Abstract
The metabolite (–)-lomaiviticin A, which contains two diazotetrahydrobenzo[b]fluorene (diazofluorene) functional groups, inhibits the growth of cultured human cancer cells at nanomolar–picomolar concentrations; however, the mechanism responsible for the potent cytotoxicity of this natural product is not known. Here we report that (–)-lomaiviticin A nicks and cleaves plasmid DNA by an ROS- and iron-independent pathway and that the potent cytotoxicity of (–)-lomaiviticin A arises from induction of DNA double-strand breaks (dsbs). In a plasmid cleavage assay, the ratio of single-strand breaks (ssbs) to dsbs is 5.3±0.6:1. Labeling studies suggest this cleavage occurs via a radical pathway. The structurally related isolates (–)-lomaiviticin C and (–)-kinamycin C, which contain one diazofluorene, are demonstrated to be much less effective DNA cleavage agents, thereby providing an explanation for the enhanced cytotoxicity of (–)-lomaiviticin A compared to other members of this family.
The bacterial metabolite (–)-lomaiviticin A (1, Figure 1A) was first isolated in 2001 and is a potent cytotoxic agent, with half-maximal inhibitory potencies (IC50) in the 0.007–72 nM range against 25 cultured human cancer cell lines.1 The union of two diazofluorene functional groups (see structure 4), four 2,6-dideoxyglycoside residues, and a highly oxygenated cyclohexenone core serve to create in 1 an impressive, C2-symmetric molecular architecture. The metabolite (–)-lomaiviticin C (2) was recently identified in the fermentation broth of the producing organism.2,3 Spectroscopic analysis revealed 2 as constitutionally and stereochemically identical to 1, save for the conversion of one diazofluorene into a hydroxyfulvene (blue in 2). This structural change diminishes cytotoxicity by two orders of magnitude.2 The kinamycins [e.g., (–)-kinamycin C (3)]4–8 were isolated in the early 1970s and are the first natural products found to contain a diazofluorene (for reviews, see refs. 9–13). The kinamycins are monomeric, do not possess carbohydrate residues, and are also two orders of magnitude less cytotoxic than 1. Collectively these data suggest that the dimeric diazofluorene structure of 1 is essential for potent activity, but the origins of this effect are not known.
Figure 1.
Structures of the metabolites employed in this study and their proposed reaction pathways. a. Structures of (–)-lomaiviticin A (1), (–)-lomaiviticin C (2), (–)-kinamycin C (3), and the diazotetrahydrobenzo[b]fluorene (diazofluorene, 4) functional group. b. Cytotoxic species proposed to form from the 1-diazo-1H-indene-4,7-dione. Red denotes the 1-diazo-1H-indene-4,7-dione functional group of 1, 2, and 3. Blue denotes the hydrofulvene functional group of 2.
Substantial effort has been directed toward delineating the reactivity of the 1-diazo-1H-indene-4,7-dione within the diazofluorene (red in 1–3) and evaluating the relevance of these reactivities to cytotoxicity. Because (–)-lomaiviticin A (1) is not widely-available, these studies have employed the kinamycins or synthetic analogs. It has been proposed that redox cycling of the naphthoquinone to generate reactive oxygen species (ROS),14,15 formation of covalent adducts (5) by nucleophilic addition,16 production of vinyl radical intermediates (6),15–21 and addition to ortho-quinone methide (7)15,18–22 or acylfulvene (8)23,24 electrophiles may contribute to cytotoxicity (Figure 1B). In principle, all of these intermediates and pathways are accessible to 1, 2, and 3, and none sufficiently account for the superior cytotoxicity of 1. Moreover, the biological target and mechanisms underlying the activity of 1 have not been established. It was indicated in the isolation report that 1 cleaved DNA under reducing conditions,1 but no primary data were disclosed. The kinamycins weakly bind DNA,25 and have demonstrated DNA damaging activity,14,15,21 but a protein target has also been implicated in their effects.25,26
We report an analysis of the DNA damaging properties of (–)-lomaiviticin A (1) and parallel experiments to elucidate these properties with (–)-lomaiviticin C (2) and (–)-kinamycin C (3). Our data indicate that the cytotoxicity of 1 arises from induction of DNA dsbs, and that this mode of DNA damage is not efficiently recapitulated by 2 or 3. We present reactivity studies demonstrating that DNA cleavage by 1 proceeds through carbon-centered free radical intermediates and that the production of these intermediates is more facile for 1 than 2 or 3.
Results
Analysis of DNA damage by 1, 2, and 3 in vitro and in tissue culture
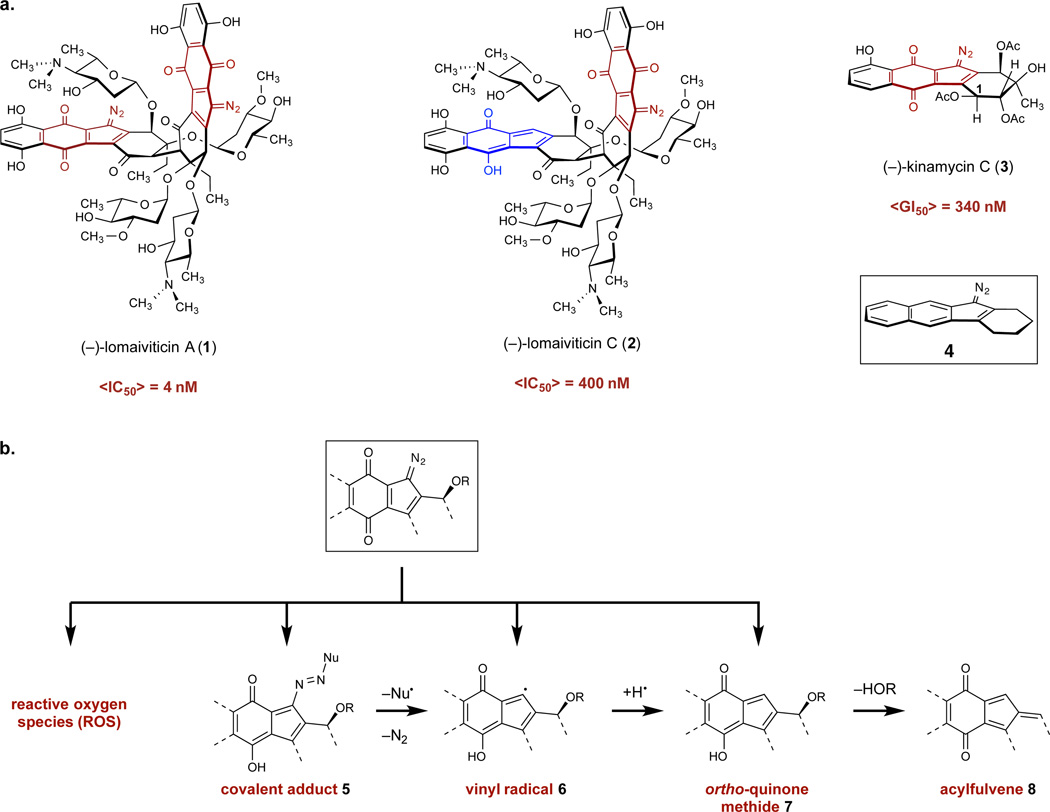
Our initial studies began with efforts to recapitulate the reported1 DNA cleavage activity of (–)-lomaiviticin A (1) in vitro. As the experimental conditions for this result (e.g., DNA substrate, concentration of 1, nature and concentration of reductant) were not disclosed, we elected to study the activity of 1 using plasmid pBR322 DNA in the presence or absence of the reducing agent dithiothreitol (DTT). Concentrations of 1 in the range of 0.5–2.0 µM were optimal for visualizing nicked (Form II) and linearized (Form III) DNA (Figure S1). Accordingly, DNA damage was assessed at 0.5 or 2.0 µM 1, with and without DTT (0 or 0.5 mM), and at varying pH (5.8, 7.4, 8.0, Figure 2A). Nicked and cleaved DNA increased with pH and addition of DTT. Form II DNA was observed on treatment with 1 alone, and large amounts of Form III DNA were observed when DTT was added. A survey of buffer conditions (Figure S2) revealed that DNA damage by 1 was slightly enhanced in tris buffer relative to the phosphate buffer used in the experiments shown in Figure 2. By comparison, (–)-lomaiviticin C (2) induced production of Form II DNA at ≥100 µM concentration in the presence or absence of DTT, but Form III DNA was not detected (Figure 2B). (–)-Kinamycin C (3) did not display DNA damaging activity at concentrations as high as 1 mM in the presence or absence of DTT (Figure 2C).
Figure 2.
Analysis of nicked and cleaved plasmid pBR322 DNA by (–)-lomaiviticin A (1), (–)-lomaiviticin C (2), and (–)-kinamycin C (3). a. Plasmid damage induced by 1, [1] = 0.5 or 2.0 µM, pH = 5.8, 7.4, or 8.0, [DTT] = 0 or 0.5 mM, 37 °C, 16 h. b. Plasmid damage induced by 2, [2] = 2–1000 µM, pH 7.4, [DTT] = 0 or 0.5 mM, 37 °C, 16 h. c. Plasmid damage induced by 3, [3] = 2–1000 µM, pH 7.4, [DTT] = 0 or 0.5 mM, 37 °C, 16 h. Top band: Form II (nicked) DNA, middle band: Form III (linearized) DNA, bottom band: Form I (supercoiled) DNA. d. Ratio of DNA dsbs () to ssbs () per DNA molecule in a plasmid cleavage assay using 1. Data points (left to right) correspond to 0.01, 0.05, 0.1, 0.2, 0.3, 0.4, 0.5, 0.6, 0.7 µM of 1. The dashed line represents the Freifelder–Trumbo relationship, defined as where h = 23.8 and L = 4361. Quantitative analyses and statistical treatments are presented in the Supporting Information.
Plasmid DNA damage by 1 was not measurably influenced by the hydroxyl radical scavengers ethanol or mannitol, the iron chelating agents diethylenetriaminepentaacetic acid or desferoxamine, or catalase, which mediates the decomposition of hydrogen peroxide (Figures S3, S4).27 Addition of superoxide dismutase, which catalyzes the disproportion of superoxide radical,27 did not measurably influence DNA cleavage by 1 in the presence of DTT.
A Freifelder–Trumbo analysis28–30 was applied to determine if the linearized DNA generated by (–)-lomaiviticin A (1) arises from an accumulation of unrelated ssbs, or from coupled strand cleavage events (Figure 2D). The fractions of Form I and III DNA remaining after treatment with 1 were quantified and the number of ssbs and dsbs per DNA molecule were calculated according to the equations fIII = n2 e−n2 and fI = e−(n1+n2) fIII and fI represent the fractions of Form III and Form I DNA, respectively, n1 represents the number of ssbs/molecule of DNA, and n2 represents the number of dsbs/molecule of DNA, and a Poisson distribution of DNA breaks is assumed. The first equation was solved for n2 by a seventh-order Taylor approximation. Over the concentration range 0.01–0.7 µM the ratio of ssbs to dsbs was constant (5.3±0.6:1) and was lower than that expected if dsbs were to arise from an accumulation of unrelated ssbs (hashed red line in Figure 2D). The latter relationship was obtained by employing a requirement of at least 23.8 base pairs28 between unrelated ssbs to prevent formation of a dsb and the 4361 base pairs-size of the plasmid.
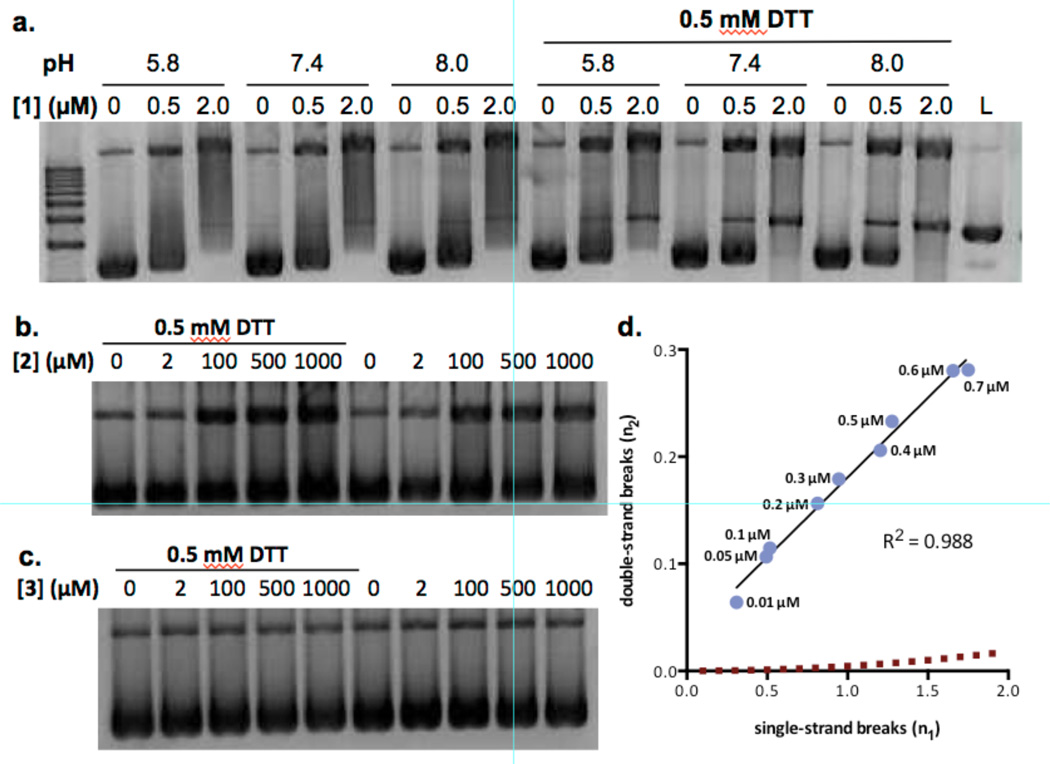
Production of phospho-SER139-H2AX (γH2AX)31,32 and translocation of p53 binding protein 1 (53BP1)33 are well-known markers of DNA dsbs. We detected the formation and colocalization of foci derived from γH2AX and 53BP1 in K562 cells treated with 0.05 or 0.5 nM (–)-lomaiviticin A (1) for 4 h (Figure 3). By comparison, 53BP1 and γH2AX foci were sparse or undetectable in cells treated with 300 nM of (–)-lomaiviticin C (2) or (–)-kinamycin C (3). We also observed foci formation and colocalization in HeLa cells treated with 1, establishing that the response is not cell line-specific (Figure S5).
Figure 3.
Immunofluorescence imaging of γH2AX and 53BP1 foci in K562 cells treated with (–)-lomaiviticin A (1), (–)-lomaiviticin C (2), or (–)-kinamycin C (3). γH2AX and 53BP1 are commonly used markers (refs. 31–33) for DNA dsbs. Immunofluorescence imaging shows that these foci are induced and colocalize in K562 cells treated with 1 (0.05–0.5 nM), but are sparse or undetectable in cells treated with 300 nM 2 or 3. Columns (left to right), 53BP1 (red), γH2AX (green), nucleus (blue), merge. Rows (top to bottom): control, 0.05 nM 1, 0.5 nM 1, 300 nM (–)-lomaiviticin C (2), 300 nM (–)-kinamycin C (3). K562 cells in exponential growth phase were incubated with 0.05 nM 1, 0.5 nM 1, 300 nM 2, or 300 nM 3 for 4 h. Immunological detection was performed using a primary antibody [rabbit polyclonal anti-53BP1 antibody (Novus Biologicals) and mouse monoclonal anti-phospho-histone H2AX (SER139) antibody (Upstate)] and visualized with Alexa 488 (goat-anti-mouse IgG) and Alexa 594 (goat-anti-rabbit IgG). Mounting medium contained DAPI to visualize nuclear DNA.
In order to quantify the γH2AX response, we conducted fluorescence-activated cell sorting analysis of K562 cells exposed to (–)-lomaiviticin A (1), (–)-lomaiviticin C (2), or (–)-kinamycin C (3) (312 nM of each). This experiment showed an increase in γH2AX by 1300% in cells treated with 1 (relative to cells treated with an anti-γH2AX antibody alone, Figure S6). γH2AX levels in cells treated with 2 or 3 were 11% and 28%, respectively, higher than control.
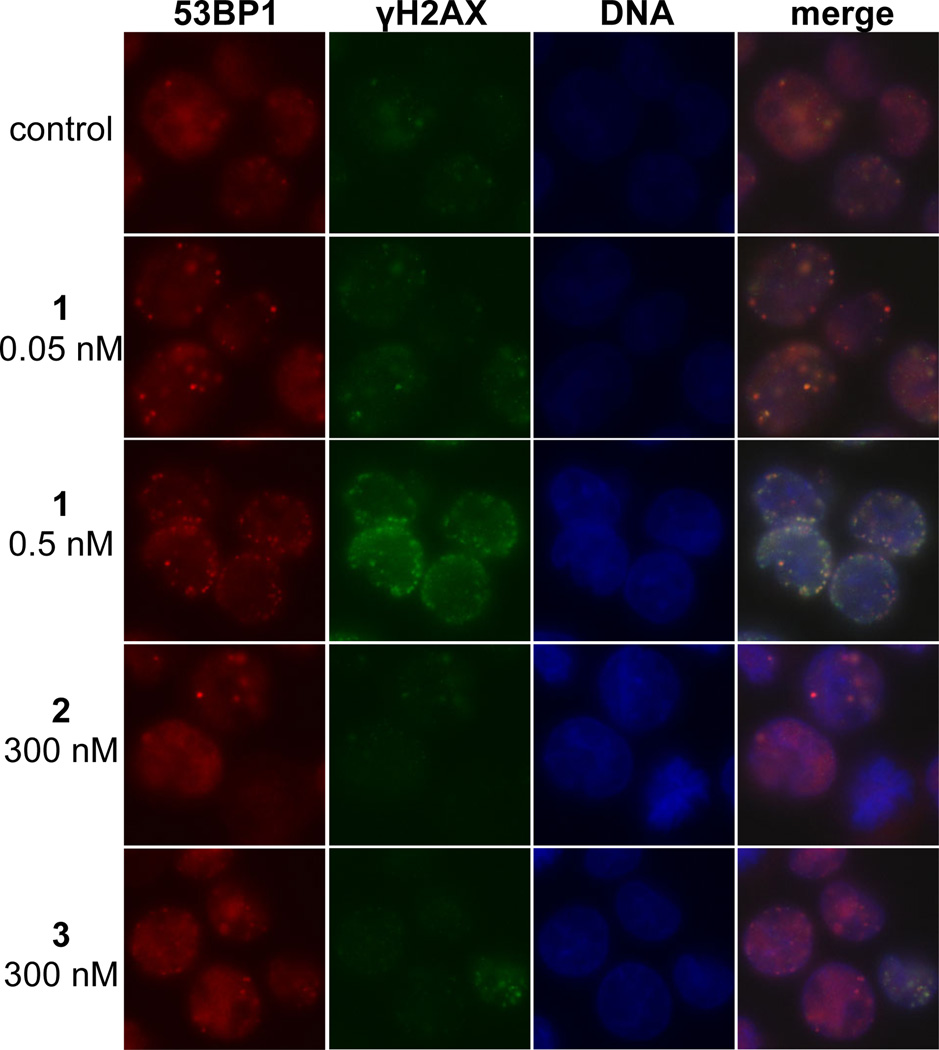
A neutral comet unwinding assay34 was employed as an independent method of dsb detection (Figure 4). K562 cells were incubated with (–)-lomaiviticin A (1, 0.5, 5, or 50 nM) for 30 min. The cells were fixed in agarose, lysed, placed in a neutral unwinding solution, and subjected to neutral electrophoresis. Visualization (SYBR Green) revealed that 1 induced production of DNA dsbs at the lowest concentration evaluated (0.5 nM). Both (–)-lomaiviticin C (2) and (–)-kinamycin C (3) displayed negligible DNA cleavage activity at 300 nM concentrations.
Figure 4.
Neutral comet unwinding assay of K562 cells treated with (–)-lomaiviticin A (1), (–)-lomaiviticin C (2), or (–)-kinamycin C (3). (–)-Lomaiviticin A (1) induces DNA dsb formation in K562 cells at 0.5–50 nM concentrations, while 2 and 3 induce minimal production of DNA double-strand breaks (dsbs) at 300 nM concentrations. Tail moment obtained in a neutral comet unwinding assay employing 1 (0.5, 5.0, or 50 nM), 2 (300 nM), or 3 (300 nM) and K562 cells. Drug exposure was 30 min. Bars represent mean tail moment (60–140 cells), error bars represent standard error of the mean. IR = ionizing radiation. Tail moment represents the extent of DNA cleavage and is defined as the product of the tail length and the fraction of DNA in the tail. The neutral comet assay is a method for the selective detection of DNA dsbs in tissue culture (see ref. 34).
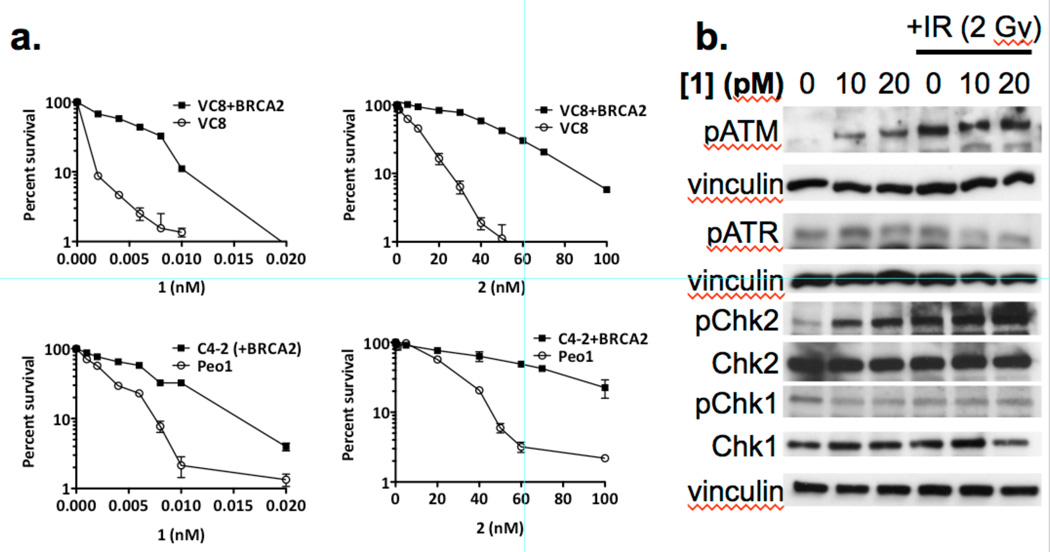
We conducted clonogenic survival assays using (–)-lomaiviticin A (1) and (–)-lomaiviticin C (2) in VC8 and Peo1 cells deficient in BRCA2 and isogenic lines transfected with and expressing the wild-type BRCA2 gene. We observed selective killing of the BRCA2-deficient cell lines for both 1 and 2, and 1 was over three orders of magnitude more potent than 2 (Figure 5A). Both BRCA2-deficient cell lines were remarkably sensitive to 1, with >98% cell killing at 10 pM 1. We detected upregulation of phospho-SER1981-ATM (pATM) and phospho-THR68-Chk2 (pChk2), but not phospho-SER428-ATR (pATR) or phospho-SER345-Chk1 (pChk1), by Western blot in MCF-7 cells treated with 1 (Figure 5B). We also detected formation of DNA dsbs in BRCA2-deficient C4-2 and Peo1 cells treated with 1 (0.2 nM) by the neutral comet unwinding assay (Figure S7, S8).
Figure 5.
Clonogenic survival curves and western blot analysis of cells treated with (–)-lomaiviticin A (1) or (–)-lomaiviticin C (2). a. Clonogenic survival curves for BRCA2-deficient VC8 and Peo1 cells and the corresponding isogenic cell lines transfected with and expressing a functional BRCA2 gene (VC8+BRCA2 and C4-2, respectively). Solid data points represent the BRCA2-proficient cell lines (VC8+BRCA2 and C4-2), unfilled data points represent the BRCA2-deficient cell lines (VC8 and Peo1). Left graphs: cells treated with 1. Right graphs: cells treated with 2. These data support DNA as the target of 1 and 2 as BRCA2 is involved in DNA repair and BRCA2-deficient cell lines are hypersensitive to DNA damaging agents (ref. 36). b. (–)-Lomaiviticin A (1) upregulates pATM and pChk2, but not pATR and pChk1, in MCF-7 cells. Western blot analysis of pATM, pATR, pChk2, Chk2, pChk1, and Chk1 in MCF-7 cells treated with 1 (10 or 20 pM). These data support production of DNA dsbs by 1 as pATM and pChk2 are involved in DNA dsb repair, while pATR and pChk1 respond primarily to replication fork stalling (ref. 37).
In vitro reactivity studies
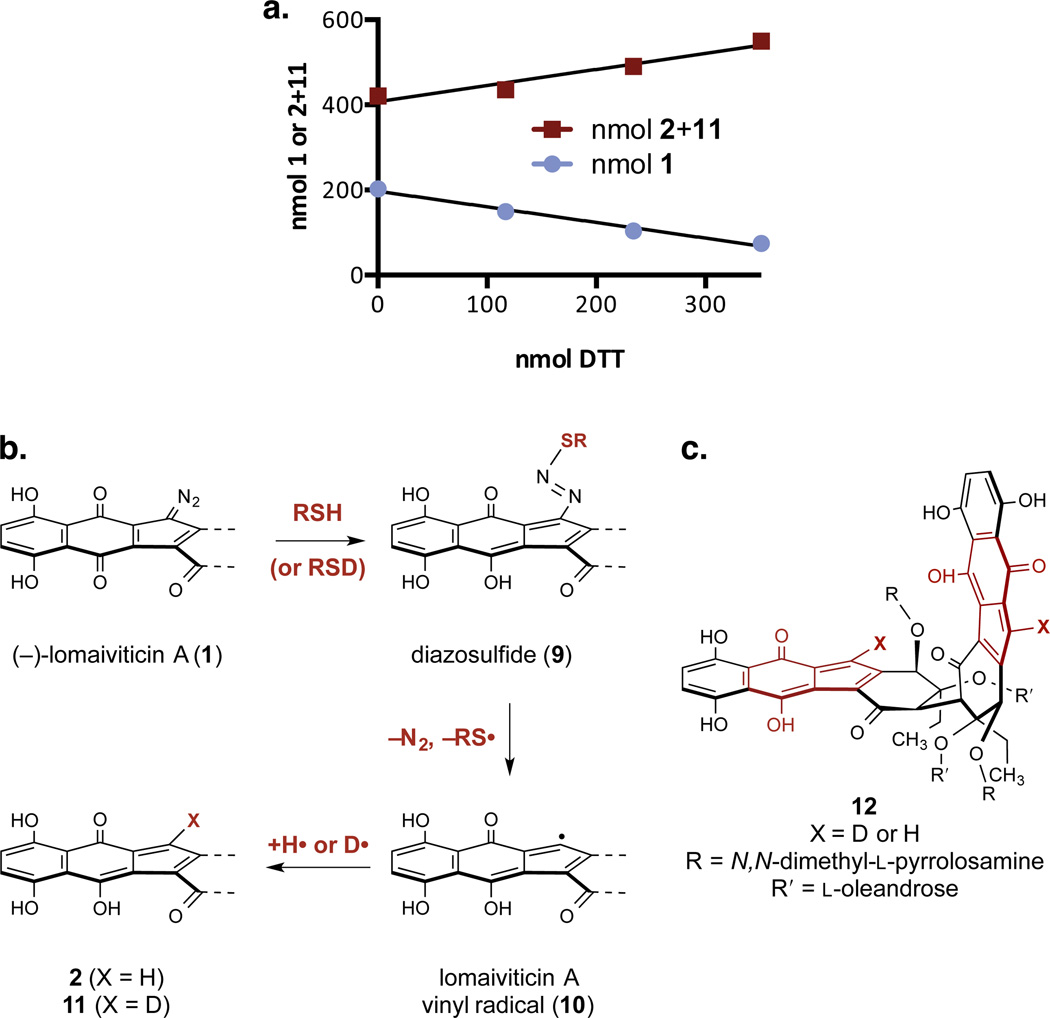
We have reported that synthetic monomeric diazofluorenes undergo hydrodediazotization on treatment with DTT in methanol, to form hydroxyfulvene products.24 Accordingly, (–)-lomaiviticin A (1) was anticipated to transform to (–)-lomaiviticin C (2) under reducing conditions. The relative rates of reduction of 1, the remaining diazofluorene of 2, and (–)-kinamycin C (3) were probed by competition experiments. A mixture of 1 (137 nmol) and 3 (125 nmol) in methanol-d4 was treated with DTT (260 nmol) and the resulting solution was analyzed by 1H NMR spectroscopy. This experiment revealed exclusive reduction of 1 to form 2, without detectable reduction of 3 (Figure S9). In a separate experiment, a mixture of 1 (202 nmol) and 2 (421 nmol) was treated with excess DTT (3 × 117 nmol) and monitored by 1H NMR spectroscopy. Under these conditions, the concentration of 1 decreased at a rate that correlated with the accumulation of 2, definitively establishing the conversion of 1 to 2 (Figure 6A). Additionally, we observed 56% deuterium atom incorporation at the vinylic position of 2 at the end of this experiment. To probe for deuterium incorporation at the vinylic position by ketone–enol tautomerization, a solution of natural 2 was warmed to 37 °C in 10% CD3OD–D2O for 4 d. Within the limits of detection, no deuterium exchange occurred at the vinylic position, establishing this site as kinetically-stable. Deuterium incorporation may be explained by a mechanism comprising nucleophilic addition of thiol to generate a diazosulfide (9), loss of dinitrogen and thiyl radical to form the vinyl radical 10, and deuterium atom abstraction (Figure 6B). The origins of incomplete deuteration are unclear but may be due to competitive hydrogen atom abstraction from residual S–H bonds in DTT.
Figure 6.
Relative reactivity studies and mechanistic pathways for reduction of the diazofluorene. a. Competition hydrodediazotization experiment between (–)-lomaiviticin A (1) and (–)-lomaiviticin C (2). 1/m = –0.01163, 0.01132 for 1, 2, respectively. Conditions: 1 (202 nmol), 2 (421 nmol), DTT (3 × 117 nmol), methanol-d4 (400 µL), 21 °C. b. Postulated pathway for the transformation of 1 to 2 and 11. c. Structure of the double hydrodediazotization product 12.
Additional experiments were conducted to further probe for the intermediacy of the vinyl radical 10, evaluate its ability to effect hydrogen atom abstraction from DNA, and test for reduction of both diazofluorenes in the presence of DNA. All exchangeable protons in calf thymus DNA and DTT were replaced with deuterium by repeated lyophilization from D2O.35 d4-DTT (1 equiv) was added to a degassed solution of calf thymus DNA and the free base of (–)-lomaiviticin A (1) in D2O, and the resulting mixture was incubated for 48 h at 37 °C. The ratio of d-(–)-lomaiviticin C (11) to (–)-lomaiviticin C (2) produced in this experiment was 1:2 (e.g., 67% hydrogen atom incorporation at the vinyl position, 1H NMR analysis), and the isotopologues 2 and 11 were isolated in 34% yield. The double hydrodediazotization product 12 (Figure 6B) was recovered in 37% yield, with 39% hydrogen atom incorporation at the vinylic positions. To remove variables arising from deuterium atom donation by DTT, the experiment was repeated without additional reductant. This experiment revealed 88% hydrogen atom incorporation at the vinyl position of the monohydrodediazotization products 11 and 2 (11:2 = 1:7, 81% recovery). We also isolated 18% of 12, containing 60% hydrogen atom incorporation at the vinylic position.
Discussion
(–)-Lomaiviticin A (1) is the most complex and cytotoxic diazofluorene-containing natural product. As the producing organism provides 1 in limited quantities (1 mg/L or less)1,2 and a total synthesis has not yet been realized, prior mechanism of action studies have employed the kinamycins or synthetic analogs.14–24 Our laboratory developed a semisynthetic route to 1 that facilitated the studies herein.2
It was noted1 that (–)-lomaiviticin A (1) cleaved DNA under reducing conditions, but no experimental parameters or primary data from this study were published. Our data show that 1 nicks and cleaves plasmid DNA at submicromolar concentrations. DNA damage by 1 is not mediated by iron, hydroxyl radical, superoxide, or hydrogen peroxide, and increases with pH. Nicking occurs in the absence of DTT, but substantial cleavage is only observed in its presence. These results were unanticipated because kinamycin-mediated DNA damage is favored at lower pH15 and is ROS-, metal-, and reductant-dependant.14,15,21 An earlier study demonstrated that (–)-kinamycin D (C-1-deacetyl-kinamycin C, see Figure 1A) cleaved and nicked pBR322 DNA at 1 mM concentration in the presence of 5.7 mM DTT,21 but (–)-kinamycin C (3) was inactive at concentrations as high as 1 mM in the presence of 0.5 mM DTT in our assay.
Our data demonstrate production of DNA dsbs in tissue culture by (–)-lomaiviticin A (1). We detected production and colocalization of γH2AX and 53BP1 foci in K562 and HeLa cells treated with ≤0.5 nM 1, but only marginal responses in cells treated with 300 nM (–)-lomaiviticin C (2) or (–)-kinamycin C (3). The neutral comet unwinding assay revealed DNA dsb formation in K562 cells treated with 0.5–50 nM 1. DNA cleavage by 0.5 nM 1 was greater than 300 nM of 2 or 3, and 5.0 nM 1 was comparable to 40 Gy of IR. In addition, BRCA2-deficient VC8 and Peo1 cells are hypersensitive to (–)-lomaiviticin A (1), and pATM/pChk2, but not pATR/pChk1, are upregulated in MCF-7 cells treated with 1. BRCA2 is involved in DNA dsb repair,36 ATM and Chk2 primarily respond to DNA dsbs, and ATR and Chk1 primarily respond to stalled replication forks.37 Thus, these data support a model involving production of DNA dsbs by 1, but not 2 or 3, in tissue culture at low nanomolar–picomolar concentrations. As dsbs are the most toxic of all DNA lesions,38 this model establishes an explanation for the increased cytotoxicity of 1.
The molecular mechanisms underlying reductive activation and DNA cleavage by (–)-lomaiviticin A (1) remain incompletely defined but our experiments provide some insights. Our plasmid cleavage assays support DNA-damage by ROS and metal-independent pathways, and our labeling experiments provide evidence for generation of vinyl radicals from both diazofluorenes of 1. Strand cleavage may initiate by hydrogen atom abstraction from the deoxyribose backbone, but further study is required to establish this. Vinyl radical intermediates were first proposed in 2005 by Feldman and Eastman, who studied the reduction of the synthetic monomeric diazofluorene dimethylprekinamycin (not shown) with tributyltin hydride,18,19 but the significance of this finding has not been supported by later studies of the kinamycins, which indicated DNA nicking is mediated by ROS and trace metals.14,15,21 Our competition experiments reveal that the first hydrodediazotization of 1 (to form 2) is faster than the second hydrodediazotization (to form 12) and is more facile than reduction of (–)-kinamycin C (3). The high mass recoveries from the hydrodediazotization experiments conducted in the presence of DNA suggest covalent adducts formed from ortho-quinone methide (7) or acylfulvene (8) intermediates are insignificant, if formed at all (Figure 1B). We speculate that nucleophilic addition to the diazo group (see structure 5, Figure 1B, and structure 9, Figure 6B) may trigger the formation of vinyl radical intermediates. Thiol addition to generate a diazosulfide (9, Figure 6B) finds precedent in the Stadler–Ziegler reaction, which involves addition of thiolates to arenediazonium ions,39,40 and is supported by an early model study, which demonstrated the addition of an aryloxide nucleophile to a diazofluorene.16 The addition of nucleotides to benzenediazonium ions, and the decomposition of these adducts to aryl radicals, has been reported.41,42 Thus, in the absence of thiol, DNA itself may behave as a nucleophile toward 1. The sequence of steps, e.g., protonation then addition, or addition followed by protonation, is not known, although the latter seems more likely given the electron-deficient nature of 1. The differing rates for the first and second hydrodediazotization, and the observation that Form III plasmid DNA is produced by 1 only in the presence of DTT, suggests DNA may be reactive toward the first diazofluorene of 1 but not toward the remaining diazofluorene of (–)-lomaiviticin C (2).
Many DNA-damaging natural products are activated by electron transfer or nucleophilic addition,43 and the enediyne antitumor agents, such as neocarzinostatin chromophore and the calicheamicins,44 provide the closest precedent for the behavior of (–)-lomaiviticin A (1). These agents undergo nucleophilic activation to generate biradical intermediates that nick and cleave dsDNA. However, the pathways leading to radical formation are distinct, and there is little doubt that the two vinyl radicals of 1 are generated stepwise. The ratio of DNA ssbs to dsbs produced by 1 (5.3±0.6:1) is below the value anticipated if dsbs arose from an accumulation of unrelated ssbs, suggesting strand cleavage occurs from a single binding event. Incidentally, this ratio is comparable to that observed for bleomycin (6:1), which mediates stepwise strand cleavage events (without dissociation),45 and is lower than that obtained for calicheamicin (2:1),46 in which strand cleavage occurs from a single reactive intermediate.44
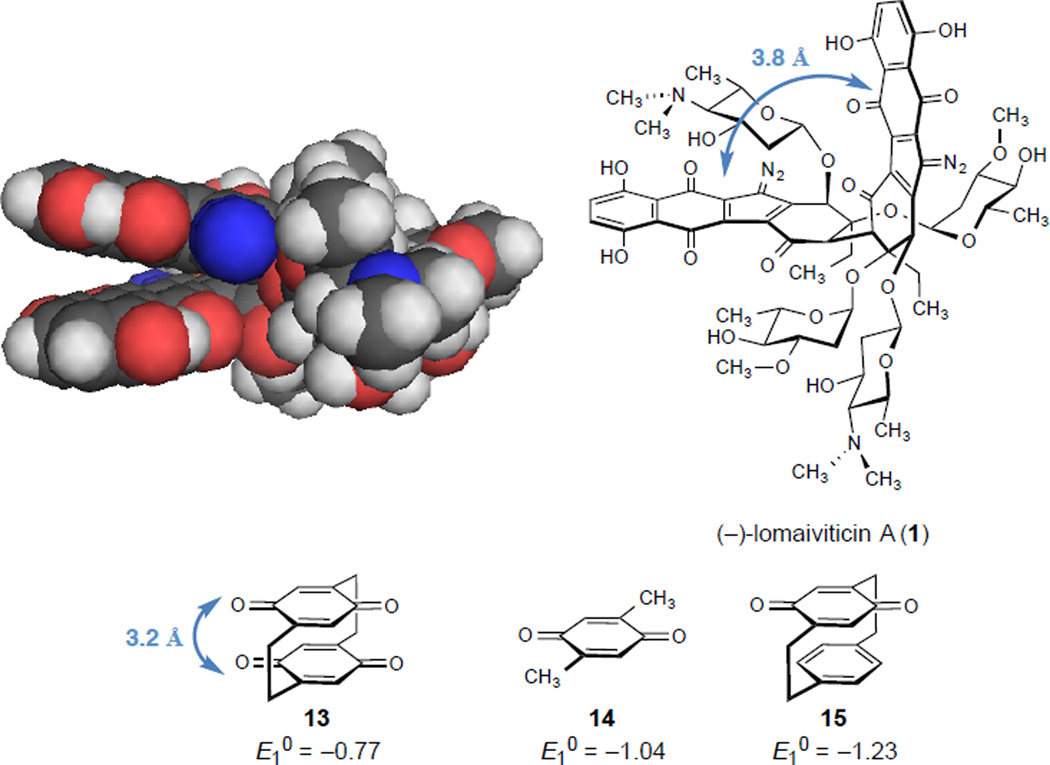
The enhanced reactivity of (–)-lomaiviticin A (1) toward reduction was unanticipated. Energy minimization of 1, conducted at the B3LYP 6-31G(d) level of theory in a water solvent continuum suggests that the diazofluorenes of 1 occupy a parallel conformation (Figure S10). The interdiazofluorene distance, as measured from the quinone carbonyl distal to the diazo and the nearest carbon atom of the opposing diazofluorene, is 3.8 Å. This arrangement minimizes nonbonded interactions between the carbohydrates of 1, which increase as the diazofluorene fragments are twisted (about the cojoining carbon–carbon bond) away from each other, and decreases the surface area exposed to water. We hypothesize that electronic delocalization between the arenes may increase electrophilicity. In support of this, the redox potential of pseudogeminal-[2.2]paracyclophane-4,7,12,15-tetrone, in which the two quinone fragments are fixed at a distance of ~3.2 Å,47 is 0.27 V more positive than 2,5-dimethylbenzoquinone,48 while the redox potential of [2.2]paracyclophane-4,7-dione, in which a benzene ring is positioned near the electrophore, is 0.19 V less positive then 2,5-dimethylbenzoquinone. The heightened reactivity of 1 vs. 3 may be due to a combination of this interaction and the additional ketone and hydroxyl49 substituents of 1.
In summary, we have shown that DNA cleavage is the primary mechanism of action of (–)-lomaiviticin A (1), provided evidence that vinyl radical species mediate this cleavage activity, and established that both diazofluorenes of 1 contribute to strand cleavage. This mode of DNA damage is not recapitulated by (–)-lomaiviticin C (2) or (–)-kinamycin C (3), and thereby accounts for the enhanced cytotoxicity of 1 relative to other diazofluorene-containing natural products. The work herein establishes a context to study the molecular mechanisms underlying reductive activation, hydrogen atom abstraction, and DNA cleavage by (–)-lomaiviticin A (1).
Methods
Cell Culture
The cells were maintained at 37 °C in a humidified atmosphere containing 5% CO2. K562 cells were cultured in RPMI 1640 medium supplemented with 10% fetal bovine serum (FBS), 1% L-glutamine, and 1% penicillin/streptomycin. HeLa cells were grown in DMEM supplemented with 10% FBS and 1% penicillin/streptomycin. MCF7 cells were grown in RPMI supplemented with L-glutamine and 10% FBS. PEO1 and PEO1 C4-2 ovarian cells and VC8 and VC8+BRCA2 Chinese hamster ovary (CHO) cells were grown in high-glucose DMEM supplemented with 10% FBS.
Plasmid cleavage assays
Reactions using supercoiled pBR322 plasmid DNA were performed in 20.0 µL of 10 mM sodium phosphate buffer/50 mM NaCl, pH 8.0, unless otherwise noted. The DNA concentration was 38 µM in DNA bp. In a typical assay, DTT (5 mM, 2.0 µL) was added to a mixture of 1 in DMSO (2.0 µL of solution in DMSO), water (final volume = 20.0 µL), buffer (100 mM sodium phosphate solution, pH 8.0, 2.0 µL), salt (500 mM NaCl, 2.0 µL), and pBR322 DNA (0.25 µg/µL in water, 2.0 µL), mixed in that order. The solution was thoroughly mixed by pipetting and incubated at 37 °C for 16 h. Solutions of 1 was prepared immediately before use. The final reaction contained 10% DMSO by volume.
Immunofluorescence
HeLa and K562 cells were grown on glass coverslips and poly-L-lysine coated glass coverslips, respectively. Following treatment, cells were washed with cold PBS, fixed with 4% paraformaldehyde at room temperature (15 min), permeabilized on ice (20 min) in 0.25% Triton X-100, and blocked with 3% bovine serum albumin in Tris-buffered saline (TBS) for at least 5 min at room temperature. Coverslips were incubated with primary mouse monoclonal anti-γH2AX (SER139) and rabbit polyclonal anti-53BP1 antibodies (1:100) for 1 h at 37 °C, washed with phosphate-buffered saline (PBS), and incubated with secondary Alexa 488 and Alexa 594 conjugated anti-mouse (1:100) and anti-rabbit (1:200) antibodies, respectively, for 1 h at 37°C. The coverslips were washed with PBS and mounted on glass slides using Vectashield mounting medium containing DAPI to counterstain DNA. Foci were visualized with a Zeiss Axiovert 200 M epifluorescence microscope equipped with a 63×/1.40 Plan-Apochromat oil immersion objective. Images were captured with a Zeiss Axiocam Mrm camera and AxioVision software.
Comet assay
Following treatment, K562 cells were immediately placed on ice. Drug was removed and cells were rinsed and re-suspended (3 × 105 cells/mL) in cold PBS prior to analysis for DNA double-strand breaks using Trevigen’s neutral comet assay reagent kit. The DNA was stained with SYBR Green I and the resulting comets were analyzed using a Zeiss Axiovert 200 M epifluorescence microscope at 10× magnification and the CometScore software. At least 50 individual comet images were analyzed from each sample for tail moment.
Reactivity Experiments (Competition Hydrodediazotization Between 1 and 3)
A solution of (–)-lomaiviticin A (1) [nominally 2.2 µM, 100 µL, containing approximately 14% (–)-lomaiviticin C (2)] and (–)-kinamycin C (3) in methanol (nominally 2.0 µM, 100 µL) were combined in a J-Young NMR tube and concentrated to dryness. The residue obtained was dissolved in methanol-d4 (100 µL) and the resulting solution was concentrated to dryness. This procedure was repeated. The residue obtained was dissolved in methanol-d4 (400 µL). A solution of 1,3,5-trimethoxybenzene in methanol-d4 (20.0 mM, 20.0 µL, 403 nmol) was added and the resulting solution was analyzed by 1H NMR spectroscopy. The molar quantity of each species in solution was determined by integrating resolved resonances of 1, 2, and 3 [1: δ 6.25 (s, 2H); 2: δ 5.43 (s, 1H); 3: δ 5.72 (s, 1H)] against the aryl resonance of 1,3,5-trimethoxybenzene [δ 6.07 (s, 3H)]. This analysis revealed the presence of 137 nmol 1, 22.2 nmol 2, and 125 nmol 3 (Table S1). A solution of DTT in methanol-d4 (4.3 mM, 20.0 µL, 86.6 nmol) was then added, and the resulting mixture was analyzed by 1H NMR spectroscopy. This process was repeated twice.
Hydrodediazotization of 1 in the Presence of Calf Thymus DNA and Dithiothreitol
Calf thymus DNA in D2O [44.6 µL, 6.85 mM/base pairs, 116 equiv, azeotroped from D2O (3 × 500 µL)] was added to a solution of 1 (3.6 mg, 2.64 µmol, 1 equiv) in 10% N,N-dimethylformamide-d7–D2O (200 µL). Dithiothreitol in D2O (1 mM, 2.64 µL, 2.64 µmol, 1.00 equiv) was added, and the resulting mixture was stirred gently for 48 h at 37 °C. The mixture was purified by reverse-phase flash-column chromatography (eluting with 10% methanol–water initially, grading to 80% methanol–water, seven steps) to afford separately a mixture of 2 and 11 (1.2 mg, 34%) and the double hydrodediazotization product 12 (orange solid, 1.1 mg, 37%). 1H NMR analysis showed 67% proton incorporation at the vinylic position for 2 and 11, and 39% total hydrogen atom incorporation at the vinylic position of 12.
Supplementary Material
Figure 7.
Acknowledgments
Financial support from the National Institute of General Medical Sciences (R01GM090000, S.B.H.), the National Institute of Environmental Health Sciences (R01ES005775, P.M.G.), the National Cancer Institute (R01CA168733, P.M.G.), the National Science Foundation (Graduate Research Fellowship to C.M.W.), the Searle Scholars Program (S.B.H.), and Yale University (S.B.H.) is gratefully acknowledged. S.B.H. acknowledges early stage investigator awards from the David and Lucile Packard Foundation, the Alfred P. Sloan Foundation, the Camille and Henry Dreyfus Foundation, and the Research Corporation for Science Advancement. We thank Dr. Tullia Dymarz, John Shen, Dr. David Spiegel, and Tina Wang for assistance.
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature
Author Contributions: L.C.C. and C.M.W. designed and performed the plasmid cleavage, immunofluorescence, comet, and flow cytometry experiments. Z.L. and C.M.W. performed the in vitro reactivity studies. P.M.G. and D.A.H. designed, performed, and analyzed the clonogenic survival assays, pATM/pChk2/pATR/pChk1 western blots, and comet assay employing BRCA2-deficient cells. S.B.H. conceived and designed the study, analyzed the data, and composed the manuscript.
The authors declare no competing financial interests.
References
- 1.He H, et al. Lomaiviticins A and B, Potent Antitumor Antibiotics from Micromonospora lomaivitiensis. J. Am. Chem. Soc. 2001;123:5362–5363. doi: 10.1021/ja010129o. [DOI] [PubMed] [Google Scholar]
- 2.Woo CM, Beizer NE, Janso JE, Herzon SB. Isolation of Lomaiviticins C–E. Transformation of Lomaiviticin C to Lomaiviticin A, Complete Structure Elucidation of Lomaiviticin A, and Structure–Activity Analyses. J. Am. Chem. Soc. 2012;134:15285–15288. doi: 10.1021/ja3074984. [DOI] [PubMed] [Google Scholar]
- 3.Kersten RD, et al. Bioactivity-Guided Genome Mining Reveals the Lomaiviticin Biosynthetic Gene Cluster in Salinispora tropica. Chembiochem. 2013;14:955–962. doi: 10.1002/cbic.201300147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ito S, Matsuya T, Ōmura S, Otani M, Nakagawa A. A New Antibiotic, Kinamycin. J. Antibiot. 1970;23:315–317. doi: 10.7164/antibiotics.23.315. [DOI] [PubMed] [Google Scholar]
- 5.Hata T, Ōmura S, Iwai Y, Nakagawa A, Otani M. A New Antibiotic, Kinamycin: Fermentation, Isolation, Purification and Properties. J. Antibiot. 1971;24:353–359. doi: 10.7164/antibiotics.24.353. [DOI] [PubMed] [Google Scholar]
- 6.Ōmura S, et al. Structure of Kinamycin C, the Structural Relation Among Kinamycin A, B, C, and D. Chem. Pharm. Bull. 1971;19:2428–2430. [Google Scholar]
- 7.Furusaki A, et al. Crystal and Molecular Structure of Kinamycin C p-Bromobenzoate. Isr. J. Chem. 1972;10:173–187. [Google Scholar]
- 8.Ōmura S, Nakagawa A, Yamada H, Hata T, Furusaki A. Structures and Biological Properties of Kinamycin A, B, C, and D. Chem. Pharm. Bull. 1973;21:931–940. doi: 10.1248/cpb.21.931. [DOI] [PubMed] [Google Scholar]
- 9.Gould SJ. Biosynthesis of the Kinamycins. Chem. Rev. 1997;97:2499–2510. doi: 10.1021/cr9600215. [DOI] [PubMed] [Google Scholar]
- 10.Marco-Contelles J, Molina MT. Naturally Occurring Diazo Compounds: The Kinamycins. Curr. Org. Chem. 2003;7:1433–1442. [Google Scholar]
- 11.Arya DP. Diazo and Diazonium DNA Cleavage Agents: Studies on Model Systems and Natural Product Mechanisms of Action. Top. Heterocycl. Chem. 2006;2:129–152. [Google Scholar]
- 12.Nawrat CC, Moody CJ. Natural Products Containing a Diazo Group. Nat. Prod. Rep. 2011;28:1426–1444. doi: 10.1039/c1np00031d. [DOI] [PubMed] [Google Scholar]
- 13.Herzon SB, Woo CM. The Diazofluorene Antitumor Antibiotics: Structural Elucidation, Biosynthetic, Synthetic, and Chemical Biological Studies. Nat. Prod. Rep. 2012;29:87–118. doi: 10.1039/c1np00052g. [DOI] [PubMed] [Google Scholar]
- 14.O'Hara KA, et al. Mechanism of the Cytotoxicity of the Diazoparaquinone Antitumor Antibiotic Kinamycin F. Free Radic. Biol. Med. 2007;43:1132–1144. doi: 10.1016/j.freeradbiomed.2007.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heinecke CL, Melander C. Analysis of Kinamycin D-Mediated DNA Cleavage. Tetrahedron Lett. 2010;51:1455–1458. [Google Scholar]
- 16.Laufer RS, Dmitrienko GI. Diazo Group Electrophilicity in Kinamycins and Lomaiviticin A: Potential Insights into the Molecular Mechanism of Antibacterial and Antitumor Activity. J. Am. Chem. Soc. 2002;124:1854–1855. doi: 10.1021/ja0167809. [DOI] [PubMed] [Google Scholar]
- 17.Arya DP, Jebaratnam DJ. DNA Cleaving Ability of 9-Diazofluorenes and Diaryl Diazomethanes: Implications for the Mode of Action of the Kinamycin Antibiotics. J. Org. Chem. 1995;60:3268–3269. [Google Scholar]
- 18.Feldman KS, Eastman KJA. Proposal for the Mechanism-of-Action of Diazoparaquinone Natural Products. J. Am. Chem. Soc. 2005;127:15344–15345. doi: 10.1021/ja053880w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Feldman KS, Eastman KJ. Studies on the Mechanism of Action of Prekinamycin, a Member of the Diazoparaquinone Family of Natural Products: Evidence for Both sp2 Radical and Orthoquinonemethide Intermediates. J. Am. Chem. Soc. 2006;128:12562–12573. doi: 10.1021/ja0642616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zeng W, et al. Mimicking the Biological Activity of Diazobenzo[b]fluorene Natural Products with Electronically Tuned Diazofluorene Analogs. Bioorg. Med. Chem. Lett. 2006;16:5148–5151. doi: 10.1016/j.bmcl.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 21.Ballard TE, Melander C. Kinamycin-Mediated DNA Cleavage Under Biomimetic Conditions. Tetrahedron Lett. 2008;49:3157–3161. [Google Scholar]
- 22.Khdour O, Skibo EB. Quinone Methide Chemistry of Prekinamycins: 13C-Labeling, Spectral Global Fitting and in vitro Studies. Org. Biomol. Chem. 2009;7:2140–2154. doi: 10.1039/b903844b. [DOI] [PubMed] [Google Scholar]
- 23.Moore HW. Bioactivation as a Model for Drug Design Bioreductive Alkylation. Science. 1977;197:527–532. doi: 10.1126/science.877572. [DOI] [PubMed] [Google Scholar]
- 24.Mulcahy SP, Woo CM, Ding WD, Ellestad GA, Herzon SB. Characterization of a Reductively-Activated Elimination Pathway Relevant to the Biological Chemistry of the Kinamycins and Lomaiviticins. Chem. Sci. 2012;3:1070–1074. [Google Scholar]
- 25.Hasinoff BB, et al. Kinamycins A and C, Bacterial Metabolites That Contain An Unusual Diazo Group, As Potential New Anticancer Agents: Antiproliferative and Cell Cycle Effects. Anti-Cancer Drugs. 2006;17:825–837. doi: 10.1097/01.cad.0000224442.78211.27. [DOI] [PubMed] [Google Scholar]
- 26.O'Hara KA, Dmitrienko GI, Hasinoff BB. Kinamycin F Downregulates Cyclin D3 in Human Leukemia K562 Cells. Chem. Biol. Interact. 2010;184:396–402. doi: 10.1016/j.cbi.2010.01.013. [DOI] [PubMed] [Google Scholar]
- 27.Halliwell B, Gutteridge JMC. In: Methods Enzymol. Alexander N, Glazer Lester Packer, editors. Vol. Volume 186. Academic Press; 1990. pp. 1–85. [DOI] [PubMed] [Google Scholar]
- 28.Freifelder D, Trumbo B. Matching of Single-Strand Breaks to Form Double-Strand Breaks in DNA. Biopolymers. 1969;7:681–693. [Google Scholar]
- 29.Povirk LF, Wübker W, Köhnlein W, Hutchinson F. DNA Double-Strand Breaks and Alkali-Labile Bonds Produced by Bleomycin. Nucleic Acids Res. 1977;4:3573–3580. doi: 10.1093/nar/4.10.3573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Boger DL, Honda T, Menezes RF, Colletti SL. Total Synthesis of Bleomycin A2 and Related Agents. 3. Synthesis and Comparative Evaluation of Deglycobleomycin A2,Epideglycobleomycin A2, Deglycobleomycin A1, and Desacetamido-, Descarboxamido-, Desmethyl-, and Desimidazolyldeglycobleomycin A2. J. Am. Chem. Soc. 1994;116:5631–5646. [Google Scholar]
- 31.Banath JP, Olive PL. Expression of Phosphorylated Histone H2AX as a Surrogate of Cell Killing by Drugs That Create DNA Double-Strand Breaks. Canc. Res. 2003;63:4347–4350. [PubMed] [Google Scholar]
- 32.Muslimovic A, Ismail IH, Gao Y, Hammarsten O. An Optimized Method for Measurement of gamma-H2AX in Blood Mononuclear and Cultured Cells. Nat. Protoc. 2008;3:1187–1193. doi: 10.1038/nprot.2008.93. [DOI] [PubMed] [Google Scholar]
- 33.Schultz LB, Chehab NH, Malikzay A, Halazonetis TD. P53 Binding Protein 1 (53bp1) Is an Early Participant in the Cellular Response to DNA Double-Strand Breaks. J. Cell Biol. 2000;151:1381–1390. doi: 10.1083/jcb.151.7.1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Collins A. The Comet Assay for DNA Damage and Repair. Mol. Biotechnol. 2004;26:249–261. doi: 10.1385/MB:26:3:249. [DOI] [PubMed] [Google Scholar]
- 35.Zein N, McGahren WJ, Morton GO, Ashcroft J, Ellestad GA. Exclusive Abstraction of Nonexchangeable Protons from DNA by Calicheamicin γ 1I. J. Am. Chem. Soc. 1989;111:6888–6890. [Google Scholar]
- 36.Roy R, Chun J, Powell SN. BRCA1 and BRCA2: Different Roles in a Common Pathway of Genome Protection. Nat Rev Cancer. 2012;12:68–78. doi: 10.1038/nrc3181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Smith J, Mun Tho L, Xu N, A. Gillespie D. In: Advances in Cancer Research. Woude George F. Vande, George Klein., editors. Vol. Volume 108. Academic Press; 2010. pp. 73–112. [Google Scholar]
- 38.Schipler A, Iliakis G. DNA Double-Strand–Break Complexity Levels and Their Possible Contributions to the Probability for Error-Prone Processing and Repair Pathway Choice. Nucleic Acids Res. 2013;41:7589–7605. doi: 10.1093/nar/gkt556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stadler O. Zur Kenntniss der Merkaptane. Ber. Dtsch. Chem. Ges. 1884;17:2075–2081. [Google Scholar]
- 40.Ziegler JH. Ueber eine Methode zur Darstellung aromatischer Sulfide von bestimmter Constitution und das Thioxanthon. Ber. Dtsch. Chem. Ges. 1890;23:2469–2472. [Google Scholar]
- 41.Gannett PM, et al. C8-Arylguanine and C8-Aryladenine Formation in Calf Thymus DNA from Arenediazonium Ions. Chem. Res. Toxicol. 1999;12:297–304. doi: 10.1021/tx980179f. [DOI] [PubMed] [Google Scholar]
- 42.Hung MH, Stock LM. Reactions of Benzenediazonium Ions with Guanine and its Derivatives. J. Org. Chem. 1982;47:448–453. [Google Scholar]
- 43.Wolkenberg SE, Boger DL. Mechanisms of in Situ Activation for DNA-Targeting Antitumor Agents. Chem. Rev. 2002;102:2477–2496. doi: 10.1021/cr010046q. [DOI] [PubMed] [Google Scholar]
- 44.Borders DB, Doyle TW. Enediyne Antibiotics as Antitumor Agents. Marcel Dekker, Inc.; 1995. [Google Scholar]
- 45.Stubbe J, Kozarich JW, Wu W, Vanderwall DE. Bleomycins: A Structural Model for Specificity, Binding, and Double Strand Cleavage. Acc. Chem. Res. 1996;29:322–330. [Google Scholar]
- 46.Drak J, Iwasawa N, Danishefsky S, Crothers DM. The Carbohydrate Domain of Calicheamicin γ I1 Determines its Sequence Specificity for DNA Cleavage. Proc. Natl. Acad. Sci. U. S. A. 1991;88:7464–7468. doi: 10.1073/pnas.88.17.7464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Irngartinger H, Acker R-D, Rebafka W, Staab HA. [2.2](2,5)Benzochinonophane: Struktur und Photochemie. Angew. Chem. 1974;86:705–706. [Google Scholar]
- 48.Wartini AR, Valenzuela J, Staab HA, Neugebauer FA. [2.2]Paracyclophane-4,7,12,15-tetrone, [2.2](1,4)Naphthalenophane-4,7,14,17-tetrone, and 1,4,8,11-Pentacenetetrone Radical Anions – A Comparative ESR Study. Eur. J. Org. Chem. 1998;1998:221–227. [Google Scholar]
- 49.Gómez M, González FJ, González I. Intra and Intermolecular Hydrogen Bonding Effects in the Electrochemical Reduction of α-Phenolic-Naphthoquinones. J. Electroanal. Chem. 2005;578:193–202. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.