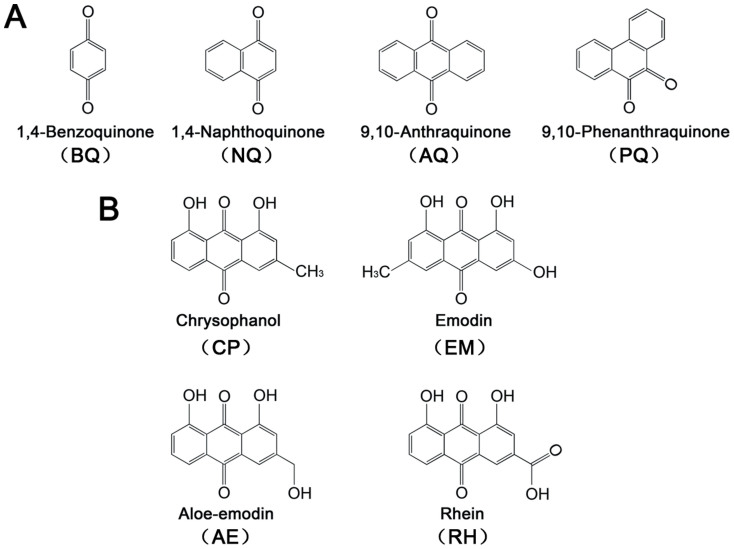
Abstract
Protein misfolding and aggregation are associated with more than twenty diseases, such as neurodegenerative diseases and metabolic diseases. The amyloid oligomers and fibrils may induce cell membrane disruption and lead to cell apoptosis. A great number of studies have focused on discovery of amyloid inhibitors which may prevent or treat amyloidosis diseases. Polyphenols have been extensively studied as a class of amyloid inhibitors, with several polyphenols under clinical trials as anti-neurodegenerative drugs. As oxidative intermediates of natural polyphenols, quinones widely exist in medicinal plants or food. In this study, we used insulin as an amyloid model to test the anti-amyloid effects of four simple quinones and four natural anthraquinone derivatives from rhubarb, a traditional herbal medicine used for treating Alzheimer's disease. Our results demonstrated that all eight quinones show inhibitory effects to different extent on insulin oligomerization, especially for 1,4-benzoquinone and 1,4-naphthoquinone. Significantly attenuated oligomerization, reduced amount of amyloid fibrils and reduced hemolysis levels were found after quinones treatments, indicating quinones may inhibit insulin from forming toxic oligomeric species. The results suggest a potential action of native anthraquinone derivatives in preventing protein misfolding diseases, the quinone skeleton may thus be further explored for designing effective anti-amyloidosis compounds.
Amyloid formation plays an important role in the pathogenesis of misfolding-based diseases such as Parkinson's disease, Alzheimer's disease and type 2 diabetes mellitus1,2,3. Under appropriate condition, proteins may form toxic aggregates by first misfolding into β-sheet-rich intermediates, which further assemble into soluble oligomers and finally to linear mature fibrils4. The aggregated species formed in this process may attach to cell membranes and cause membrane permeabilization, which induce cell dysfunction and apoptosis5,6. It is generally believed that interruption of amyloid aggregation may provide a plausible therapeutic approach for protein misfolding diseases7.
Insulin is a hypoglycemic hormone that widely used as anti-diabetic drug. Insulin amyloidosis was commonly found at the site of repeated insulin injection in diabetic patients as well as in the clinic formulations8,9,10,11. Different from the natively unstructured amyloidogenic proteins such as islet amyloid polypeptide, amyloid-β and α-synuclein12,13,14, insulin belongs to the category of natively structured amyloidogenic proteins, such as prion and lysozyme15,16,17, and is also commonly used as a classical model to probe the mechanisms of protein fibrillation since insulin amyloids share the same typical cross-β structure as the mature fibrils of the disease relevant amyloids4,18,19, as well as an amyloid model system for studying aggregation inhibitors20,21.
Many aggregation inhibitors have been identified to affect aggregation process, for example, small molecules, peptides and oligomer- or amyloid-specific antibodies22. Among them, native polyphenols from food or herbal medicines, such as epigallocatechin gallate (EGCG), curcumin and resveratrol, have been well-studied as amyloid inhibitors23,24,25. The tea catechin polyphenol, EGCG, can inhibit the aggregation of various proteins and is being assessed in clinical trials for Alzheimer's disease26. Quinones, the oxidation products of polyphenols that include benzoquinones (BQ), naphthoquinones (NQ), anthraquinones (AQ) and phenanthraquinones (PQ), are widely found in nature. Recently, several quinones have been reported to inhibit amyloid aggregation efficiently. For example, 1,4-benzoquinone, the simplest quinone, inhibits lysozyme fibrillization more efficiently than the corresponding phenol27. Similarly, 9,10-anthraquinone strongly inhibits the aggregation of β-amyloid protein28. In addition, several derivatives of 1,4-naphthoquinone have been explored as drug candidates for amyloidosis diseases29. These evidences all implicate quinones as a class of compounds with inhibitory effect on protein misfolding and aggregation.
A number of bioactive derivatives of AQ can be found in rhubarb extract (Rheum rhabarbarum L.), a traditional herbal medicine which may be used for treating amyloidosis related diseases such as Alzheimer's disease, with a possible mechanism that some of its effective components may affect β-amyloid protein related pathogenic pathways30,31. One of the anthraquinones, emodin (EM), has been reported to effectively inhibit tau (a microtubule-associated protein) aggregation and show neuroprotective effect32. Previous reports on natural quinones mainly focus on the mechanisms behind their anti-cancer or anti-bacterial activities33,34, their exact effects and mechanisms on protein misfolding diseases remain limited. Here, we propose that rhubarb anthraquinones may affect protein misfolding and aggregation. To test this hypothesis, the amyloid inhibitory effects of a set of simple quinones and natural AQ derivatives from rhubarb (Figure 1) were compared, with insulin used as a model amyloid system.
Figure 1. Chemical structures of quinones investigated in this study.
(A) Simple quinones: 1,4-benzoquinone (BQ), 1,4-naphthoquinone (NQ), 9,10-anthraquinone (AQ) and 9,10-phenanthraquinone (PQ); (B) Anthraquinone derivatives from rhubarb: chrysophanol (CP), emodin (EM), aloe-emodin (AE) and rhein (RH).
Results
Quinones inhibit amyloid formation of insulin
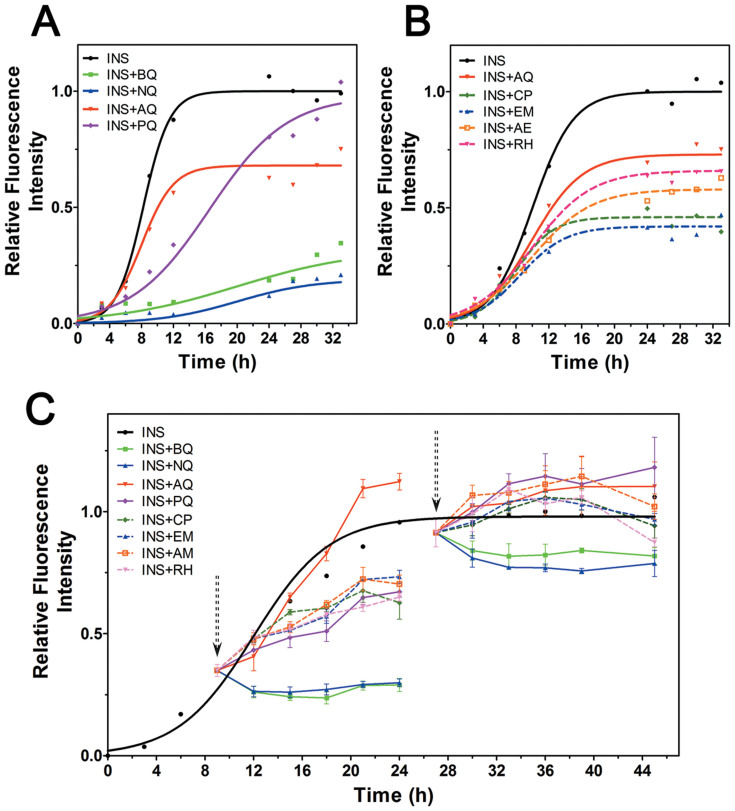
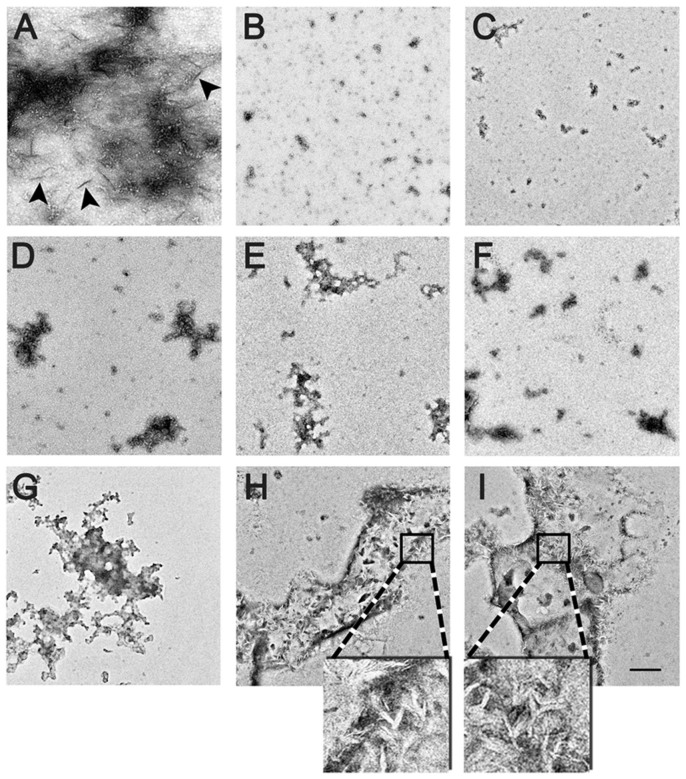
Thioflavin-T (ThT) based fluorescence was used to monitor the aggregation process of 100 μM insulin that co-incubated with 50 μM of various quinones at 65°C. The morphology of insulin aggregates in the absence or presence of quinones was observed by transmission electronic microscopy (TEM). Control insulin gave a strong ThT emission with a lag time of 4.52 ± 0.45 h and reached a plateau within 14 h (Figure 2A). After incubation for 24 h, TEM image showed a mesh of insulin aggregates characterized as linear fibrils (Figure 3A). Except AQ, the other three simple quinones all prolonged the aggregation lag time to different extent. BQ and NQ showed the strongest inhibitory effect accompanied with significantly decreased fluorescence intensities (Figure 2A), consistently, only small dispersed amorphous aggregates were identified in TEM (Figure 3B & 3C). In contrast, PQ (9,10-phenanthraquinone) treatment prolongs the lag time of insulin to 6.97 ± 0.48 h (P < 0.05), but the maximum fluorescence intensity after 24 h incubation was similar to that of insulin alone (Figure 2A). Although AQ did not extend the lag time (4.51 ± 0.23 h), it nevertheless significantly reduced the maximum fluorescence intensity by 40% (Figure 2A). Despite the detectable increased fluorescence intensity after prolonged incubation, classic fibrils were not found in samples co-incubated with AQ and PQ, only scattered amorphous aggregates were observed (Figure 3D & 3E) similar to a previous report35.
Figure 2. Effects of quinones on amyloid formation of insulin.
(A) Relative ThT fluorescence of insulin in the presence or absence of simple quinones; (B) Relative ThT fluorescence of insulin in the presence or absence of anthraquinone derivatives from rhubarb, AQ was used as a control; (C) Relative ThT fluorescence of pre-incubated insulin with quinones treatments start at 9 h or 27 h, the arrows indicate the quinones adding points.
Figure 3. TEM images of amyloid formed in different sample groups.
(A) Insulin; (B)–(I) insulin co-incubated with BQ, NQ, AQ, PQ, CP, EM, AE, RH, respectively. The enlarged frames (4x) show details of tiny protofibril-like structures buried in the amorphous aggregates. Scale bar represents 200 nm.
Compared to AQ, four AQ derivatives from rhubarb showed no significant effects on lag time; however, slightly stronger inhibitory effects in terms of maximum fluorescence intensities were observed (Table 1), with CP (chrysophanol) and EM showed the greatest inhibition effects (Figure 2B). Large amount of amorphous aggregates was observed by TEM for samples co-incubated with the rhubarb anthraquinone derivatives (Figure 3F–3I); however, for AE (aloe-emodin) and and RH (rhein), tiny short protofibril-like structures were found buried in the amorphous aggregates (Figure 3H & 3I, enlarged frames).
Table 1. Effects of quinones on the amyloidogenecity of insulin.
| Lag time (h) | T50 (h) | Maximum intensity[2] | |
|---|---|---|---|
| INS[1] | 4.52 ± 0.45 | 8.17 ± 0.85 | 1.04 ± 0.05 |
| INS + BQ | N.D.[3] | N.D. | 0.19 ± 0.04 |
| INS + NQ | N.D. | N.D. | 0.12 ± 0.02 |
| INS + AQ | 4.51 ± 0.23 | 7.83 ± 1.03 | 0.63 ± 0.04 |
| INS + PQ | 6.97 ± 0.48 | 13.00 ± 0.82 | 0.77 ± 0.06 |
| INS + CP | 5.01 ± 0.32 | 8.50 ± 0.41 | 0.50 ± 0.09 |
| INS + EM | 4.73 ± 0.27 | 8.67 ± 0.94 | 0.42 ± 0.05 |
| INS + AE | 4.67 ± 0.54 | 9.67 ± 1.70 | 0.50 ± 0.19 |
| INS + RH | 4.75 ± 0.43 | 9.45 ± 0.43 | 0.64 ± 0.10 |
[1]The concentration of insulin was 100 μM and all assays were repeated three times.
[2]The maximum fluorescence intensity of insulin was set as 1.
[3]N.D., the lag time and T50 were too long to be accurately determined during the test.
To investigate whether quinones can affect the aggregation of already formed amyloid intermediates, each quinone was added to pre-incubated insulin (9 h). BQ and NQ suppressed the increase of fluorescence intensity in the following incubation, indicating blockage to the aggregation of already formed amyloid intermediates (Figure 2C). In contrast, PQ and AQ derivatives showed little inhibitory effects to pre-formed amyloid intermediates since the fluorescence continued to increase (Figure 2C). We further studied whether quinones can disaggregate preformed fibrils, quinones were added to insulin amyloids pre-incubated for 27 h. Partial disaggregation effects were detected in BQ and NQ treated samples, while no disaggregation effects were found in all other samples according to ThT fluorescence assays (Figure 2C).
Quinones delay the secondary structural transformation of insulin
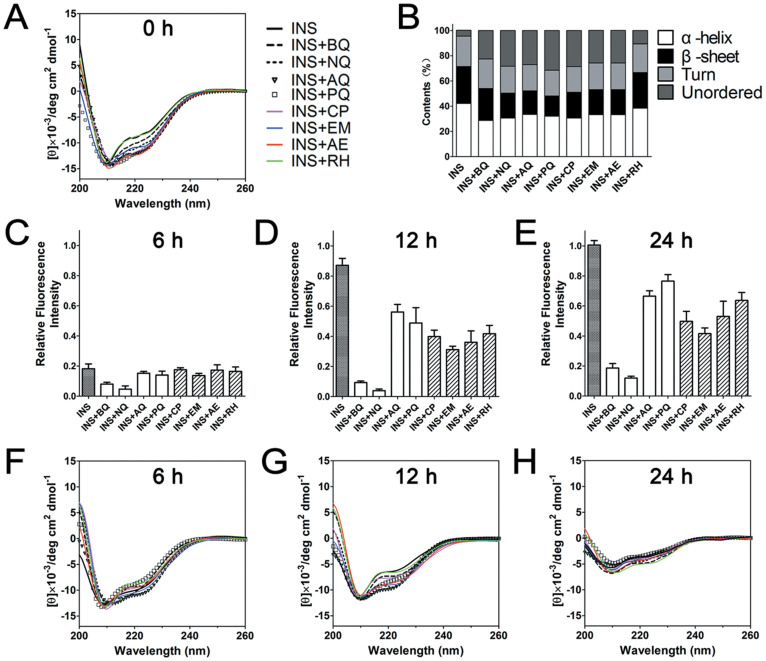
Far-UV CD spectroscopy was applied to analyze the effects of quinones on the secondary structure transition of insulin during amyloid formation. For native insulin, which is a highly structured molecule rich in α-helix, two characteristic minima at 208 and 222 nm were observed in CD spectrum (Figure 4A). During incubation, these two minima were decreased, indicating decreased content of α-helix (Figure 4A). The incubation also caused reduced ellipticity near 200 nm that represents formation of random coil conformation during incubation (Figure 4A), which is similar to a previous research studied under extreme temperatures36. β-structure formation is regarded as a generic structural conversion in protein aggregation37, typical β-structure signature at 215 nm was not obvious until the fluorescence intensity of insulin reached its maximum at 24 h.
Figure 4. Secondary structures of insulin in the presence and absence of quinones.
Far-UV circular dichroism spectra of sample groups at 0 h (A), and percentages of secondary structure contents (B). Relative thioflavin-T fluorescence intensities at 6 h (C), 12 h (D). 24 h (E) and corresponding far-UV circular dichroism spectra (F–H). The fluorescence signals of all samples were normalized against that of insulin incubated for 24 h, which was set as 1.
For quinones treated samples, it was interesting to note that the addition of quinones immediately decreased the positive signal at 200 nm and increased the negative band near 220 nm which refer to a reduction of α-helix content (Figure 4A & 4B), suggesting there may be a direct interaction between quinones and insulin. After co-incubated with quinones for 6 h and 12 h, the spectral changes at 200 and 220 nm were significantly less compared to those of the insulin control, indicating quinones delay the structural transformation (Figure 4F & 4G), which was consistent with the accompanying ThT-fluorescence measurements (Figure 4C & 4D). At the end of 24 h incubation, although the fluorescence still showed notable inhibitory effects of quinones (Figure 4E), the CD spectra nevertheless suggested the secondary structures of all quinones treated samples were similar to that of control insulin (Figure 4H).
Quinones inhibit the oligomerization and fibrillation of insulin
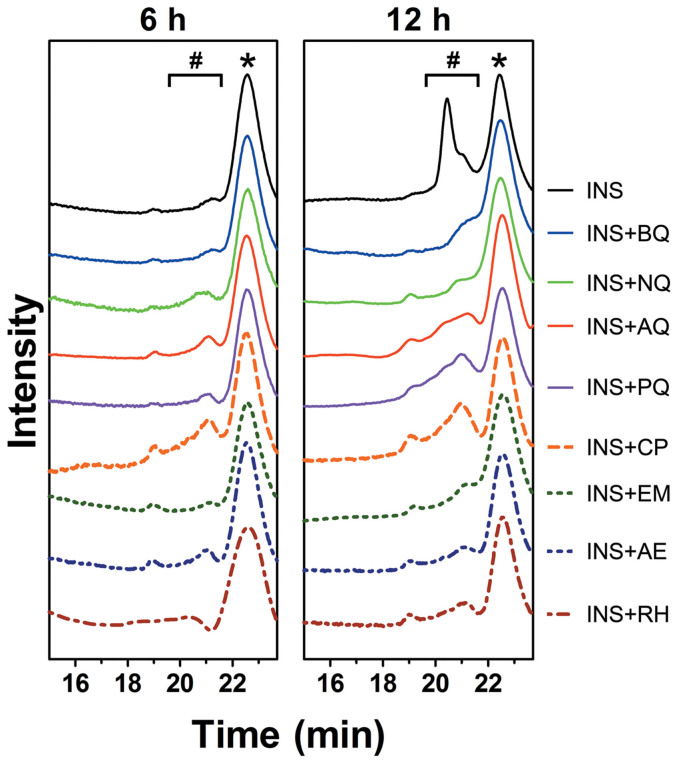
Size-exclusion chromatography (SEC) was first used to analyze the oligomerization states. For insulin, the monomer showed a single peak eluting at 22.4 min with a calculated molecular weight (MW) of 5.8 kD, while the addition of quinones caused no change to the monomeric status of insulin at the beginning of the incubation (Figure 5 & Figure S1). After incubating for 6 h, small peaks appeared with elution time between 19.5–21.5 min (ca. 10–16 kD), suggesting the formation of dimers and trimers (Figure 5). The intensities of these dimer and trimer peaks rose sharply after 12 h incubation (Figure 5), whereas quinones strongly inhibited the oligomerization of insulin (Figure 5 & Figure S2). This result agrees with those obtained from the CD spectrometry and ThT fluorescence assay, which was further corroborated by an immuno dot-blot assay, which probes the formation of oligomers and fibrils in the incubated samples with the oligomer- or fibril-specific antibodies (Figure S3).
Figure 5. Size-exclusion gel filtration profiles of insulin co-incubated with quinones.
* insulin monomers; # insulin oligomers.
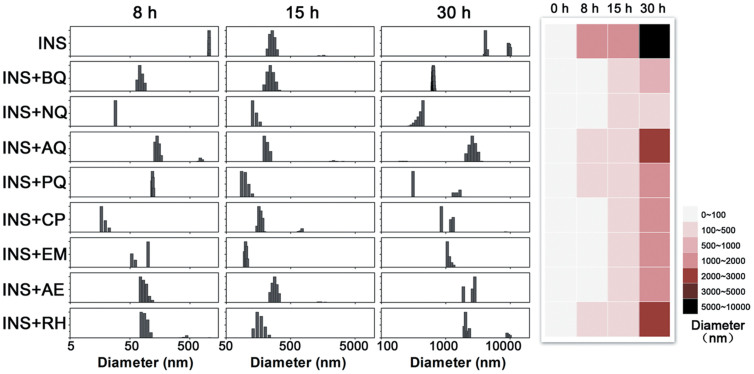
Dynamic light scattering (DLS) measurements were further performed to monitor particle size distributions (Figure 6). At 0 h, the average diameter of each sample was less than 100 nm (data not shown). For insulin, oligomers with diameters over 1000 nm were observed after 8 h incubation, and fibrils with size greater than 5000 nm were detected after 30 h. In contrast, at each time point, the average particle sizes of all quinones treated samples were less than those of insulin alone, with BQ- and NQ-treatment showed the strongest effects in reducing the average particle sizes of insulin. Consistent with the ThT results, rhubarb anthraquinone derivatives also showed inhibitory effects greater than that of AQ (Figure 6). Among all eight quinones tested, NQ has the highest inhibitory effect as shown in a colour map (Figure 6).
Figure 6. Particle size distributions detected by dynamic light scattering.
The colour map on the right summarized the size distributions of all experimental groups measured at indicated time points.
Quinones attenuate membrane disruption effects that induced by toxic insulin aggregates
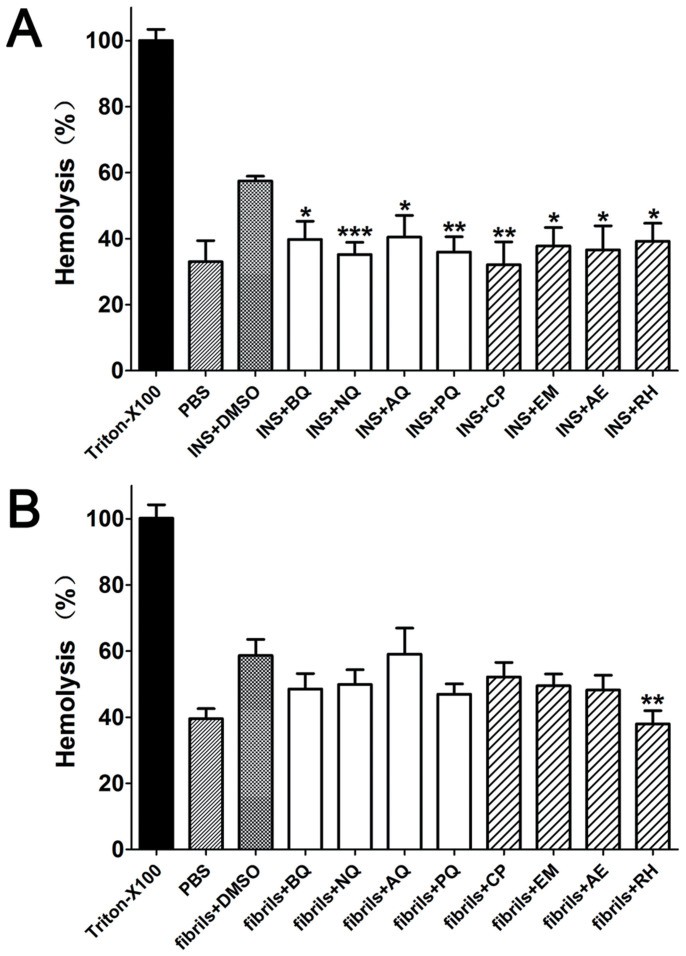
Oligomers and mature amyloids have been shown to disintegrate cell membrane and cause cell death38,39. Erythrocyte and insulin aggregates were co-incubated to simulate cell membrane disruption. After 24 h incubation, insulin fibrils induced a hemolysis rate of 57.5 ± 2.5%, which was higher than that of PBS-treated controls (33.6 ± 8.3%, P < 0.05). In quinones-treated samples, all hemolysis rates were decreased to ca. 40% of that of insulin (P < 0.05; Figure 7A). To investigate whether quinones by themselves interrupt the binding between insulin and membrane, a control study was performed in which quinones were added to completely formed insulin fibrils, and then mixed with erythrocyte for further incubation, the results showed quinones could not attenuate the membrane disruption effect of insulin fibrils except for RH (Figure 7B). Another control study suggested quinones by themselves had no protective effect on erythrocyte hemolysis, some compounds even slightly induced hemolysis (Figure S4). Unlike ThT data, no difference was found between simple quinones and rhubarb anthraquinone derivatives comparing their anti membrane disruption effects.
Figure 7. The hemolysis rates of insulin aggregates in the presence or absence of quinones.
(A) Hemolysis levels of pre-prepared quinones-coincubated insulin solution; (B) Hemolysis rates of already formed insulin fibrils with quinones added then. *, P < 0.05; **, P < 0.01 and ***, P < 0.001 compared with the group of protein with DMSO.
Discussion and Conclusion
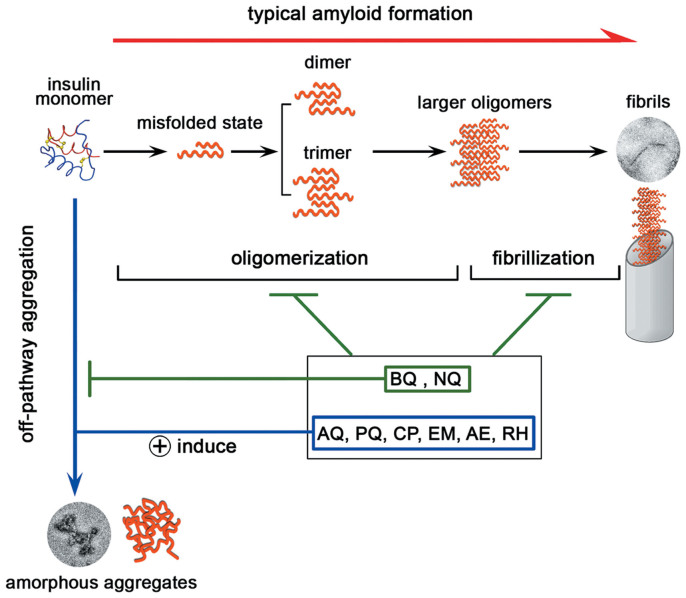
According to classic amyloidogenesis model, insulin amyloid formation may be divided into two stages, in the initial oligomerization stage, insulin monomers misfold and assemble into small molecular weight oligomers including dimers,trimers and other low MW oligomers; in the second fibrillization stage, low order oligomers assemble into large oligomers which grow into linear fibrils40,41 (Figure 8). Recent studies also reported the existence of “off-pathway” oligomerization pathways during protein misfolding42 (Figure 8). For example, it was revealed that α-synuclein (an amyloidogenic protein associated with Parkinson's disease) may form nontoxic and unstructured non-amyloid aggregates43. In another elegant recent study, EGCG was found to redirect the amyloid formation of Aβ into off-pathway nontoxic aggregates44. Here, we found all eight tested quinones inhibit the classic amyloid formation pathways of insulin, both in the oligomerization and fibrillization stages. Since for the samples treated with AQ, PQ and AQ derivatives all showed un-classic fibrillation characters including the complete or partial formation of amorphous aggregates that unlike typical linear amyloid morphology, decreased ThT fluorescence intensities, decreased average particle sizes as detected by DLS, and significantly attenuated toxicities as the hemolysis assays indicated. All these phenomena suggested that quinones may drive insulin undergoing the “off-pathway” aggregation while inhibiting the classic amyloid formation pathway (Figure 8). Although the exact mechanisms of quinones' inhibitory effects remain unclear, we reason that quinones may directly interact with insulin as the CD and hemolysis results suggested and thus disrupt oligomerization, similar to what EGCG does to the amyloid formation of Aβ and α-synuclein43,44. It will be of great future interest to further probe the mechanism with the nuclear magnetic resonance (NMR) spectrometry and molecular dynamics simulation approaches, which have been successfully applied in mapping the interaction sites between amyloidogenic proteins and small ligands45,46, and also in studying insulin and monosaccharides interaction47.
Figure 8. A schematic representation of how quinones interfere the aggregation pathway of insulin.
It is worth mentioning that BQ and NQ, two simplest quinones used in this study, showed the highest efficacy in inhibiting insulin aggregation; whereas AQ and PQ, which containing more benzene rings, showed lower inhibitory effects. The amyloid inhibitory effects of the eight quinones follow the rank order of NQ > BQ > PQ > EM > CP > AE > RH > AQ. It is generally accepted that aromatic-intereactions and hydrophobic interactions are two major known driving forces responsible for amyloid formation48,49. For quinones, molecular simulations have implicated that quinones may destabilize the intermolecular cross-β strand hydrogen bonds of oligomers and mature fibrils via more favorable polar interactions with the protein backbones, the quinonic moiety may bind to peptide backbone carbonyl oxygens and amide hydrogens tightly through π+δ− interactions and hydrogen bonds, which allow the intercalation of the quinone molecule into the protein oligomers28,50. We speculate that the aromatic ring of monocyclic and dicyclic quinones (BQ and NQ) may also competitively interact with aromatic residues and thus intercalate into quino-protein oligomers which can not further aggregate into fibrils. For tricyclic quinones, the extra benzene ring may increase the steric hindrance of the molecule and interfere the binding of compound and protein. For rhubarb derived AQ derivatives, the inhibitory effect may be influenced by substituent groups. It has been reported that aromatic amyloid inhibitors with strong electron withdrawing group on monocyclic benzene may increase the inhibitory potency51. For tri- and tetra-substituted AQ with more than two electron donating phenolic hydroxyl groups, the regulator may be more complex than that of monocyclic compounds. The phenolic hydroxyl groups may bind with hydrophobic residues to prevent hydrophobic interaction caused oligomerization, which may explain why AQ derivatives inhibit aggregation more efficiently than AQ. In addition, different factors such as metals and peptides may also affect the aggregation properties52,53, several small molecules have been reported to inhibit amyloid aggregation by chelating metal ions from metal-Aβ species54,55. Since quinones and their derivatives have also been reported as chelators56, it will be interesting to investigate whether quinones can also interrupt the ligands (such as metal ions) induced misfolding and aggregation.
In β-amyloid model, natural quinones such as pyrroloquinoline quinone and α-tocopherol quinone were reported as effective amyloid inhibitors on aggregation35,57. Simple quinones in our study, including the first time studied 9,10-phenanthraquinone, may act as functional skeleton entities that contribute to the inhibitory effect of natural quinones. Moreover, anthraquinone derivatives from rhubarb further indicate that natural quinones with similar configuration may provide a huge resource of amyloid inhibitors. Although some quinones from plants have been thought to be allergens58, quinones are still important sources for deriving new anti-amyloid inhibitors. And more extensive conditions such as metal ions,crowding, pressure and shear force may be considered to validate the present results. It will also be interesting to extent this research to traditional natural medicines containing more complex quinones as well as to other pathogenic amyloid systems.
Methods
Materials
Porcine insulin was obtained from Wanbang Biopharmaceutics INC. (Xuzhou, China). 1,4-benzoquinone, 1,4-naphthoquinone, 9,10-anthraquinone, 9,10-phenanthraquinone, chrysophanol (1,8-dihydroxy-3-methyl-anthraquinone), emodin (1,3,8-Trihydroxy-6- methyl-anthraquinone), aloe-emodin (1,8-dihydroxy-3-hydroxymethyl- anthraquinone) and rhein (1,8-dihydroxy-3-carboxy-anthraxquinone) were obtained from Aladdin-Reagents (Shanghai, China). Thioflavin-T (ThT) was from Sigma–Aldrich (St. Louis, USA). Anti-oligomer antibody (A-11), anti-fibril antibody (OC) and anti-rabbit IgG were from Merck Millipore (Billerica, USA). All other chemicals were of the highest grade available. For hemolytic assays, fresh blood was collected from healthy volunteers with approval by the ethics committees of the Third Hospital of Wuhan.
Amyloid formation and thioflavin-T (ThT) fluorescence assays
Insulin was dissolved in 50 mM PBS (100 mM NaCl, pH 7.4) to a final concentration of 100 μM. Freshly prepared solution was mixed with different quinones in a molar ratio of protein:compound = 2:1 and sonicated for 2 min. DMSO was used to prepare the stock solutions of quinones with a final solution DMSO concentration of 0.5% (control study demonstrated that 0.5% DMSO has no significant effect on aggregation, data not shown). All groups were static incubated at 65°C to form aggregates. The aggregation status of designated time points was determined by ThT fluorescence (a final ThT concentration of 20 μM) assay recorded on a Hitachi FL-2700 fluorometer (Hitachi, Japan) at 482 nm with an excitation wavelength of 450 nm as we previously described59. The data were normalized against the DMSO or quinone controls background measured at the corresponding concentrations. For the aggregation intermediate disruption and fibril disaggregation experiments, each quinone was added to pre-incubated insulin at 9 h and 27 h respectively and the fluorescence was monitored in the following incubation. Each experiment was performed at least three times.
Far-UV circular dichroism (CD) and data analysis
A JASCO-810 circular dichroism spectropolarimeter (JASCO, Japan) was used to monitor secondary structures. Incubated samples were diluted to a final concentration of 20 μM and detected in a 1 mm path length at 25°C under a constant flow of N2. CD spectra were obtained from 260 to 190 nm at a 50 nm/min scanning speed and a 2 nm bandwidth. All samples were measured in triplicates and the averages were taken. The data were converted to mean residue ellipticity [θ] and were further calculated with the software CDPro60.
Size-exclusion chromatography (SEC)
Incubated samples of designated time points were centrifuged and the supernatant was injected into a Tosoh TSK GW2000 column to perform SEC analysis on an L-2000 HPLC system (Hitachi, Japan). Elution buffer was 20% acetonitrile (0.003% TFA) with a flow rate of 0.3 ml/min and a detection wavelength of 215 nm.
Dynamic light scattering (DLS) analysis
The sizes of insulin aggregates were measured by dynamic light scattering in a zeta pals potential analyzer (Brookhaven Instruments, USA). Incubated samples were vortexed and detected at room temperature. The scattering angle was set at 90°. Each measurement was repeated three times and the average mean particle size was recorded. The data was analyzed by the multimodal size distribution (MSD) software as we previously described61.
Dot blot assay
Incubated samples were prepared as described. 3 μL of sample solution aliquoted at indicated time points was blotted onto a nitrocellulose membrane (Biorad, USA). Dried membrane was blocked with 5% non-fat milk for 1 h at room temperature and then incubated with A-11 (oligomer-specific antibody; 1:2500) or OC antibody (fibril-specific antibody; 1:2500) at 4°C overnight. The membrane was incubated with anti-rabbit IgG (1:5000) for 2 h at room temperature later. An ECL chemiluminescence kit (Advansta, USA) was used for the final blot development.
Transmission electronic microscopy (TEM)
Samples for TEM assay were prepared as we previously described62. Incubated solution was applied onto a 300-mesh formvar-carbon coated copper grid and sit for 5 min. 1% freshly prepared uranyl formate was then dropwise added for staining. Dried samples were observed under a transmission microscope (Hitachi, Japan) operating at an accelerating voltage of 200 kV.
Hemolytic assay
Erythrocytes were separated from fresh blood through centrifugation at 1000 g and washed three times with isotonic phosphate buffered saline (pH 7.4) as described63. Insulin with or without quinones were first incubated at 65°C for 24 h, after the formation of aggregates were confirmed with the ThT assays, the incubated samples were mixed with cell suspensions (the final sample contains 2% hematocrit and 90 μM insulin fibrils) and shaked at 200 rpm for 24 h at 37°C. To investigate the possible effects of quinones on fibril-induced membrane disruption, mature insulin fibrils were first mixed with erythrocytes, then quinones were added and incubated for another 24 h. The absorption of supernatant was tested at 540 nm. The hemolytic rate was calculated in relation to the hemolysis of erythrocytes with 1% Triton-100, which was taken as 100%. Erythrocytes treated with DMSO and quinones, but without insulin were used as controls.
Statistical analysis
Each experiment was repeated three times and the data were expressed as the mean ± SD. The Kruskal-Wallis test and the Mann-Whitney test were used to evaluate statistical significance. Difference was considered statistically significant at P < 0.05.
Author Contributions
Study concept and design: H.G., L.Z. and K.H. Acquisition of data: H.G., Z.H., B.C., A.P., X.Z. and Y.S. Drafting the manuscript and critical revision: H.G., A.P., L.Z. and K.H. All authors have approved the final version of the paper.
Supplementary Material
Supplementary Information
Acknowledgments
The authors thank the Analytical and Testing Center of Huazhong University of Science and Technology, and the Research Core Facility of College of Life Sciences, Wuhan University for technical supports. This work was supported by the National Basic Research Program of China (2012CB524901), the Natural Science Foundation of China (Nos 31271370, 81100687, 81172971 and 81222043), the Program for New Century Excellent Talents in University (NECT11-0170), the Municipal Key Technology Program of Wuhan (Wuhan Bureau of Science & Technology, No. 201260523174), the Health Bureau of Wuhan (WX12B06) and the Natural Science Foundation of Hubei Province (2011BCB012 and 2013CFB359).
References
- Cao P., Abedini A. & Raleigh D. P. Aggregation of islet amyloid polypeptide: from physical chemistry to cell biology. Curr Opin Struct Biol 23, 82–9 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dikiy I. & Eliezer D. Folding and misfolding of alpha-synuclein on membranes. Biochim Biophys Acta 1818, 1013–8 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nussbaum J. M., Seward M. E. & Bloom G. S. Alzheimer disease: a tale of two prions. Prion 7, 14–9 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes D. H. et al. Mechanism of islet amyloid polypeptide fibrillation at lipid interfaces studied by infrared reflection absorption spectroscopy. Biophys J 93, 3132–41 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oren Z. & Shai Y. Mode of action of linear amphipathic alpha-helical antimicrobial peptides. Biopolymers 47, 451–63 (1998). [DOI] [PubMed] [Google Scholar]
- Mattson M. P. & Goodman Y. Different amyloidogenic peptides share a similar mechanism of neurotoxicity involving reactive oxygen species and calcium. Brain Res 676, 219–24 (1995). [DOI] [PubMed] [Google Scholar]
- Naiki H. & Nagai Y. Molecular pathogenesis of protein misfolding diseases: pathological molecular environments versus quality control systems against misfolded proteins. J Biochem 146, 751–6 (2009). [DOI] [PubMed] [Google Scholar]
- Yumlu S., Barany R., Eriksson M. & Rocken C. Localized insulin-derived amyloidosis in patients with diabetes mellitus: a case report. Hum Pathol 40, 1655–1660 (2009). [DOI] [PubMed] [Google Scholar]
- Okamura S., Hayashino Y., Kore-Eda S. & Tsujii S. Localized Amyloidosis at the Site of Repeated Insulin Injection in a Patient With Type 2 Diabetes. Diabetes Care 36, E200–E200 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brange J., Andersen L., Laursen E. D., Meyn G. & Rasmussen E. Toward understanding insulin fibrillation. J Pharm Sci 86, 517–25 (1997). [DOI] [PubMed] [Google Scholar]
- Huang K., Dong J., Phillips N. B., Carey P. R. & Weiss M. A. Proinsulin is refractory to protein fibrillation - Topological protection of a precursor protein from cross-beta assembly. Journal of Biological Chemistry 280, 42345–42355 (2005). [DOI] [PubMed] [Google Scholar]
- Seeliger J. & Winter R. Islet amyloid polypeptide: aggregation and fibrillogenesis in vitro and its inhibition. Subcell Biochem 65, 185–209 (2012). [DOI] [PubMed] [Google Scholar]
- Eisele Y. S. From soluble abeta to progressive abeta aggregation: could prion-like templated misfolding play a role? Brain Pathol 23, 333–41 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marques O. & Outeiro T. F. Alpha-synuclein: from secretion to dysfunction and death. Cell Death Dis 3, e350 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan K. M. et al. Conversion of alpha-helices into beta-sheets features in the formation of the scrapie prion proteins. Proc Natl Acad Sci U S A 90, 10962–6 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill S. E., Miti T., Richmond T. & Muschol M. Spatial Extent of Charge Repulsion Regulates Assembly Pathways for Lysozyme Amyloid Fibrils. PLoS One 6 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen L., Frokjaer S., Brange J., Uversky V. N. & Fink A. L. Probing the mechanism of insulin fibril formation with insulin mutants. Biochemistry 40, 8397–8409 (2001). [DOI] [PubMed] [Google Scholar]
- Nielsen L., Frokjaer S., Brange J., Uversky V. N. & Fink A. L. Probing the mechanism of insulin fibril formation with insulin mutants. Biochemistry 40, 8397–409 (2001). [DOI] [PubMed] [Google Scholar]
- Wan Z. L. et al. Diabetes-associated mutations in human insulin: crystal structure and photo-cross-linking studies of a-chain variant insulin Wakayama. Biochemistry 44, 5000–16 (2005). [DOI] [PubMed] [Google Scholar]
- Kachooei E. et al. Inhibition study on insulin fibrillation and cytotoxicity by paclitaxel. J Biochem 155, 361–73 (2014). [DOI] [PubMed] [Google Scholar]
- Mishra N. K., Joshi K. B. & Verma S. Inhibition of human and bovine insulin fibril formation by designed peptide conjugates. Mol Pharm 10, 3903–12 (2013). [DOI] [PubMed] [Google Scholar]
- Cheng B. et al. Inhibiting toxic aggregation of amyloidogenic proteins: a therapeutic strategy for protein misfolding diseases. Biochim Biophys Acta 1830, 4860–71 (2013). [DOI] [PubMed] [Google Scholar]
- Bieschke J. et al. EGCG remodels mature alpha-synuclein and amyloid-beta fibrils and reduces cellular toxicity. Proc Natl Acad Sci U S A 107, 7710–5 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono K., Hasegawa K., Naiki H. & Yamada M. Curcumin has potent anti-amyloidogenic effects for Alzheimer's beta-amyloid fibrils in vitro. J Neurosci Res 75, 742–50 (2004). [DOI] [PubMed] [Google Scholar]
- Mishra R., Sellin D., Radovan D., Gohlke A. & Winter R. Inhibiting islet amyloid polypeptide fibril formation by the red wine compound resveratrol. Chembiochem 10, 445–9 (2009). [DOI] [PubMed] [Google Scholar]
- Nie Q., Du X. G. & Geng M. Y. Small molecule inhibitors of amyloid beta peptide aggregation as a potential therapeutic strategy for Alzheimer's disease. Acta Pharmacol Sin 32, 545–51 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng S., Song X. H. & Zeng C. M. Inhibition of amyloid fibrillation of lysozyme by phenolic compounds involves quinoprotein formation. FEBS Lett 586, 3951–5 (2012). [DOI] [PubMed] [Google Scholar]
- Convertino M., Pellarin R., Catto M., Carotti A. & Caflisch A. 9,10-Anthraquinone hinders beta-aggregation: how does a small molecule interfere with Abeta-peptide amyloid fibrillation? Protein Sci 18, 792–800 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bermejo-Bescos P. et al. In vitro antiamyloidogenic properties of 1,4-naphthoquinones. Biochem Biophys Res Commun 400, 169–74 (2010). [DOI] [PubMed] [Google Scholar]
- Misiti F. et al. Protective effect of rhubarb derivatives on amyloid beta (1-42) peptide-induced apoptosis in IMR-32 cells: a case of nutrigenomic. Brain Res Bull 71, 29–36 (2006). [DOI] [PubMed] [Google Scholar]
- Liu J. et al. Rhein lysinate decreases the generation of beta-amyloid in the brain tissues of Alzheimer's disease model mice by inhibiting inflammatory response and oxidative stress. J Asian Nat Prod Res 15, 756–63 (2013). [DOI] [PubMed] [Google Scholar]
- Pickhardt M. et al. Anthraquinones inhibit tau aggregation and dissolve Alzheimer's paired helical filaments in vitro and in cells. J Biol Chem 280, 3628–35 (2005). [DOI] [PubMed] [Google Scholar]
- Siripong P. et al. Antitumor activity of liposomal naphthoquinone esters isolated from Thai medicinal plant: Rhinacanthus nasutus KURZ. Biol Pharm Bull 29, 2279–83 (2006). [DOI] [PubMed] [Google Scholar]
- Dey D., Ray R. & Hazra B. Antitubercular and Antibacterial Activity of Quinonoid Natural Products Against Multi-Drug Resistant Clinical Isolates. Phytother Res (2013). [DOI] [PubMed]
- Kim J. et al. Pyrroloquinoline quinone inhibits the fibrillation of amyloid proteins. Prion 4, 26–31 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arora A., Ha C. & Park C. B. Insulin amyloid fibrillation at above 100 degrees C: new insights into protein folding under extreme temperatures. Protein Sci 13, 2429–36 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobson C. M. Protein misfolding, evolution and disease. Trends Biochem Sci 24, 329–32 (1999). [DOI] [PubMed] [Google Scholar]
- Wang J. B., Wang Y. M. & Zeng C. M. Quercetin inhibits amyloid fibrillation of bovine insulin and destabilizes preformed fibrils. Biochem Biophys Res Commun 415, 675–9 (2011). [DOI] [PubMed] [Google Scholar]
- Grudzielanek S. et al. Cytotoxicity of insulin within its self-assembly and amyloidogenic pathways. J Mol Biol 370, 372–84 (2007). [DOI] [PubMed] [Google Scholar]
- Kelly J. W. Alternative conformations of amyloidogenic proteins govern their behavior. Curr Opin Struct Biol 6, 11–7 (1996). [DOI] [PubMed] [Google Scholar]
- Gillam J. E. & MacPhee C. E. Modelling amyloid fibril formation kinetics: mechanisms of nucleation and growth. J Phys Condens Matter 25, 373101 (2013). [DOI] [PubMed] [Google Scholar]
- Uversky V. N. Mysterious oligomerization of the amyloidogenic proteins. FEBS J 277, 2940–53 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrnhoefer D. E. et al. EGCG redirects amyloidogenic polypeptides into unstructured, off-pathway oligomers. Nat Struct Mol Biol 15, 558–66 (2008). [DOI] [PubMed] [Google Scholar]
- Zhang T., Zhang J., Derreumaux P. & Mu Y. Molecular mechanism of the inhibition of EGCG on the Alzheimer Abeta(1-42) dimer. J Phys Chem B 117, 3993–4002 (2013). [DOI] [PubMed] [Google Scholar]
- Brender J. R. et al. Zinc stabilization of prefibrillar oligomers of human islet amyloid polypeptide. Chem Commun (Camb) 49, 3339–41 (2013). [DOI] [PubMed] [Google Scholar]
- Hyung S. J. et al. Insights into antiamyloidogenic properties of the green tea extract (-)-epigallocatechin-3-gallate toward metal-associated amyloid-beta species. Proc Natl Acad Sci U S A 110, 3743–3748 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falconi M. et al. Spectroscopic and molecular dynamics simulation studies of the interaction of insulin with glucose. Int J Biol Macromol 29, 161–8 (2001). [DOI] [PubMed] [Google Scholar]
- Ahmad E. et al. A mechanistic approach for islet amyloid polypeptide aggregation to develop anti-amyloidogenic agents for type-2 diabetes. Biochimie 93, 793–805 (2011). [DOI] [PubMed] [Google Scholar]
- Wu C., Lei H., Wang Z., Zhang W. & Duan Y. Phenol red interacts with the protofibril-like oligomers of an amyloidogenic hexapeptide NFGAIL through both hydrophobic and aromatic contacts. Biophys J 91, 3664–72 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer E. A., Castellano R. K. & Diederich F. Interactions with aromatic rings in chemical and biological recognition. Angew Chem Int Ed Engl 42, 1210–50 (2003). [DOI] [PubMed] [Google Scholar]
- Levy-Sakin M., Shreberk M., Daniel Y. & Gazit E. Targeting insulin amyloid assembly by small aromatic molecules: toward rational design of aggregation inhibitors. Islets 1, 210–5 (2009). [DOI] [PubMed] [Google Scholar]
- DeToma A. S., Salamekh S., Ramamoorthy A. & Lim M. H. Misfolded proteins in Alzheimer's disease and type II diabetes. Chem Soc Rev 41, 608–21 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savelieff M. G., Lee S., Liu Y. & Lim M. H. Untangling amyloid-beta, tau, and metals in Alzheimer's disease. ACS Chem Biol 8, 856–65 (2013). [DOI] [PubMed] [Google Scholar]
- Mancino A. M., Hindo S. S., Kochi A. & Lim M. H. Effects of clioquinol on metal-triggered amyloid-beta aggregation revisited. Inorg Chem 48, 9596–8 (2009). [DOI] [PubMed] [Google Scholar]
- Hindo S. S. et al. Small molecule modulators of copper-induced Abeta aggregation. J Am Chem Soc 131, 16663–5 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulaboski R. et al. Hydroxylated derivatives of dimethoxy-1,4-benzoquinone as redox switchable earth-alkaline metal ligands and radical scavengers. Scientific Reports 3 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S. G. et al. alpha-Tocopherol quinone inhibits beta-amyloid aggregation and cytotoxicity, disaggregates preformed fibrils and decreases the production of reactive oxygen species, NO and inflammatory cytokines. Neurochem Int 57, 914–22 (2010). [DOI] [PubMed] [Google Scholar]
- Lepoittevin J. P. & Benezra C. Allergic contact dermatitis caused by naturally occurring quinones. Pharm Weekbl Sci 13, 119–22 (1991). [DOI] [PubMed] [Google Scholar]
- Zhang X. et al. Porcine islet amyloid polypeptide fragments are refractory to amyloid formation. FEBS Lett 585, 71–7 (2011). [DOI] [PubMed] [Google Scholar]
- Huang K., Maiti N. C., Phillips N. B., Carey P. R. & Weiss M. A. Structure-specific effects of protein topology on cross-beta assembly: studies of insulin fibrillation. Biochemistry 45, 10278–93 (2006). [DOI] [PubMed] [Google Scholar]
- Gong H. et al. Bisphenol A accelerates toxic amyloid formation of human islet amyloid polypeptide: a possible link between bisphenol A exposure and type 2 diabetes. PLoS One 8, e54198 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng B. et al. Silibinin inhibits the toxic aggregation of human islet amyloid polypeptide. Biochem Biophys Res Commun 419, 495–9 (2012). [DOI] [PubMed] [Google Scholar]
- Li Y. et al. Dissecting the role of disulfide bonds on the amyloid formation of insulin. Biochem Biophys Res Commun 423, 373–8 (2012). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information