Abstract
Background:
Aberrant activation of Wnt/β-catenin has been implicated in various cancer-related processes, for example, proliferation or tumour cell survival. However, the exact mechanism by which β-catenin provides liver tumour cells with a selective advantage is still unclear. This study was aimed to analyse growth behaviour and survival of β-catenin-driven mouse liver tumours after β-catenin ablation.
Methods:
Transgenic mice with a controllable hepatocyte-specific knockout of Ctnnb1 (encoding β-catenin) were generated and liver tumours were induced by means of a N-nitrosodiethylamine/phenobarbital tumour initiation/promotion protocol, which leads to the outgrowth of hepatocellular tumours with activated β-catenin. Cre recombinase was activated and the effects of the knockout in the tumours were studied.
Results:
Activation of Cre recombinase led to the knockout of Ctnnb1 in a fraction of tumour cells, thus resulting in the formation of two different tumour cell subpopulations, with or without β-catenin. Comparative analysis of the two subpopulations revealed that cell proliferation was significantly decreased in Ctnnb1-deleted hepatoma cells, compared with the corresponding non-deleted cell population, whereas no increased rate of apoptosis after knockout of Ctnnb1 was observed.
Conclusions:
β-catenin-dependent signalling is an important regulator of hepatoma cell growth in mice, but not a crucial factor in the regulation of tumour survival.
Keywords: Wnt/β-catenin signalling, liver, hepatocellular tumour, Connexin 32, proliferation, Ctnnb1, human hepatocellular carcinomas
Up to 30% of human hepatocellular carcinomas and 80% of hepatoblastomas display aberrant activation of the Wnt/β-catenin signalling pathway (de La Coste et al, 1998; Lopez-Terrada et al, 2009; Schmidt et al, 2011). This is mainly caused by genetic alterations in the CTNNB1 gene, encoding β-catenin, an important transcription factor in canonical Wnt signalling. Mutations in CTNNB1 lead to amino acid exchanges near the N terminus of the β-catenin protein where important phosphorylation sites are located, which are essential for its proteasomal degradation. On accumulation in the cytoplasm, mutant β-catenin translocates into the nucleus to associate with T-cell factor/lymphoid enhancer factor proteins and to promote the expression of target genes (Lustig and Behrens, 2003).
In mouse liver, tumours harbouring a mutationally activated Wnt/β-catenin signalling pathway can be induced by an initiation/promotion regimen according to Moennikes et al (2000): 6-week-old mice are given a single dose of the liver carcinogen N-nitrosodiethylamine (DEN) followed by chronic treatment with phenobarbital (PB). Using this regimen, the tumour promoter PB selects for the outgrowth of Ctnnb1-mutated hepatoma cells (Ctnnb1 is the mouse orthologue to CTNNB1 in humans) and the frequency of liver tumours mutated in Ctnnb1 is about 80% (Aydinlik et al, 2001).
There is evidence that aberrant β-catenin-dependent signalling promotes cell proliferation and inhibits apoptosis, thus providing cancer cells with a selective advantage. Expression of an oncogenic form of β-catenin leads to hepatomegaly in transgenic mice due to increased hepatocyte proliferation (Cadoret et al, 2001). Furthermore, several known target genes of β-catenin are implicated in cell cycle progression, such as cyclin D1 and c-myc (He et al, 1998; Shtutman et al, 1999), or in the regulation of apoptosis (Zhang et al, 2001). Activation of β-catenin in different cell lines results in increased proliferation, while at the same time inhibition of apoptosis is observed (Orford et al, 1999; Shang et al, 2004). Aberrant Wnt signalling is sufficient to generate intestinal lesions and adenomas (Shibata et al, 1997; Romagnolo et al, 1999) and is also required for maintenance of colorectal tumour xenografts in mice (Scholer-Dahirel et al, 2011). In liver, β-catenin-positive cell islets, which have evaded the hepatocyte-specific knockout of Ctnnb1 in respective transgenic mice, show a higher proliferative index in presence of PB and in livers with a pre-cirrhotic phenotype, as compared with surrounding hepatocytes lacking β-catenin (Braeuning et al, 2010). This suggests that, at least under certain conditions, β-catenin provides a growth advantage to hepatocytes, thereby contributing to liver carcinogenesis. Respective cell islets also exhibit reduced levels of the gap junction-forming protein connexin 32 (Cx32), similar to what had been observed earlier in Ctnnb1-mutated liver tumours (Moennikes et al, 2000; Marx-Stoelting et al, 2008) Thus, inhibition of cell–cell communication might be involved in the regulation of hepatoma cell proliferation by β-catenin.
In a study conducted by Malanchi et al (2008), transgenic mice (referred to as K14-creERT2:β-cateninlox/lox mice) were used to investigate the role of β-catenin in established chemically induced skin tumours with increased Wnt/β-catenin signalling. Expression of Cre recombinase under the control of the keratin 14 (K14) locus resulted in the deletion of Ctnnb1 in epidermal cells and subsequent complete tumour regression within 5–6 weeks, clearly indicating the important role of the signalling protein for tumour cell survival in this model.
Despite the impressive results from the above-mentioned experiment, the relevance of β-catenin activation for proliferation and survival of tumour cells in liver is not well understood. We have therefore now performed an experiment similar to that of Malanchi et al (2008), using a genetically modified mouse line that enabled us to study the effect of a tamoxifen-induced, hepatocyte-specific knockout of Ctnnb1 during chemical hepatocarcinogenesis. The results of our study show that Ctnnb1 ablation negatively affects liver tumour cell proliferation but has no significant influence on their survival.
Materials and methods
Animal breeding
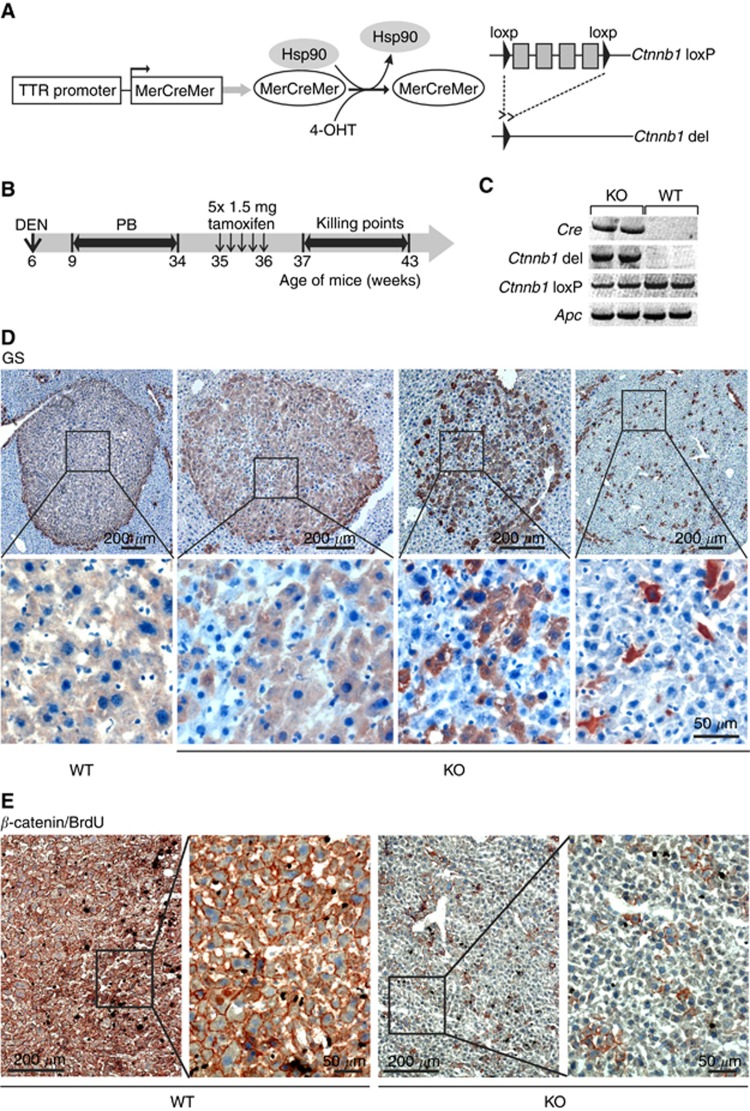
Transgenic Ctnnb1loxP/loxP mice carrying loxP sites flanking the exons 3 and 6 in the Ctnnb1 gene (Huelsken et al, 2001) were interbred with TTR-Cre-Tam mice expressing a tamoxifen-inducible modified Cre recombinase (MerCreMer) under the control of the hepatocyte-specific transthyretin promoter (Tannour-Louet et al, 2002; Anson et al, 2012) to obtain Ctnnb1loxP/loxP, TTR-Cre-Tam mice (Figure 1A). Genotyping of mice for Ctnnb1loxP/loxP and Cre was performed by standard PCR as recently described (Braeuning et al, 2009). Mice carrying Ctnnb1loxP/loxP and one Cre allele are referred to as ‘Ctnnb1 KO mice' in the following text, the respective Cre-null mice are called ‘Ctnnb1 WT mice', as they are phenotypically normal. Mice were kept on a 12-h dark/light cycle and had access to food and tap water ad libitum. Animals received humane care and protocols complied with institutional guidelines.
Figure 1.
Deletion of β-catenin in β-catenin-activated mouse liver tumours. (A) Schematic representation of the Ctnnb1 gene in livers of transgenic mice. Expression of the modified Cre protein MerCreMer is under the control of the liver-specific transthyretin (TTR) promoter. In the presence of the tamoxifen metabolite 4-hydroxytamoxifen (4-OHT), Cre recombinase, which is flanked by modified ligand-binding domains of the mouse oestrogen receptor (Mer), is released from its chaperones (heat shock protein 90, Hsp90) and targets loxP sites flanking the exons 3 and 6 within the Ctnnb1 gene. Cre-mediated recombination results in a deleted allele encoding a non-functional β-catenin protein. (B) Schematic delineation of experimental setup: mice received a single intraperitoneal (i.p.) injection of N-DEN (90 μg g−1 body weight) at 6 weeks of age. Subsequently, mice were continuously fed a diet containing 0.05% PB to select for Ctnnb1-mutated hepatomas. After a treatment-free period of 1 week, deletion of the Ctnnb1 gene in KO mice was achieved by i.p. injection of 1.5 mg tamoxifen for 5 consecutive days. Animals were then killed at different time points after application. (C) PCR analysis of tumour DNA from two representative Ctnnb1 KO and WT mice. The recombined, deleted Ctnnb1 gene was detected exclusively in Cre-expressing KO mice after tamoxifen treatment. Non-recombined Ctnnb1loxP/loxP was detected in both Ctnnb1 KO and WT mice but with lower amounts in Ctnnb1 KO mice. The Apc gene was amplified as a reference gene. (D) Immunostaining for GS, a marker for β-catenin activation. Ctnnb1-mutated tumours from WT animals show uniform expression of GS (left images). After KO of Ctnnb1, GS-negative hepatoma cells are observed within the tumours. The number of residual GS-positive cells varies between different tumours. (E) Tumours from Ctnnb1 WT and KO mice double stained for β-catenin and BrdU. β-catenin is present at the cell membrane of all tumour cells from WT mice, whereas only single cells possess β-catenin in tumour tissue from a respective KO animal.
Animal experiment
For the induction of liver tumours (Figure 1B), we followed a standard initiation/promotion protocol (Moennikes et al, 2000). Six-week-old male C3H Ctnnb1 WT (n=16) and KO (n=35) mice were given a single intraperitoneal injection of DEN (90 μg g−1 body weight; dissolved at 9 mg ml−1 in 0.9% NaCl solution, injected volume 10 μl g−1 body weight). After a treatment-free interval of 3 weeks, mice were kept on a diet containing 0.05% PB (Ssniff, Soest, Germany) for a time period of 25 weeks. After 1 week on a PB-free control diet, mice were then given five intraperitoneal injections of each 1.5 mg tamoxifen for 5 consecutive days. Tamoxifen (Sigma, Taufkirchen, Germany) was dissolved in ethanol (67 mg ml−1) and further diluted in corn oil to a final concentration of 10 mg ml−1 as previously described (Ganzenberg et al, 2013). Mice were killed 1–7 weeks after the last tamoxifen treatment. 5-bromodeoxyuridine (BrdU; AppliChem, Darmstadt, Germany) was dissolved in the drinking water (1 mg ml−1) and given orally to the animals for 72 h before killing. Killing was always between 0900 and 1100 h to avoid circadian influences. Livers were excised and the number and size of macroscopically visible tumours were recorded. Liver aliquots were frozen on dry ice and stored at −70 °C for further analyses or were fixed in 4% paraformaldehyde and embedded in paraffin for the preparation of TUNEL (TdT-mediated dUTP nick end labelling) stainings (see section below).
Immunohistochemical staining
Slices from frozen livers (10 μm thickness) were prepared and immunohistochemically stained as previously described (Braeuning et al, 2010), using antibodies against glutamine synthetase (GS; 1 : 1000, Sigma), β-catenin (1 : 50, Cell Signaling, Danvers, MA, USA) or BrdU (1 : 50, Dako, Glostrup, Denmark) in combination with horseradish peroxidase-conjugated secondary antibodies directed against rabbit (1 : 100, Dako) or mouse (1 : 20, Sigma) immunoglobulins with 3-amino-9 ethylcarbazole/H2O2 as substrates. For double staining of GS and Cx32, GS was visualised by the use of β-galactosidase-conjugated secondary antibodies (1 : 50, American Qualex, San Clemente, CA, USA) in combination with an antibody against Cx32 (1 : 250, Invitrogen/Zymed, Darmstadt, Germany) and horseradish peroxidase-conjugated secondary antibodies directed against rabbit (1 : 100, Dako) immunoglobulins with 3-amino-9-ethylcarbazole/H2O2 as substrates.
Tumour analyses
An Axio Imager light microscope (Imager.M1, Zeiss, Göttingen, Germany) with Axiovision Rel. 4.5 software (Zeiss) was used for acquisition of photographs and for further analysis of tumours. Counting of GS- and BrdU-positive cells within tumour subpopulations, grading of Cx32 levels and determination of tumour area fractions, which are equivalent to the tumour volume fractions, were performed using images of stained liver sections.
PCR analyses
Liver slices were stained for GS and respective tumour areas were punched out using a sharpened cannula. The tissue samples were digested with proteinase K and DNA was amplified by standard PCR methods. Tamoxifen treatment-induced Cre-mediated recombination was verified by screening for deletion of the loxP-flanked parts within the Ctnnb1 gene of four representative tumours (Figure 1C; for further details see Huelsken et al (2001)). Hot spot mutations in exon 3 of the Ctnnb1 gene in GS-positive tumours were detected by standard sequencing (Braeuning et al, 2010).
TUNEL assay
Apoptotic cell death was determined by TUNEL staining using the In Situ Cell Death Detection Kit, POD (Roche, Mannheim, Germany) according to the manufacturer's instructions for paraffin-embedded tissue sections. To induce DNA strand breaks in positive controls, sections were incubated with benzonase nuclease (Sigma) before labelling procedures.
Statistical analyses
The percentages of BrdU-labelled tumour cells were determined for the GS-negative and -positive tumour cell subpopulations within each tumour and the paired Student's T-test was used for statistical analyses. Differences were considered significant when P<0.05. Statistical significance is indicated by asterisks (*P<0.05; ***P<0.001).
Results
Tamoxifen-induced recombination of Ctnnb1 in transgenic mice
Following the induction of Cre recombinase by tamoxifen according to Ganzenberg et al (2013), PCR analyses of tumour tissue samples demonstrated Ctnnb1 deletion exclusively in the Cre-positive mice. Accordingly, the levels of non-recombined floxed Ctnnb1 were reduced in these mice (Figure 1C). Residual floxed Ctnnb1 in tumour cells from KO animals may derive from the non-parenchymal cells not expressing Cre, or from incomplete recombination in the hepatoma cells. Hepatic tumour burden (measured as the tumour volume fraction) at the time point of tamoxifen injection was ∼3%, as can be estimated from the observed tumour volume determined 1 week later at the time of killing of the first study group (compare Figure 2B).
Figure 2.
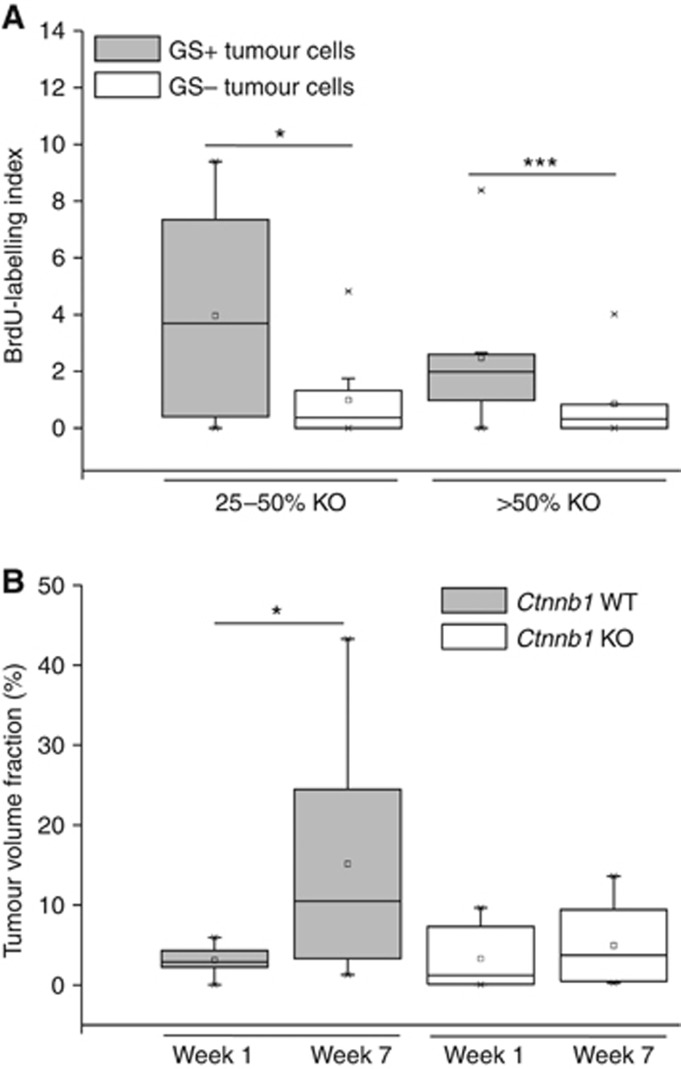
β-catenin deletion diminishes hepatoma cell proliferation. (A) BrdU-labelling index of GS-positive and -negative subpopulations in tumours of Ctnnb1 KO mice after tamoxifen administration. The labelling index is expressed as the percentage of BrdU-positive nuclei in a whole tumour section. On average, 759 nuclei per tumour were counted. Tumours double stained for BrdU and GS are stratified into two groups according to their level of Ctnnb1 knockout as determined by the percentage of GS-negative tumour cells (25–50% KO, n=9; >50% KO, n=13). GS-positive tumour subpopulations show a higher proliferation index compared with the GS-negative ones within the same tumour. (B) Tumour volume fraction from Ctnnb1 KO and WT mice 1 and 7 weeks after tamoxifen application. Livers from WT mice show an increase in tumour burden over time, whereas the tumour volume fraction in livers from Ctnnb1 KO mice is not significantly altered during the 6 weeks' time period. Group sizes: n=8 (WT groups; Ctnnb1 KO 7 weeks); and n=7 (Ctnnb1 KO 1 week). Statistical differences are indicated by one (*P<0.05) or three (***P<0.001) asterisks.
Tumour phenotype after tamoxifen-induced KO of Ctnnb1
Morphologically, the majority (∼90% in number and size) of liver tumours were eosinophilic, well-differentiated hepatocellular adenoma. Expression of the direct β-catenin target GS is one of the most reliable markers for β-catenin activation in murine liver. It is homogeneously expressed in chemically induced mouse liver tumours and there is an almost 100% concordance between activated β-catenin and GS expression (Loeppen et al, 2002; Chafey et al, 2009). Furthermore, GS is absent from Ctnnb1 KO hepatocytes (Braeuning et al, 2010). To confirm the correlation between high GS expression and activating mutations in the Ctnnb1 gene, tumour mutation analyses were performed. Twelve out of 13 analysed GS-positive tumours (92.3%) from Ctnnb1 WT (5 out of 5 tumours Ctnnb1 mutated) or KO (7 out of 8) animals were point mutated in the hot spot regions of the Ctnnb1 gene.
Immunostains of tumours from tamoxifen-treated Ctnnb1 WT mice revealed homogeneous GS expression throughout the tumours (Figure 1D, left image), indicative of active β-catenin-dependent signalling. Accordingly, high levels of the β-catenin protein were present in the respective GS-positive tumour cells (Figure 1E). In contrast, tumours from KO animals were composed of a mixture of GS-negative and GS-positive tumour cell subpopulations (Figure 1D, images 2–4). Of note, immunostaining for β-catenin was mostly membranous, as can be expected from chemically induced mouse liver tumours that contain rather low levels of nuclear β-catenin (Devereux et al, 1999; Aydinlik et al, 2001; Braeuning et al, 2007). The proportion of GS-negative and -positive cells was at great variance between individual tumours, and no clear-cut correlation between the time point of killing and the proportion of GS-negative hepatocytes was apparent. A comparable scattered pattern of expression was seen in tumours stained for β-catenin (Figure 1E). Obviously, the recombination of floxed Ctnnb1 alleles by Cre was incomplete, leading to a situation where one fraction of tumour cells is Ctnnb1 KO and therefore GS negative, whereas the other fraction of cells still possesses a non-recombined, mutationally activated Ctnnb1 allele that drives the expression of the marker protein GS. This heterogeneous phenotype allowed us to investigate two corresponding cell populations within one and the same tumour.
Effect of β-catenin ablation on tumour cell proliferation and apoptosis
For the analysis of tumour cell proliferation, the tumours were first categorised into two groups based on the efficiency of the β-catenin KO, evidenced by loss of the marker protein GS (25–50% or >50% β-catenin-/GS-negative cells, respectively). Tumour by tumour, the fractions of BrdU-incorporating cells were determined separately for both the GS-positive and the GS-negative hepatoma cell subpopulations. Very small tumours and tumours with a poor recombination rate (that is, <25%) were omitted from the analysis, as a too low number of cells in either of the two subpopulations would lead to inconclusive results. The results of this analysis clearly demonstrated that tumour cells negative for GS exhibit a lower proliferative index as compared with their GS-positive counterparts (Figure 2A). For comparison, GS-positive cells from Ctnnb1 WT tumours had a mean BrdU-labelling index of 10.3% (data not shown), significantly higher (P<0.05; Student's t-test) than the values obtained with the GS-positive sub-populations from partially Ctnnb1-ablated tumours (4.0% for 25–50% Ctnnb1 KO; 2.2% for >50% Ctnnb1 KO). BrdU labelling in surrounding normal tissue was always <1%, irrespective of the genotype. In addition, the tumour volume fraction of Ctnnb1 WT and KO mice 1 and 7 weeks after tamoxifen treatment was determined (Figure 2B). After 7 weeks, tumour burden in livers from Ctnnb1 WT mice was significantly increased, as compared with the tumour burden 6 weeks earlier. In contrast, Ctnnb1 KO mice showed no obvious alteration in tumour volume fraction over time.
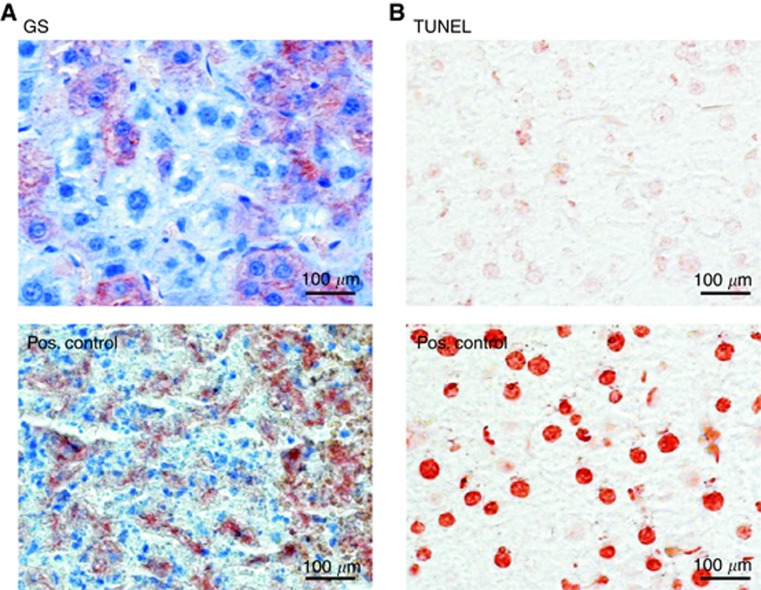
Furthermore, the impact of Ctnnb1 KO on tumour cell death was investigated 7 weeks after β-catenin ablation. At this time point, the tumour tissue showed intact hepatoma cell nuclei with no obvious signs of necrosis or inflammation. Apoptosis was also almost entirely absent from the tumours in our experiment. TUNEL-stained liver sections revealed no increase in apoptotic cell nuclei after tamoxifen-induced deletion of Ctnnb1. Figure 3 shows representative non-apoptotic tumour tissue with residual GS-positive cells from an animal killed 3 weeks after tamoxifen treatment.
Figure 3.
Absence of necrosis and inflammation from Ctnnb1 KO livers. (A) Intact GS-stained tumour tissue taken from a Ctnnb1 KO animal versus positive ‘Pos.' control showing tissue from a highly necrotic tumour infiltrated by immune cells and with remnants of former GS-positive hepatoma cells (sample taken from a previous study by Singh et al (2013)). (B) Tumour tissue from A stained by TUNEL technology reveals the absence of apoptotic cell nuclei after KO of Ctnnb1. Positive control was pre-treated with benzonase nuclease to generate free DNA ends. Liver was taken from a Cre-positive, Ctnnb1 KO animal 3 weeks after tamoxifen treatment.
Cx32 levels in different tumour subpopulations
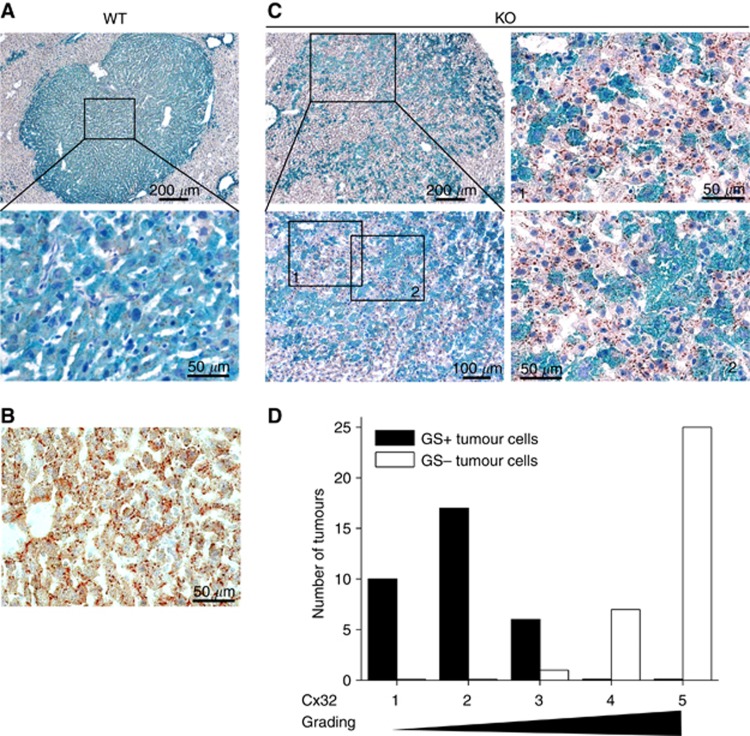
Immunohistochemical stainings were performed to analyse the localisation of Cx32, the major gap junction-forming protein in mouse liver, mediating gap junctional intercellular communication (GJIC) (Kumar and Gilula, 1996). Clearly reduced membranous Cx32 was found in GS-positive hepatomas from WT mice (Figure 4A), whereas hepatocytes from normal liver tissue exhibited physiological immunoreactivity for Cx32 in plaque-like structures at the cell membranes (Figure 4B). Interestingly, Cx32 appeared at cell membranes of GS-negative tumour subpopulations after KO of Ctnnb1 (Figure 4C and D).
Figure 4.
Appearance of Cx32 at membranes of Ctnnb1 KO tumour cells. (A) Reduced Cx32 levels are observable in the membranes of Ctnnb1-mutated, GS-positive tumour cells in livers from Ctnnb1 WT animals. (B) In contrast, normal liver tissue shows high membranous Cx32 levels. (C) After KO of Ctnnb1, Cx32 reappears in the GS-negative tumour cell subpopulations (image details are referred to as 1 and 2). The Ctnnb1 KO animal was killed 7 weeks after β-catenin ablation. (D) Cx32 grading of GS-positive and -negative tumour cells in hepatomas of KO mice revealed higher Cx32 levels in tumour populations lacking GS expression. Tissue slices were double stained for Cx32 and GS and analysed by light microscopy for the presence of Cx32 plaques at the cell membranes of hepatoma cells. For each tumour (in total, n=33 tumours were analysed), the degree of Cx32 expression in the two hepatoma subpopulations was classified from grade 1 (low Cx32 levels) to 5 (high Cx32 levels, comparable to surrounding normal tissue). The numbers of tumours in each class are given in the diagram for the GS-positive and GS-negative sub-populations, respectively.
Discussion
In the present work, we investigated the impact of a conditional KO of Ctnnb1 on the growth behaviour of mouse liver tumour cells with an activated Wnt/β-catenin signalling pathway.
In a recent study conducted by Ganzenberg et al (2013), tamoxifen-induced, Cre-mediated recombination of the Ctnnb1 gene led to β-catenin ablation in >99% of hepatocytes in livers from transgenic mice. Using the same mouse strain and treatment protocol, tamoxifen application was less effective in our experiment and the degree of Ctnnb1 KO varied considerably between different tumours. The reason for this phenomenon is not known. Transthyretin, the gene whose promoter was used to drive the expression of the Cre recombinase in our transgenic system, is not significantly altered in expression in Ctnnb1-mutated mouse liver tumours (Stahl et al, 2005) and hence altered expression of Cre is most likely not the underlying reason for incomplete recombination. Rather, we suspect that the different age of the mice at the time point of treatment might have had a role. Even more likely, pre-treatment with PB, an inducer of cytochrome P450 3A family enzymes involved in xenobiotic metabolism, might have contributed to the observed effect by inducing tamoxifen metabolism (Kivistö et al, 1998).
Nonetheless, although certainly of disadvantage for analyses in tissue homogenates, the coexistence of β-catenin-positive and -negative tumour cells allowed us a direct comparison of the two populations in individual tumours. Our results show that the proliferative index of GS-negative tumour cells was significantly lower than that of the neighbouring GS-positive tumour cells (Figure 2A). This suggests that β-catenin-dependent signalling has a role in regulating cell proliferation in these tumours. Malanchi et al (2008) showed that the survival of skin tumours crucially depends on the presence of active β-catenin: the KO of Ctnnb1 in murine skin tumours with aberrant β-catenin activation resulted in complete tumour regression within 6 weeks. By contrast, however, no indication for the death of hepatoma cells was observed in our analyses (Figure 3). Rather, the loss of β-catenin slowed down the proliferation of the tumour cells. Subsequently, the tumour volume fraction in livers from Ctnnb1 KO mice did not significantly change within a time period of 6 weeks (Figure 2B). This result points towards differences in the role of β-catenin-dependent signalling for the survival of tumour cells in liver and skin.
An important process involved in the regulation of cell proliferation is Cx32-mediated GJIC. Downregulation of GJIC often occurs during tumorigenesis and triggers proliferation of cancer cells (Chipman et al, 2003). Cx32, the major gap junction-forming protein in mouse liver, is absent from the membranes of Ctnnb1-mutated tumour cells (Moennikes et al, 2000; Marx-Stoelting et al, 2008). Our present results confirm that membranous Cx32 protein is decreased in GS-positive tumour cells (Figure 4A). After KO of Ctnnb1, Cx32 reappeared at the cell membrane of GS-negative subpopulations (Figure 4C). This indicates that β-catenin-dependent signalling has a role in Cx32-mediated GJIC. Furthermore, it has been reported that the inhibition of GJIC can make tumour cells independent from growth-restraining factors from neighbouring cells (Yamasaki and Naus, 1996; Chipman et al, 2003). This raises the possibility that there is a relationship between cell–cell communication and cell proliferation in the examined tumours. Reduced proliferation in the GS-negative hepatoma cell population could be a consequence of recurring Cx32-mediated GJIC. In tumours with more Cx32-positive cells, the remaining β-catenin-positive cells might receive stronger growth-restricting signals from surrounding cells, possibly leading to a stronger inhibition of cell proliferation.
In summary, the present results show that the ablation of β-catenin in mouse liver tumours leads to a significant attenuation of tumour cell proliferation but not to an induction of tumour cell death and hence not to tumour regression. Therefore, β-catenin-dependent signalling seems to be an important factor involved in the regulation of cell proliferation in this experimental system, but its loss is not sufficient to trigger cell death in those tumours.
In addition, GS-positive tumours from WT mice showed massive BrdU incorporation several weeks after the animals were set on a PB-free diet and the tumour volume fraction in respective livers significantly increased over time. This indicates that PB, as a proliferative stimulus, is only required at early stages of tumorigenesis but is no longer needed for maintenance of tumour growth in established Ctnnb1-mutated mouse liver tumours.
Acknowledgments
This work was supported by the Deutsche Forschungsgemeinschaft (grant SFB773). We acknowledge the excellent technical assistance by Johanna Mahr and Elke Zabinsky. We also thank Dr Sabine Colnot (Paris, France) who kindly provided the ApcloxP/loxP/TTR-Cre-Tam mice and Dr Joerg Huelsken (Lausanne, Switzerland) for the kind gift of Ctnnb1loxP/loxP mice.
The authors declare no conflict of interest.
Footnotes
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 3.0 Unported License.
References
- Anson M, Crain-Denoyelle AM, Baud V, Chereau F, Gougelet A, Terris B, Yamagoe S, Colnot S, Viguier M, Perret C, Couty JP. Oncogenic beta-catenin triggers an inflammatory response that determines the aggressiveness of hepatocellular carcinoma in mice. J Clin Invest. 2012;122:586–599. doi: 10.1172/JCI43937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aydinlik H, Nguyen TD, Moennikes O, Buchmann A, Schwarz M. Selective pressure during tumor promotion by phenobarbital leads to clonal outgrowth of beta-catenin-mutated mouse liver tumors. Oncogene. 2001;20:7812–7816. doi: 10.1038/sj.onc.1204982. [DOI] [PubMed] [Google Scholar]
- Braeuning A, Menzel M, Kleinschnitz EM, Harada N, Tamai Y, Köhle C, Buchmann A, Schwarz M. Serum components and activated Ha-ras antagonize expression of perivenous marker genes stimulated by beta-catenin signaling in mouse hepatocytes. FEBS J. 2007;274:4766–4777. doi: 10.1111/j.1742-4658.2007.06002.x. [DOI] [PubMed] [Google Scholar]
- Braeuning A, Sanna R, Huelsken J, Schwarz M. Inducibility of drug-metabolizing enzymes by xenobiotics in mice with liver-specific knockout of Ctnnb1. Drug Metab Dispos. 2009;37:1138–1145. doi: 10.1124/dmd.108.026179. [DOI] [PubMed] [Google Scholar]
- Braeuning A, Singh Y, Rignall B, Buchmann A, Hammad S, Othman A, von Recklinghausen I, Godoy P, Hoehme S, Drasdo D, Hengstler JG, Schwarz M. Phenotype and growth behavior of residual beta-catenin-positive hepatocytes in livers of beta-catenin-deficient mice. Histochem Cell Biol. 2010;134:469–481. doi: 10.1007/s00418-010-0747-1. [DOI] [PubMed] [Google Scholar]
- Cadoret A, Ovejero C, Saadi-Kheddouci S, Souil E, Fabre M, Romagnolo B, Kahn A, Perret C. Hepatomegaly in transgenic mice expressing an oncogenic form of beta-catenin. Cancer Res. 2001;61:3245–3249. [PubMed] [Google Scholar]
- Chafey P, Finzi L, Boisgard R, Cauzac M, Clary G, Broussard C, Pegorier JP, Guillonneau F, Mayeux P, Camoin L, Tavitian B, Colnot S, Perret C. Proteomic analysis of beta-catenin activation in mouse liver by DIGE analysis identifies glucose metabolism as a new target of the Wnt pathway. Proteomics. 2009;9:3889–3900. doi: 10.1002/pmic.200800609. [DOI] [PubMed] [Google Scholar]
- Chipman JK, Mally A, Edwards GO. Disruption of gap junctions in toxicity and carcinogenicity. Toxicol Sci. 2003;71:146–153. doi: 10.1093/toxsci/71.2.146. [DOI] [PubMed] [Google Scholar]
- de La Coste A, Romagnolo B, Billuart P, Renard CA, Buendia MA, Soubrane O, Fabre M, Chelly J, Beldjord C, Kahn A, Perret C. Somatic mutations of the beta-catenin gene are frequent in mouse and human hepatocellular carcinomas. Proc Natl Acad Sci USA. 1998;95:8847–8851. doi: 10.1073/pnas.95.15.8847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devereux TR, Anna CH, Foley JF, White CM, Sills RC, Barrett JC. Mutation of beta-catenin is an early event in chemically induced mouse hepatocellular carcinogenesis. Oncogene. 1999;18:4726–4733. doi: 10.1038/sj.onc.1202858. [DOI] [PubMed] [Google Scholar]
- Ganzenberg K, Singh Y, Braeuning A. The time point of beta-catenin knockout in hepatocytes determines their response to xenobiotic activation of the constitutive androstane receptor. Toxicology. 2013;308:113–121. doi: 10.1016/j.tox.2013.03.019. [DOI] [PubMed] [Google Scholar]
- He TC, Sparks AB, Rago C, Hermeking H, Zawel L, da Costa LT, Morin PJ, Vogelstein B, Kinzler KW. Identification of c-MYC as a target of the APC pathway. Science. 1998;281:1509–1512. doi: 10.1126/science.281.5382.1509. [DOI] [PubMed] [Google Scholar]
- Huelsken J, Vogel R, Erdmann B, Cotsarelis G, Birchmeier W. beta-Catenin controls hair follicle morphogenesis and stem cell differentiation in the skin. Cell. 2001;105:533–545. doi: 10.1016/s0092-8674(01)00336-1. [DOI] [PubMed] [Google Scholar]
- Kivistö KT, Villikka K, Nyman L, Anttila M, Neuvonen PJ. Tamoxifen and toremifene concentrations in plasma are greatly decreased by rifampin. Clin Pharmacol Ther. 1998;64:648–654. doi: 10.1016/S0009-9236(98)90055-8. [DOI] [PubMed] [Google Scholar]
- Kumar NM, Gilula NB. The gap junction communication channel. Cell. 1996;84:381–388. doi: 10.1016/s0092-8674(00)81282-9. [DOI] [PubMed] [Google Scholar]
- Loeppen S, Schneider D, Gaunitz F, Gebhardt R, Kurek R, Buchmann A, Schwarz M. Overexpression of glutamine synthetase is associated with beta-catenin-mutations in mouse liver tumors during promotion of hepatocarcinogenesis by phenobarbital. Cancer Res. 2002;62:5685–5688. [PubMed] [Google Scholar]
- Lopez-Terrada D, Gunaratne PH, Adesina AM, Pulliam J, Hoang DM, Nguyen Y, Mistretta TA, Margolin J, Finegold MJ. Histologic subtypes of hepatoblastoma are characterized by differential canonical Wnt and Notch pathway activation in DLK+ precursors. Hum Pathol. 2009;40:783–794. doi: 10.1016/j.humpath.2008.07.022. [DOI] [PubMed] [Google Scholar]
- Lustig B, Behrens J. The Wnt signaling pathway and its role in tumor development. J Cancer Res Clin Oncol. 2003;129:199–221. doi: 10.1007/s00432-003-0431-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malanchi I, Peinado H, Kassen D, Hussenet T, Metzger D, Chambon P, Huber M, Hohl D, Cano A, Birchmeier W, Huelsken J. Cutaneous cancer stem cell maintenance is dependent on beta-catenin signalling. Nature. 2008;452:650–653. doi: 10.1038/nature06835. [DOI] [PubMed] [Google Scholar]
- Marx-Stoelting P, Mahr J, Knorpp T, Schreiber S, Templin MF, Ott T, Buchmann A, Schwarz M. Tumor promotion in liver of mice with a conditional Cx26 knockout. Toxicol Sci. 2008;103:260–267. doi: 10.1093/toxsci/kfn043. [DOI] [PubMed] [Google Scholar]
- Moennikes O, Buchmann A, Romualdi A, Ott T, Werringloer J, Willecke K, Schwarz M. Lack of phenobarbital-mediated promotion of hepatocarcinogenesis in connexin32-null mice. Cancer Res. 2000;60:5087–5091. [PubMed] [Google Scholar]
- Orford K, Orford CC, Byers SW. Exogenous expression of beta-catenin regulates contact inhibition, anchorage-independent growth, anoikis, and radiation-induced cell cycle arrest. J Cell Biol. 1999;146:855–868. doi: 10.1083/jcb.146.4.855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romagnolo B, Berrebi D, Saadi-Keddoucci S, Porteu A, Pichard AL, Peuchmaur M, Vandewalle A, Kahn A, Perret C. Intestinal dysplasia and adenoma in transgenic mice after overexpression of an activated beta-catenin. Cancer Res. 1999;59:3875–3879. [PubMed] [Google Scholar]
- Schmidt A, Braeuning A, Ruck P, Seitz G, Armeanu-Ebinger S, Fuchs J, Warmann SW, Schwarz M. Differential expression of glutamine synthetase and cytochrome P450 isoforms in human hepatoblastoma. Toxicology. 2011;281:7–14. doi: 10.1016/j.tox.2011.01.006. [DOI] [PubMed] [Google Scholar]
- Scholer-Dahirel A, Schlabach MR, Loo A, Bagdasarian L, Meyer R, Guo R, Woolfenden S, Yu KK, Markovits J, Killary K, Sonkin D, Yao YM, Warmuth M, Sellers WR, Schlegel R, Stegmeier F, Mosher RE, McLaughlin ME. Maintenance of adenomatous polyposis coli (APC)-mutant colorectal cancer is dependent on Wnt/beta-catenin signaling. Proc Natl Acad Sci USA. 2011;108:17135–17140. doi: 10.1073/pnas.1104182108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang XZ, Zhu H, Lin K, Tu Z, Chen J, Nelson DR, Liu C. Stabilized beta-catenin promotes hepatocyte proliferation and inhibits TNFalpha-induced apoptosis. Lab Invest. 2004;84:332–341. doi: 10.1038/labinvest.3700043. [DOI] [PubMed] [Google Scholar]
- Shibata H, Toyama K, Shioya H, Ito M, Hirota M, Hasegawa S, Matsumoto H, Takano H, Akiyama T, Toyoshima K, Kanamaru R, Kanegae Y, Saito I, Nakamura Y, Shiba K, Noda T. Rapid colorectal adenoma formation initiated by conditional targeting of the Apc gene. Science. 1997;278:120–123. doi: 10.1126/science.278.5335.120. [DOI] [PubMed] [Google Scholar]
- Shtutman M, Zhurinsky J, Simcha I, Albanese C, D'Amico M, Pestell R, Ben-Ze'ev A. The cyclin D1 gene is a target of the beta-catenin/LEF-1 pathway. Proc Natl Acad Sci USA. 1999;96:5522–5527. doi: 10.1073/pnas.96.10.5522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh Y, Braeuning A, Schmid A, Pichler BJ, Schwarz M. Selective poisoning of Ctnnb1-mutated hepatoma cells in mouse liver tumors by a single application of acetaminophen. Arch Toxicol. 2013;87:1595–1607. doi: 10.1007/s00204-013-1030-8. [DOI] [PubMed] [Google Scholar]
- Stahl S, Ittrich C, Marx-Stoelting P, Kohle C, Altug-Teber O, Riess O, Bonin M, Jobst J, Kaiser S, Buchmann A, Schwarz M. Genotype-phenotype relationships in hepatocellular tumors from mice and man. Hepatology. 2005;42:353–361. doi: 10.1002/hep.20768. [DOI] [PubMed] [Google Scholar]
- Tannour-Louet M, Porteu A, Vaulont S, Kahn A, Vasseur-Cognet M. A tamoxifen-inducible chimeric Cre recombinase specifically effective in the fetal and adult mouse liver. Hepatology. 2002;35:1072–1081. doi: 10.1053/jhep.2002.33164. [DOI] [PubMed] [Google Scholar]
- Yamasaki H, Naus CC. Role of connexin genes in growth control. Carcinogenesis. 1996;17:1199–1213. doi: 10.1093/carcin/17.6.1199. [DOI] [PubMed] [Google Scholar]
- Zhang T, Otevrel T, Gao Z, Ehrlich SM, Fields JZ, Boman BM. Evidence that APC regulates survivin expression: a possible mechanism contributing to the stem cell origin of colon cancer. Cancer Res. 2001;61:8664–8667. [PubMed] [Google Scholar]