Abstract
SUN proteins accelerate the pathological progression of laminopathies. Although the mechanisms remain to be elucidated, an intriguing possibility is that high levels of SUN proteins lead to a hyperactive DNA damage response.
Lamins are a class of intermediate filaments that underlie the inner nuclear membrane and provide mechanical structure and stiffness to the nuclear envelope. Over a dozen human diseases, termed laminopathies, are associated with mutations in the gene encoding lamin A, including muscular dystrophies associated with cardiac myopathy, lipodystrophies, diabetes, peripheral neuropathies, and premature aging syndromes [1]. The mechanisms of how mutations in lamin A lead to the progression of the phenotypically diverse laminopathies are not understood.
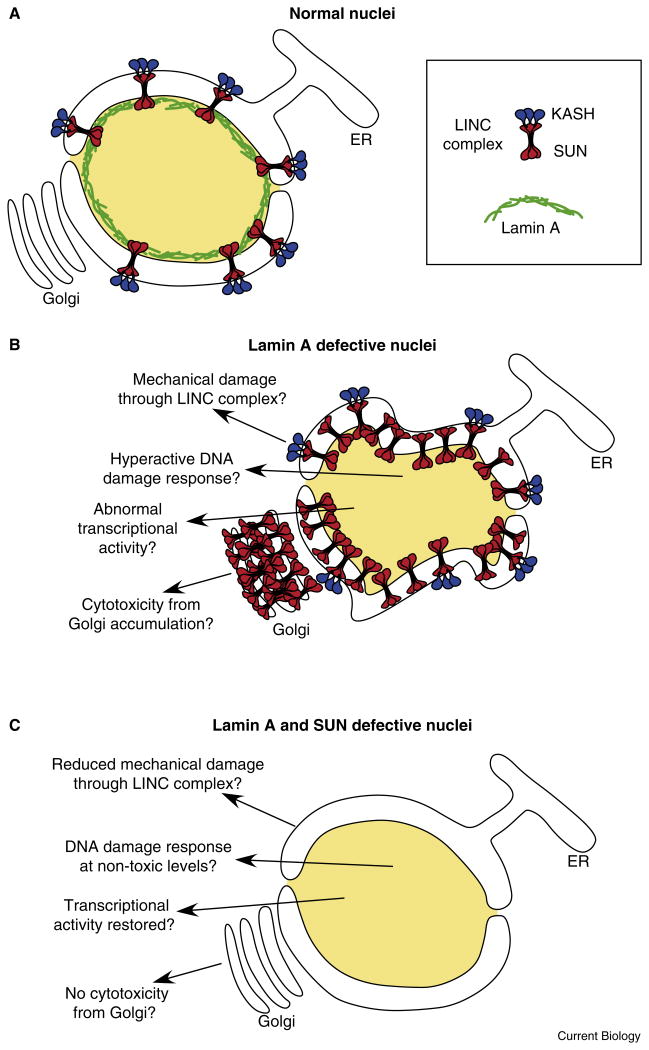
The field has focused on two models for how defects in lamin A contribute to disease [2] (Figure 1). In the first, lamin A plays a mechanical role; defects in lamin A make nuclei less stiff and subject to physical damage. The second model postulates that mutations in lamin A disrupt the transcriptional profile of important differentiation genes. Two recent papers report exciting new findings about the role of SUN proteins at the nuclear envelope. Each proposes an alternative model for how SUN proteins and lamin A together contribute to the pathology of laminopathies (Figure 1). Chen et al. [3] propose that an abnormal accumulation of SUN proteins at the Golgi leads to cellular toxicity, while Lei et al. [4], as reported in this issue of Current Biology, propose that high levels of SUN proteins at the nuclear envelope in lamin A mutant cells lead to toxicity through hyperactivity of the DNA damage response.
Figure 1.
Models for how lamin A and SUN proteins contribute to pathologies of laminopathies.
(A) A normal nucleus with lamin A (green), SUN (red), and KASH (blue) proteins at the nuclear envelope. (B) A deformed nucleus lacking lamin A with an over-accumulation of SUN proteins at the nuclear envelope. (C) The knockdown of SUN proteins rescues many of the pathologies of a lamin A mutant nucleus. See text for discussion of the four models.
Lamins are mechanically coupled to the cytoskeleton by a complex of SUN and KASH proteins termed the LINC complex (for linker of the nucleoskeleton and the cytoskeleton) [5]. SUN proteins localize to the inner nuclear membrane where their nucleoplasmic domains interact with lamins and their luminal domains recruit KASH proteins to the outer nuclear membrane. The SUN–KASH interaction was recently characterized at the structural level [6]. The cytoplasmic domains of KASH proteins then associate with various components of the cytoskeleton, completing bridges across the nuclear envelope [5]. SUN proteins play important roles in a wide variety of cellular and developmental functions, mostly through regulating the intracellular position of the nucleus. In mice, SUN proteins are required for gametogenesis, neurogenesis, muscle development, and retinogenesis; Sun1-/- Sun2-/- double knockout mice die shortly after birth as they are unable to breathe [7–9]. It has long been hypothesized that defects in SUN proteins might lead to progression of laminopathies [10–13].
To test the role of SUN proteins in the progression of laminopathies, Chen et al. [3] engineered mice mutant for both lamin A and Sun1. Surprisingly, knockout of Sun1 reduced the severity of phenotypes associated with the lamin A-/- (Muscular Dystrophy) or lamin AΔ9 (Progeria) mutant mice. While far short of being healthy, the Sun1, lamin A double mutant mice lived longer and grew larger than the lamin A single mutants. Knockout of Sun1 also ameliorated cellular pathologies associated with laminopathies in the lamin A mutant mice, including defects in bone structure, muscle formation, senescence, heterochromatin marks, and the shape of nuclei. Similar results were found when Sun1 was knocked down in cells from patients with Hutchinson-Gilford Progeria Syndrome. This led to the surprising conclusion that Sun1 protein enhances the defects associated with lamin A mutations in disease.
As a possible mechanism, Sun1 protein is more stable and over accumulates at the nuclear envelope in lamin A mutant mouse and Progeria patient cultured fibroblasts. Moreover, Sun1 abnormally accumulates in the Golgi of lamin A mutant mouse cells, leading to the proposal that large amounts of Sun1 at the Golgi are toxic to the cell and contribute to the pathology of laminopathies [3]. While this is an intriguing and novel model, no Sun1 was observed at the Golgi in cells from Hutchinson-Gilford patients [3,10], and the role of Sun2, which does not over-accumulate in a lamin A mutant background, is not clear. Moreover, the experiments implicating the Golgi rely on drug or overexpression treatments that could have many non-specific targets.
An alternative model is that SUN proteins mediate the DNA damage response. Lei et al. [4] isolated embryonic fibroblasts from Sun1-/-Sun2-/- double knockout mice to study the role of SUN proteins in the stability of the genome. The Sun1-/- Sun2-/-fibroblasts arrested in S-phase of the cell cycle, displayed a slightly higher incidence of apoptosis, exhibited excessive DNA damage, and had decreased levels of perinuclear heterochromatin. While examining markers of the DNA damage response, Lei et al. [4] found that levels of phosphorylated histone 2A.X and Chk1, both markers of an active DNA damage checkpoint, were much lower in the Sun1-/- Sun2-/- cells. Most significantly, treatment with DNA-damaging chemicals failed to activate the DNA damage checkpoint; ATM was not activated and cells did not arrest in G2 in damaged Sun1-/-Sun2-/- mutant cells [4]. Thus, either Sun1 or Sun2 is required for the DNA damage response in normal cells.
Lei et al. [4] attempted to find a mechanism for how SUN proteins might mediate the DNA damage response by identifying interacting proteins. They found that both Sun1 and Sun2 bound to the catalytic subunit of DNAPK, a protein known to function in DNA repair. However, knocking down DNAPK only led to a small reduction in activated ATM or histone 2A.X, suggesting that this could only be part of the mechanism [4]. It therefore remains to be addressed exactly how SUN proteins mediate the DNA damage response. Nonetheless, given these results, and previous results showing a role for the SUN protein Mps3 in repair of DNA double strand breaks in yeast [14], it is fairly clear that SUN proteins are involved in DNA repair.
How might a role in DNA repair be linked to the progression of laminopathies? Mutations in lamin A result in an excess of DNA damage [15,16] and an increase in the levels of Sun1 protein [3]. Since SUN proteins participate in the DNA damage response, Lei et al. [4] propose that extra Sun1 in the lamin A mutant background could contribute to disease pathologies by inducing hyperactivity in the DNA damage response. Knockdown of Sun1 could then partially neutralize the effect of lamin A mutations by reducing the DNA damage response.
These two new reports, along with an established literature, suggest multiple potential mechanisms for how SUN proteins might link lamin A mutations to the pathologies of laminopathies (Figure 1). In a normal nucleus, lamin A forms a network providing mechanical strength to the nuclear envelope. The LINC complex, consisting of SUN and KASH proteins, then links lamin to the cytoskeleton. Mutations in lamin A lead to an over-accumulation of SUN proteins at the nuclear envelope and Golgi complex. Various defects then lead to the diverse phenotypes of laminopathies. However, knockdowns of SUN protein activity can rescue many of the pathologies associated with lamin A mutations.
While it still remains to be elucidated how SUN proteins contribute to the progression of laminopathies, a combination of the four potential models may contribute to the disease. In an established model, an excess of SUN proteins could lead to more mechanical forces on the nuclear envelope, which is already weakened by the disruption of lamin A. Alternatively, mutations in lamin A and/or overexpression of SUN could affect the transcriptional activities of important differentiation genes. Two new models proposed here suggest that an excess of SUN proteins could lead to cellular toxicity by disrupting the Golgi complex or inducing hyperactivity of the DNA damage response. Future experiments by geneticists, cell biologists, and clinicians are required to determine the molecular mechanisms and relative merits of these four models in the progression of laminopathies.
References
- 1.Worman HJ. Nuclear lamins and laminopathies. J Pathol. 2012;226:316–325. doi: 10.1002/path.2999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Prokocimer M, Davidovich M, Nissim-Rafinia M, Wiesel-Motiuk N, Bar D, Barkan R, Meshorer E, Gruenbaum Y. Nuclear lamins: key regulators of nuclear structure and activities. J Cell Mol Med. 2009;13:1059–1085. doi: 10.1111/j.1582-4934.2008.00676.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen CY, Chi YH, Mutalif RA, Starost MF, Myers TG, Anderson SA, Stewart CL, Jeang KT. Accumulation of the inner nuclear envelope protein sun1 is pathogenic in progeric and dystrophic laminopathies. Cell. 2012;149:565–577. doi: 10.1016/j.cell.2012.01.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lei K, Zhu X, Xu R, Shao C, Xu T, Zhuang Y, Han M. Inner nuclear envelope proteins SUN1 and SUN2 play a prominent role in the DNA damage response. Curr Biol. 2012;22:1609–1615. doi: 10.1016/j.cub.2012.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Starr DA, Fridolfsson HN. Interactions between nuclei and the cytoskeleton are mediated by SUN-KASH nuclear-envelope bridges. Annu Rev Cell Dev Biol. 2010;26:421–444. doi: 10.1146/annurev-cellbio-100109-104037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sosa BA, Rothballer A, Kutay U, Schwartz TU. LINC complexes form by binding of three KASH peptides to domain interfaces of trimeric SUN proteins. Cell. 2012;149:1035–1047. doi: 10.1016/j.cell.2012.03.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang X, Lei K, Yuan X, Wu X, Zhuang Y, Xu T, Xu R, Han M. SUN1/2 and Syne/Nesprin-1/2 complexes connect centrosome to the nucleus during neurogenesis and neuronal migration in mice. Neuron. 2009;64:173–187. doi: 10.1016/j.neuron.2009.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ding X, Xu R, Yu J, Xu T, Zhuang Y, Han M. SUN1 is required for telomere attachment to nuclear envelope and gametogenesis in mice. Dev Cell. 2007;12:863–872. doi: 10.1016/j.devcel.2007.03.018. [DOI] [PubMed] [Google Scholar]
- 9.Yu J, Lei K, Zhou M, Craft CM, Xu G, Xu T, Zhuang Y, Xu R, Han M. KASH protein Syne-2/Nesprin-2 and SUN proteins SUN1/2 mediate nuclear migration during mammalian retinal development. Hum Mol Genet. 2011;20:1061–1073. doi: 10.1093/hmg/ddq549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Haque F, Mazzeo D, Patel JT, Smallwood DT, Ellis JA, Shannahan CM, Shackleton S. Mammalian SUN protein networks at the inner nuclear membrane and their role in laminopathy disease processes. J Biol Chem. 2010;285:3487–3498. doi: 10.1074/jbc.M109.071910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kandert S, Luke Y, Kleinhenz T, Neumann S, Lu W, Jaeger VM, Munck M, Wehnert M, Muller CR, Zhou Z, et al. Nesprin-2 giant safeguards nuclear envelope architecture in LMNA S143F progeria cells. Hum Mol Genet. 2007;16:2944–2959. doi: 10.1093/hmg/ddm255. [DOI] [PubMed] [Google Scholar]
- 12.Zhang Q, Bethmann C, Worth NF, Davies JD, Wasner C, Feuer A, Ragnauth CD, Yi Q, Mellad JA, Warren DT, et al. Nesprin-1 and -2 are involved in the pathogenesis of Emery Dreifuss muscular dystrophy and are critical for nuclear envelope integrity. Hum Mol Genet. 2007;16:2816–2833. doi: 10.1093/hmg/ddm238. [DOI] [PubMed] [Google Scholar]
- 13.Puckelwartz MJ, Kessler E, Zhang Y, Hodzic D, Randles KN, Morris G, Earley JU, Hadhazy M, Holaska JM, Mewborn SK, et al. Disruption of nesprin-1 produces an Emery Dreifuss muscular dystrophy-like phenotype in mice. Hum Mol Genet. 2009;18:607–620. doi: 10.1093/hmg/ddn386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Oza P, Jaspersen SL, Miele A, Dekker J, Peterson CL. Mechanisms that regulate localization of a DNA double-strand break to the nuclear periphery. Genes Dev. 2009;23:912–927. doi: 10.1101/gad.1782209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gonzalez-Suarez I, Gonzalo S. Nurturing the genome: A-type lamins preserve genomic stability. Nucleus. 2010;1:129–135. doi: 10.4161/nucl.1.2.10797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu B, Wang J, Chan KM, Tjia WM, Deng W, Guan X, Huang JD, Li KM, Chau PY, Chen DJ, et al. Genomic instability in laminopathy-based premature aging. Nat Med. 2005;11:780–785. doi: 10.1038/nm1266. [DOI] [PubMed] [Google Scholar]