [DOI] [PubMed] [Google Scholar]
- Nimmagadda A., Thurston K., Nollert M. U. & McFetridge P. S. F. Chemical modification of SWNT alters in vitro cell-SWNT interactions. J. Biomed. Mat. Res. Part A 76, 614–625 (2006). [DOI] [PubMed] [Google Scholar]
- Zumft W. G. Cell biology and molecular basis of denitrification. Microbiol. Mol. Biol. Rev. 61, 533–616 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Zhu H., Colvin V. L. & Alvarez P. J. Cellular and transcriptional response of Pseudomonas stutzeri to quantum dots under aerobic and denitrifying conditions. Environ. Sci. Technol. 45, 4988–4994 (2011). [DOI] [PubMed] [Google Scholar]
- Zheng X., Wu R. & Chen Y. Effects of ZnO nanoparticles on wastewater biological nitrogen and phosphorus removal. Environ. Sci. Technol. 45, 2826–2832 (2011). [DOI] [PubMed] [Google Scholar]
- Zheng X., Su Y. & Chen Y. Acute and chronic responses of activated sludge viability and performance to silica nanoparticles. Environ. Sci. Technol. 46, 7182–7188 (2012). [DOI] [PubMed] [Google Scholar]
- Chen Y., Su Y., Zheng X., Chen H. & Yang H. Alumina nanoparticles-induced effects on wastewater nitrogen and phosphorus removal after short-term and long-term exposure. Water Res. 46, 4379–4386 (2012). [DOI] [PubMed] [Google Scholar]
- Desvergne B., Michalik L. & Wahli W. Transcriptional regulation of metabolism. Physiol. Rev. 86, 465–514 (2006). [DOI] [PubMed] [Google Scholar]
- Siciliano S. D., Roy R. & Greer C. W. Reduction in denitrification activity in field soils exposed to long term contamination by 2,4,6-trinitrotoluene (TNT). FEMS Microbiol. Ecol. 32, 61–68 (2000). [DOI] [PubMed] [Google Scholar]
- Saltiel A. R. & Kahn C. R. Insulin signalling and the regulation of glucose and lipid metabolism. Nature 414, 799–806 (2001). [DOI] [PubMed] [Google Scholar]
- Yoshida M., Muneyuki E. & Hisabori T. ATP synthase - A marvellous rotary engine of the cell. Nat. Rev. Mol. Cell Biol. 2, 669–677 (2001). [DOI] [PubMed] [Google Scholar]
- Philippot L. et al. Characterization and transcriptional analysis of Pseudomonas fluorescens denitrifying clusters containing the nar, nir, nor and nos genes. Biochim. Biophys. Acta 1517, 436–440 (2001). [DOI] [PubMed] [Google Scholar]
- Colvin V. L. The potential environmental impact of engineered nanomaterials. Nat. Biotechnol. 21, 1166–1170 (2003). [DOI] [PubMed] [Google Scholar]
- Magrez A. et al. Cellular toxicity of carbon-based nanomaterials. Nano Lett. 6, 1121–1125 (2006). [DOI] [PubMed] [Google Scholar]
- Nel A., Xia T., Madler L. & Li N. Toxic potential of materials at the nanolevel. Science 311, 622–627 (2006). [DOI] [PubMed] [Google Scholar]
- Bottini M. et al. Multi-walled carbon nanotubes induce T lymphocyte apoptosis. Toxicol. Lett. 160, 121–126 (2006). [DOI] [PubMed] [Google Scholar]
- Stelling J., Klamt S., Bettenbrock K., Schuster S. & Gilles E. D. Metabolic network structure determines key aspects of functionality and regulation. Nature 420, 190–193 (2002). [DOI] [PubMed] [Google Scholar]
- Covert M. W., Knight E. M., Reed J. L., Herrgard M. J. & Palsson B. O. Integrating high-throughput and computational data elucidates bacterial networks. Nature 429, 92–96 (2004). [DOI] [PubMed] [Google Scholar]
- Khanna K. K. & Jackson S. P. DNA double-strand breaks: Signaling, repair and the cancer connection. Nat. Genet. 27, 247–254 (2001). [DOI] [PubMed] [Google Scholar]
- Gon S. et al. A novel regulatory mechanism couples deoxyribonucleotide synthesis and DNA replication in Escherichia coli. EMBO J. 25, 1137–1147 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan A. & Reichard P. Ribonucleotide reductases. Annu. Rev. Biochem. 67, 71–98 (1998). [DOI] [PubMed] [Google Scholar]
- Blaszczyk M. Effect of medium composition on the denitrification of nitrate by Paracoccus denitrificans. Appl. Environ. Microbiol. 59, 3951–3953 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghafari P. et al. Impact of carbon nanotubes on the ingestion and digestion of bacteria by ciliated protozoa. Nat. Nanotechnol. 3, 347–351 (2008). [DOI] [PubMed] [Google Scholar]
- Yang C., Mamouni J., Tang Y. & Yang L. Antimicrobial activity of single-walled carbon nanotubes: Length effect. Langmuir 26, 16013–16019 (2010). [DOI] [PubMed] [Google Scholar]
- Li R. et al. SOAP2: An improved ultrafast tool for short read alignment. Bioinformatics 25, 1966–1967 (2009). [DOI] [PubMed] [Google Scholar]
- Mortazavi A., Williams B. A., Mccue K., Schaeffer L. & Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 5, 621–628 (2008). [DOI] [PubMed] [Google Scholar]
- Conesa A. et al. Blast2GO: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21, 3674–3676 (2005). [DOI] [PubMed] [Google Scholar]
- Kristjansson J. K. & Hollocher T. C. First practical assay for soluble nitrous oxide reductase of denitrifying bacteria and a partial kinetic characterization. J. Biol. Chem. 255, 704–707 (1980). [PubMed] [Google Scholar]
- Zhu X. & Chen Y. Reduction of N2O and NO generation in anaerobic-aerobic (low dissolved oxygen) biological wastewater treatment process by using sludge alkaline fermentation liquid. Environ. Sci. Technol. 45, 2137–2143 (2011). [DOI] [PubMed] [Google Scholar]
- APHA. Standard Methods for the Examination of Water and Wastewater, 20th ed.(American Public Health Association: Washington, DC, 1998). [Google Scholar]
- Rosamond M. S., Thuss S. J. & Schiff S. L. Dependence of riverine nitrous oxide emissions on dissolved oxygen levels. Nat. Geosci. 5, 715–718 (2012). [Google Scholar]
- Wang Y. et al. Effect of anaerobic reaction time on denitrifying phosphorus removal and N2O production. Bioresour. Technol. 102, 5674–5684 (2011). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information