Abstract
Purpose
To describe the process and assess outcomes for the first 2 years of newborn screening for severe combined immunodeficiency (SCID NBS) in New York State (NYS).
Methods
The NYS algorithm utilizes a first-tier molecular screen for TRECs (T-cell receptor excision circles), the absence of which is indicative of increased risk of immunodeficiency.
Results
During the first 2 years, 485,912 infants were screened for SCID. Repeat specimens were requested from 561 premature and 746 non-premature infants with low or borderline TRECs. A total of 531 infants were referred for diagnostic evaluation leading to identification of 10 infants with SCID and 87 with a clinically significant non-SCID abnormality based on flow cytometry or CBC results (positive predictive value 20.3 %). Nine infants were diagnosed with typical SCID and one with leaky SCID. SCID diagnoses included two patients with adenosine deaminase deficiency, three patients with typical and one with leaky IL2RG-related SCID, one patient with IL7Rα-related SCID, and three cases of typical SCID, etiology unknown. TRECs were undetectable in eight of the nine babies with typical SCID. Infants with other non-SCID conditions included 27 patients with a syndrome that included T-cell impairment, 18 of which had DiGeorge syndrome. Seventeen infants had T-cell impairment secondary to another clinically significant condition, and 13 were classified as ‘other’. Among 30 infants classified as idiopathic T-cell lymphopenia, 11 have since resolved, and the remainder continues to be followed. One infant with undetectable TRECs had normal follow-up studies. Molecular studies revealed the presence of two changes in the infant’s DNA.
Conclusions
Overall, ten infants with SCID were identified during the first 2 years of screening in NYS, yielding an incidence of approximately 1 in 48,500 live births, which is consistent with the incidence observed by other states screening for SCID. The incidence of any clinically significant laboratory abnormality was approximately 1 in 5,000; both estimates are higher than estimates prior to the onset of newborn screening for SCID. Improvements to the NYS algorithm included the addition of a borderline category that reduced the proportion of infants referred for flow cytometric analysis, without decreasing sensitivity. We identified a large number of infants with abnormal TRECs and subsequent idiopathic T-cell lymphopenia. Long-term follow-up studies are needed to determine the prognosis and optimal treatment for this group of patients, some of whom may present with previously unrecognized, transient lymphopenia of infancy.
Keywords: Severe Combined Immunodeficiency, newborn screening, DiGeorge syndrome, idiopathic T-cell lymphopenia
Introduction
In January 2010, the Secretary’s Advisory Committee on Heritable Disorders in Newborns and Children (SACHDNC) voted unanimously to recommend the addition of severe combined immunodeficiency (SCID) as a core condition and related T-cell deficiency as a secondary condition to the Recommended Uniform Newborn Screening Panel (RUSP), based on the outcome of an evidence review. On May 21, 2010, the Secretary of Health and Human Services, Kathleen Sebelius added SCID to the RUSP [1]. SCID is a genetically heterogeneous disorder with a common finding of low or absent functional T-cells [1, 2]. Depending on the causative gene, B- and natural killer (NK)-cells may also be low or absent. The specific clinical course varies; however, untreated, essentially all infants with SCID will contract life-threatening bacterial, fungal or viral infections [3].Without treatment, the mortality rate is high and the majority of infants will expire before 1 year of age [3]. Treatment is dependent on the type of SCID and usually includes hematopoietic stem cell transplant (HSCT), enzyme replacement therapy, or gene therapy [1]. Newborn screening (NBS) allows for early diagnosis and treatment, which is associated with increased survival rates [4]. The long-term survival rate is at least 94 % when infants with SCID are treated with HSCT by 3.5 months of age [2, 4–6]. When HSCT is performed after 3.5 months of age, the survival rate is as low as 66 % [2, 7, 8].
Early diagnosis is best achieved through newborn screening because newborns with SCID are typically asymptomatic at birth and usually do not have a positive family history [8, 9]. In 2005, Chan and Puck described a method for SCID NBS using quantification of T-cell receptor excision circles (TRECs) and demonstrated its reliability in identification of infants with T-cell lymphopenia [10]. Wisconsin and Massachusetts initiated population-wide SCID screening using the TREC assay in 2008 and 2009, respectively [11–14]. The first infant with combined immunodeficiency (RAC2 mutation) identified via screening and requiring transplant was identified in Wisconsin in 2008 [14].
TRECs are a unique, DNA by-product formed during the normal process of T-cell maturation in the thymus. Low or absent TRECs in a dried blood spot (DBS) may be indicative of an underlying T-cell deficiency. Differential diagnoses include typical SCID, hypomorphic mutations leading to leaky SCID with or without an Omenn syndrome phenotype, complete DiGeorge anomaly, secondary causes of T-cell lymphopenia and idiopathic T-cell lymphopenia [15–17]. Non-pathological low TREC levels may be associated with prematurity [13].
In September 2010, New York became the fourth state to screen newborns for SCID using the TREC assay. The TREC assay is the first DNA-based first-tier newborn screening test performed in NYS. We describe our method for high-throughput, automated DNA extraction and TREC analysis to accommodate cost effective SCID screening in a high birthrate state [18]. In this report, the laboratory method and subsequent follow-up process are described in detail. These data build on the current knowledge base regarding screening and outcomes of NBS for SCID.
Methods
DNA Extraction
Eighty-seven DBS are punched into 96-well V-bottom plates (Axygen Scientific) along with one blank punch to monitor possible contamination during the extraction process. High quality DNA is extracted from a single 3-mm DBS using an automated lab-developed method on the Beckman Coulter NXp liquid handling system [18]. Briefly, an initial wash with molecular grade water is followed by a 10-min incubation in 100 µL red blood cell lysis buffer per sample and two additional 5 min water washes (80 µL/sample). All eluates are removed and discarded. Buffer A (50 µL) is added to each well and plates are incubated at 70 °C for 12 min. Buffer B (50 µL) is added to each Buffer A/DBS eluate and incubated at 99 °C for 12 min, to bring to a final extracted eluate volume of 100 µL. All extraction steps are performed on a shaking Peltier automated laboratory positioner (Thermo) unit on the NXp surface at 500 rpm at room temperature. Plates containing the DNA and the cleaned DBS are sealed by aluminum film (Axygen Scientific) and stored overnight at 4 °C prior to testing.
Calibration Curves
Concentrated plasmid containing the δRec-ψJα TREC (henceforth, SJ-TREC) sequence for the calibration curve was kindly provided by Dr. Anne Comeau from the New England Newborn Screening Program with permission of Dr. Daniel Douek [12]. Plasmid concentration was determined using a Nanodrop spectrophotometer. A 2× serial dilution of the stock plasmid was prepared in Tris-EDTA (TE) buffer to prepare an eight point calibration curve. TREC plasmid concentrations of 1,000, 500, 250, 125, 62.5, 31.2, 15.6, 7.8 copies per µL were empirically determined during assay validation to fall within the clinically relevant range. All calibration curve aliquots are stored at −80 °C.
Multiplex qPCR Reaction
Amultiplex quantitative real-time PCR assay (qPCR) that utilizes TaqMan® chemistry is used to detect TREC copy number and RNaseP levels in a single 10 µL reaction volume. The TREC primers amplify a 62-bp product that spans the signal joint of the SJ-TREC, with the TREC probe lying across the splice junction; therefore, only circularized DNA resulting from T-cell receptor rearrangement can be amplified. The reaction also includes primers/probe to co-amplify the RNaseP gene RPPH1, to provide real-time information about the extraction robustness and to monitor for contamination in the no template controls. The 10 µL qPCR reaction is set up in a 384-well optical plate (Applied Biosystems) and is comprised of the following components: 5 µL Environmental Master Mix (Applied Biosystems), 0.5 µL TaqMan RNaseP VIC Control Reagent (Applied Biosystems), 0.5 µL of a Custom TaqMan TREC FAM Assay (Applied Biosystems), 2 µL of molecular grade water and 2 µL of extracted DNA. The Custom TaqMan TREC FAMAssay consists of an SJ-TREC forward primer (5′ tgacacctctggtttttgtaaagg 3′), an SJTREC reverse primer (5′ tgcaggtgcctatgcatca 3′) and the SJTREC TaqMan MGB (minor groove binder) Probe (5′-FAMcccactcctgtgcacg- NFQ-3′). The assay quantifies the amount of FAM and VIC signal generated on an Applied Biosystems 7900HT Fast Real-Time PCR system with a 384-well block by using the following thermal profile: 1 cycle at 95 °C (10 min) followed by 45 cycles of 95 °C (15 s) and 60 °C (1 min). All DNA and reaction mixes were added to the 384-well plates using a Beckman Biomek NXp liquid handling system.
Data Collection and Analysis
Absolute qPCR data was collected using Applied Biosystems SequenceDetection System software version 2.3. Threshold and baseline (ΔRn) values were manually corrected for both targets for each run. Threshold and baseline values for TREC were 0.12 and 25 and 0.1 and 23 for RNaseP, respectively. Following the completion of each 384-well plate run, the calibration points constructing the curve were analyzed and up to 5 points out of 24 were allowed exclusion as long as one calibration point from the lowest concentration TREC copy calibrator was included. Calibration curves with slopes not between −3.0 and −3.6, an r2 <0.95 or a y-intercept of <36 or >43 were considered fails [19].
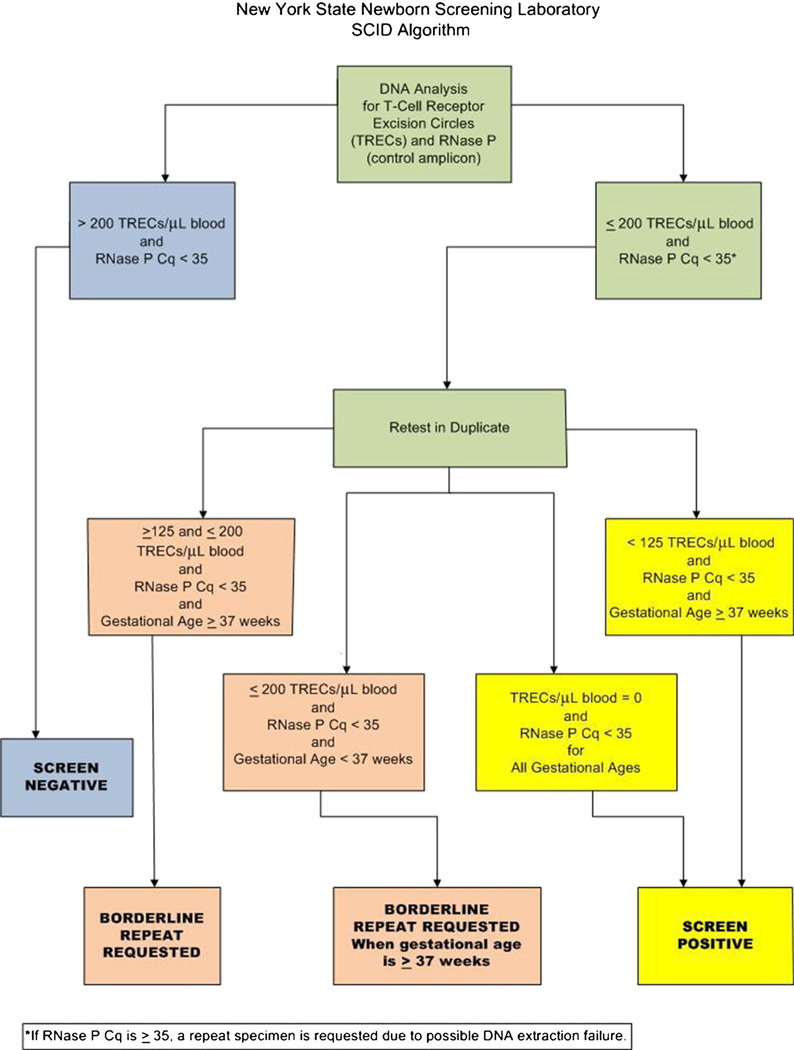
TREC copy number was estimated by linear regression analysis of individual TREC Cqs on the calibration curve. A comparison of TREC to RNaseP Cqs was used to validate specimen results by correlating DNA extraction quality (RNaseP) and TREC values. Samples with >200 TRECs and an RNaseP Cq value <35 were considered to be within acceptable limits (screen negative). During the validation process, it was empirically ascertained that an RNaseP Cq value of 35 equated to ~0.1 ng/ µL of extracted DNA. We determined that TRECs could not be reliably detected at DNA concentrations <0.1 ng/µL, therefore, samples with RNaseP Cq ≥35 were considered assay fails that were likely due to problematic DNA extractions. Samples with ≤200 TRECs and/or an RNaseP Cq value <35 were considered abnormal. Abnormal samples were retested in duplicate using a fresh DBS punch and a manual version of the same DNA extraction. The average of the three calculated TREC values determined whether further action was required (see Appendix).
Data Accrual and Analyses Leading to Changes in Reporting Algorithm
Originally samples were considered within acceptable limits if the average of the three retests was >200 or if two of the three retests had >200 TRECs. All samples with average TRECs ≤200 were considered abnormal and referred for follow-up diagnostic and clinical testing. A threshold of 200 TRECs was originally selected as the cutoff because it corresponded to approximately 10 % of the daily TREC mean in NYS and was two to three times higher than any TREC value determined in any sample from an infant with confirmed SCID tested during the assay validation process. A review of the data after the first 3.5 months of screening showed that TRECs were undetectable in the single infant confirmed with SCID. Infants with TREC levels 150–200 were found to have no evidence of immune deficiency, except for one infant with 22q11.2 deletion syndrome (Table V, Case Number 17). Therefore, In January 2011, a borderline category was added. In these instances, a repeat specimen was requested for infants with 150–200 TRECs. Non-premature infants with ≤200 TRECs on the repeat specimen were referred for a diagnostic evaluation. In July, 2011, the borderline category was expanded further to 125–200 TRECs.
Table V.
Patients with 22q11 deletion syndrome; N=18
| Case number |
Gender | TREC average on initial specimen (TRECs/µL) |
Molecular | Immunologic findingsa | Other features of DiGeorge syndrome (based on reports from Center Directors) |
|---|---|---|---|---|---|
| 11 | Female | 36 | 22q11.2 deletionb | Low CD3 (1,046), CD4 (791), CD8 (267), and CD19 (260) | Ventral septal defect (VSD) and interrupted aortic arch, history of hypocalcemia, seizures and unilateral kidney |
| 12 | Female | 96 | arr15q13.3(32,003,537-32,444,044x3,22q11.21 (18,644,790-21,041,014)x1)c | low CD3 (600), CD4 (474), CD8 (131), CD19(188) and CD16CD56 (106) | Unknown |
| 13 | Female | Undetectable | ish del (22)(q11.2q11.2)c | low CD3 (797), CD4 (590), CD8 (198), CD19 (231), CD16CD56 (140) | Patent ductus arteriosus, bicuspid aortic valve, criss-crossed pulmonary arteries, ventral septal defect, interrupted aortic arch, history of hypocalcemia, complete heart block w/temporary pacemaker placement |
| 14 | Male | 113 | 22q11.2 deletion | Original flow cytometry had slightly low CD3; CD3 (2,050), CD19 (734), CD16CD56 (621) CD4 (1,477), CD8 (547) | Diagnosed at 10 months when evaluated for feeding problems and developmental delay |
| 15 | Male | 170 | 22q11.2 deletion | CD3 (2,311), CD4 (1,782), CD8 (481), CD19 (942) | Recurrent candidal urinary tract infections, for which he takes prophylaxis. Failure to thrive, absent left kidney, right renal hydronephrosis |
| 16 | Female | 904 pre-thymectomy; 48 post-thymectomy | ish del (22)(q11.2q11.2) | Not applicable (thymectomy) | Cardiac disease, thymectomy |
| 17 | Female | 188 | Unknown | CD3 (2,189), CD4 (1,555), CD8 (547), CD19 (547), CD16CD56 (86) | Unknown |
| 18 | Female | 26 | 22q11.2 deletion | CD3 (1,134), CD4 (994), CD8 (146), CD19 (729), CD16CD56 (517) | Tetralogy of Fallot and absent PDA, s/p Blalock taussig Shunt and post balloon valvuloplasty |
| 19 | Female | Undetectable | 22q11.2 deletion | CD3 (809), CD4 (379), CD8 (236), CD19 (389), CD16CD56 (440) | Unknown |
| 20 | Female | Undetectable | Unknown | CD3 (1,347), CD4 (699), CD8 (675), CD19 (748), CD16CD56 not available | Congenital heart defect and hypocalcemia |
| 21 | Male | 43 | 22q11.2 deletion | CD4 lymphopenia (data not available) | Complex congenital heart defect, hypoplasia of parathyroid gland |
| 22 | Female | 54 | ish del (22)(q11.2q11.2) | Tcell lymphopenia with normal B and NK cells; CD3 (991), CD4 (722), CD8 (237)CD19 (1,780), CD16CD56 (821) | Bactrim prophylaxis until T cell function demonstrated by normal mitogen response, complex congenital heart defect, hypocalcemia with seizures |
| 23 | Male | 81 | Unknown | T cell lymphopenia, normal B cells, mild decrease of NK cells; CD3 (1,379), CD4 (824), CD8 (383), CD19 (975), CD16CD56 (875) | Complex congenital heart defect |
| 24 | Male | 104 | 22q11.2 deletion | Normal T cell numbers; CD3 (2,653), CD4 (1,629), CD8 (991); CD19 and CD16CD56 not done | VSD |
| 25 | Male | 46 | ish del(22)(q11.2)(q11.2) | Unknown; data not available | Truncus arteriosus, pulmonary artery stenosis branch |
| 26 | Male | 77 | ish del(22)(q11.2)(q11.2) | Mildly low CD3, CD4, CD8, normal LSM; CD3 (2,294), CD4 (1,498), CD8 (762), CD19 (1,264), CD16CD56 (734) | Moderate-sized VSD |
| 27 | Male | 51 | ish del(22)(q11.2)(q11.2) | CD3 (1,300), CD4 (1,018), CD8 (255), CD19 (2,017), CD16CD56 (105) | No cardiac defect, Remainder of medical history unknown |
| 28 | Female | 129 | ish del(22)(q11.2)(q11.2) | Adequate number of B, T and NK lymphocytes, flow cytometry data not available | Truncus arteriosus, AV canal with large VSD |
(Flow cytometry) is provided in cells/µL; reference values are not indicated since the flow analysis was performed in multiple testing laboratories
Denotes clinical diagnosis by Center Director
Denotes nomenclature provided from molecular cytogenetic report
Premature Infants
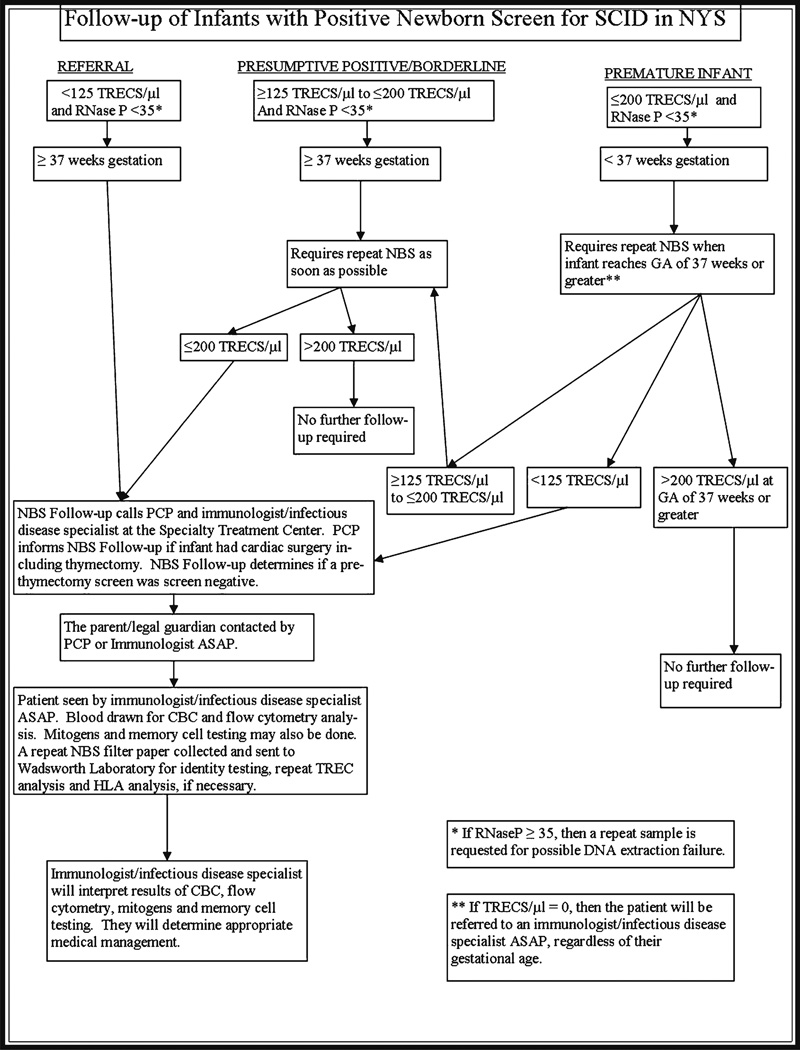
An exception in the algorithm was made for premature infants based on the experience of the Wisconsin and Massachusetts Programs of identifying lower TREC values in this population [6, 11]. For infants with ≤200 TRECs who were born prematurely (defined as <37 weeks gestation), a repeat specimen was requested at an age equivalent to at least 37 weeks gestation. However, if a repeat specimen was received prior to 37 weeks and >200 TRECs were present, the specimen was considered to be within acceptable limits and no further follow-up was requested. Infants with undetectable TRECs were referred for a diagnostic evaluation, regardless of gestational age (see Appendix).
Follow-Up Process
Infants with low TRECs, as defined above, were referred to an immunologist or infectious disease specialist for diagnostic evaluation at one of eight Specialty Care Centers in NYS. Follow-up clinical testing included complete blood count (CBC) and flow cytometry studies to assess the number of lymphocytes and T-cells (see Appendix). If clinically indicated, T-cell activation with mitogens, chromosome analysis and genetic testing were performed as appropriate. At the time of referral, a repeat newborn screening specimen was requested, to verify identity (i.e., that the sample with low TRECs came from the baby undergoing diagnostic evaluation) and to repeat the TREC analysis. The diagnosis was determined by the treating physician and categorized using the guidelines developed by the Newborn Screening Translational Research Network (NBSTRN) [20, 21]. The treating physician reported the outcome as no evidence of disease or a clinically significant laboratory abnormality defined as any finding on CBC or flow cytometry that required treatment or monitoring. For newborns without SCID, a birth defect or another syndrome, who required ongoing monitoring or treatment for a deficiency of T-cells, the consensus of the Center Directors was to use the diagnosis of idiopathic T-cell lymphopenia (ITCL). For infants with a SCID or leaky SCID diagnosis, a strict laboratory cut-off value was not used as a case definition. National case definitions had not been developed when SCID NBS started in NYS. The precedent for other disorders on the NY newborn screening panel is to rely on the clinical expertise of the specialists to diagnose and categorize cases. Therefore, a similar approach was used when SCID NBS was implemented.
Sequencing TREC Probe Region
DNA extracted (2 µL) as described previously [18] was used to sequence the TREC probe region of one infant with undetectable TRECs on multiple specimens but normal confirmatory studies. Primers up and downstream TREC amplicon were designed and a 329 bp PCR product was amplified using standard conditions (Primers: SJTREC_8F [5′ - GAA GAA GGC TCT GTC TAG TGT GA - 3] and SJTREC_7R [5′ - GCA ACT CGT GAG AAC GGT GA - 3′]). BigDye® Sanger sequencing was performed on an ABI 3730 DNA Analyzer.
Results
The average TREC counts and associated 95 % confidence intervals (CI) were estimated over a 3 month period using initial specimens from the overall newborn screening population stratified by gender and race/ethnicity. The mean TRECs/µL in the population was 1,832 (95 % CI=1,823–1,841). Gender and race/ethnicity distributions both differed between the screen negative and screen positive referral populations. Among infants referred, the male to female ratio was 1.72 (336:195; p<0.001; chi-square test). Male infants had fewer TRECs than females (average in males 1,700 [95 % CI=1,689–1,712] versus females 1,971 [95 % CI=1,958–1,984]). Race/ethnicity also differed between screen negative and screen positive infants (p<0.001; chi-square test), and appeared to be primarily driven by an increased frequency of Black infants in the screen positive referral population, who had lower TRECs than other race/ethnicities (Table I).
Table I.
TREC and referral distribution by race/ethnicity
| Race/ethnicity | Proportion screen negative infants |
Proportion screen positive infants |
N infantsa | Mean TREC copies/µl (95 % confidence interval)a |
|---|---|---|---|---|
| White | 48.4 % | 29.6 % | 31,315 | 1,885 (1,873–1,898) |
| Black | 16.4 % | 41.6 % | 10,227 | 1,649 (1,628–1,669) |
| Hispanic | 17.7 % | 14.7 % | 9,899 | 1,844 (1,821–1,866) |
| Asian | 8.0 % | 2.4 % | 5,606 | 1,861 (1,833–1,888) |
| Other | 9.5 % | 11.3 % | 6,512 | 1,819 (1,793–1,846) |
Includes screen negative (not referred) and screen positive infants tested over a 3 month period
Screening Outcomes
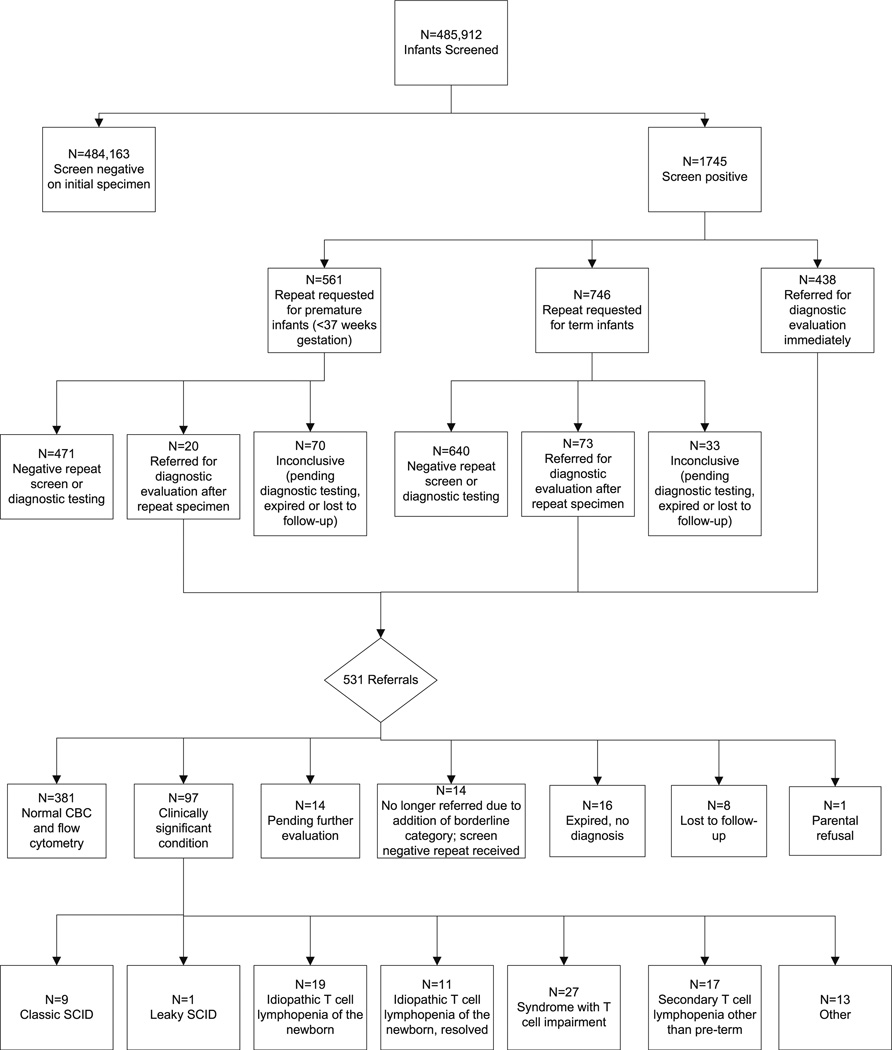
From September 29, 2010 to September 28, 2012, 485,912 infants were screened for severe T-cell defects by the New York State Newborn Screening Program. Overall, 99.6 % of infants screened negative, e.g. normal number of TRECs. During this 2 year period, 0.36 % (1 in 278) infants had an abnormal result and were referred for a diagnostic evaluation or a repeat specimen was requested (Table II; Fig. 1). Addition of the borderline category to the screening algorithm, where a repeat specimen was requested instead of immediate referral, reduced the overall referral rate from 0.2 to 0.1 %.
Table II.
Patients referred for a diagnostic evaluation; N=531
| Number of infants |
Percent (%) | Outcome |
|---|---|---|
| 381 | 71.8 | Referred to specialist; normal diagnostic evaluation |
| 97 | 18.3 | Referred to specialist; clinically significant condition |
| 14 | 2.6 | Referred to specialist; pending further evaluation |
| 14 | 2.6 | No longer referral due to addition of borderline category; screen negative repeat specimen received |
| 16 | 3.0 | Expired, no diagnosis |
| 8 | 1.5 | Lost to follow-up |
| 1 | 0.19 | Parental refusal |
Fig. 1.
The total number screen negative and screen positive are shown in boxes followed by the number of infants in each category (i.e. referral, borderline, premature). The final disposition of screen positive infants is also included
Overall, 97 infants with a clinically significant condition were identified via screening. Nine infants with typical SCID and one infant with leaky SCID were identified (Table III). Thirty infants were diagnosed with idiopathic T-cell lymphopenia (newborns without SCID, a birth defect or another syndrome, who required ongoing monitoring or treatment for a deficiency of T-cells). Twenty-seven infants were diagnosed with a non-SCID syndrome with T-cell impairment. Seventeen infants were diagnosed with secondary T-cell lymphopenia as a complication of a major birth defect or surgical thymectomy. Other laboratory abnormalities were identified in 13 infants. Absolute T-cell counts (CD3) were normal on flow cytometry; however, these infants exhibited other immune abnormalities including two with neutropenia, one with hypogammaglobulinemia, one with a low absolute lymphocyte count, one with selective IgA deficiency, seven with low CD19 and one with a low CD8 of unknown etiology.
Table III.
Patients with abnormal flow cytometry and/or CBC; N=97
| Number of infants |
Diagnosis |
|---|---|
| 9 | Typical SCID |
| 1 | Leaky SCID |
| 19 | Idiopathic T cell lymphopenia of the newborn |
| 11 | Idiopathic T cell lymphopenia of the newborn, resolved |
| 27 | Syndrome with T cell impairment |
| 17 | Secondary T cell lymphopenia other than pre-term |
| 13 | Other |
Overall Statistics 2010–2012
For typical and leaky SCID, the PPV was 2.1 % (10/478; 468 false positives) and the negative predictive value was 100 % (no known false negatives). Borderline and premature infants that resolved on a repeat newborn screen were excluded from this calculation. Pending, lost-to-follow-up and expired infants were also excluded. For typical and leaky SCID only, the PPV was 0.6 % before and 2.7 % after addition of the borderline category, respectively. The incidence of SCID in NYS during the first 2 years of screening was approximately 1 in 48,500.
The overall positive predictive value (PPV) using the current algorithm was 20.3 % (97/478; 381 were false positives) when all infants with a confirmed abnormal flow cytometry or other abnormal immunologic test result were considered. The PPV for all abnormal immunologic test results increased from 11.0 to 24.0 % after the addition of the borderline category. The incidence of any clinically significant immune disorder after a positive TREC screen was approximately 1 in 5,000. The classifications described below are based on the recently published CLSI guidelines [21].
Typical and Leaky SCID
Seventy percent of all SCID cases were male. Two were classified as White, two as Black, three as Hispanic, two as Asian and one as “Other” on the Guthrie card. The majority of cases (N=7) underwent HSCT and are doing well (Table IV). Themost common clinical phenotype was an absence of T-cells (T-B+NK+; N=5), including the one infant with leaky SCID. Three babies, two with ADA deficiency and one with common gamma chain deficiency, also had an absence of B- and natural killer cells (T-B−NK−). Two cases with IL2RG mutations had B cells but no NK cells (T-B+NK−). One case with typical SCID and multiple congenital anomalies underwent extensive genetic testing but the disease-causing gene was not identified; this patient expired. Eight of nine babies with typical SCID repeatedly had undetectable TRECs. One baby with typical SCID had a single assay with >100 TRECs, resulting in an average of 55. The baby with leaky SCID had an average of 65 TRECs and a single assay with >100 TRECs.
Table IV.
Patients with typical and leaky SCID; N=10a
| Case number |
Gender | TREC average on initial specimen (TRECs/µL) |
Genotype | Phenotypeb | Outcome and treatment |
|---|---|---|---|---|---|
| 1 | Female | Undetectable | IL7R (CD127-), c.353G>A, c.83-2A>T | T-B+NK+ Typical SCID CD3 (33), CD4 (22), CD8 (16), CD19 (866), CD16CD56 (409) |
Transplant at age 5 months, unrelated cord blood donor with (6 out 8 match) conditioning, patient currently doing well |
| 2 | Male | Undetectable | Etiology unknown; chromosome 3p13-p14.2 deletion (no known association with immunodeficiency) (arr 3p13p14.2 62465455-72819352x1); negative for ADA, AK2, CD3D, CD3E, CD3Z, DCLRE1C, IL2RG, IL7R, JAK3, LIG4, NHEJ1, PNP, PTPRC, RAC2, RAG1, RAG2, RMRP, ZAP70 mutations | T-B+NK+ CD3 (334), CD4 (108), CD8 (248), CD19 (378), CD16CD56 (882), CD45RA (<1), CD45RO (463), Absent antigen responses to Candida and tetanus toxoid, low to virtually absent mitogen responses |
Patient expired, MCA (cleft lip/palate, blepharophimosis, left iris coloboma, ASD, malrotation of small bowel, respiratory distress, thymic atrophy on autopsy) |
| 3 | Male | 65 | IL2RG mutation | T-B+NK+ Leaky SCID CD3 (235), CD4 (176), CD8 (22), CD19 (559), CD16CD56 (516) |
S/p HSCT, patient currently doing well |
| 4 | Female | Undetectable | ADA deficiency | T-B-NK- Typical SCID ALC (0.34), flow cytometry not done due to small number of lymphocytes in specimen |
Profound lymphopenia, Treatment: bactrim and fluconazole prophylaxis; Adagen started about 3 weeks after diagnosis |
| 5 | Male | Undetectable | IL2RG mutation | T-B+NK- Typical SCID CD3 (0), CD4 (0), CD8 (0), CD19 (3,519), CD16CD56 (71) |
Older brother with SCID, s/p HSCT |
| 6 | Male | Undetectable | ADA deficiency | T-B-NK- Typical SCID ALC (0.6), flow cytometry not done due to small number of lymphocytes in specimen |
Profound lymphopenia, Adagen bi-weekly |
| 7 | Male | Undetectable | IL2RG mutation | T-B+NK- Typical SCID CD3 (3), CD4 (1), CD8 (<1), CD19 (323), CD16CD56 (39) |
S/p HSCT |
| 8 | Male | Undetectable | Unknown; genetic testing not done | T-B+NK+ Typical SCID CD3 (28), CD4 (0), CD8 (0), CD19 (384), CD16CD56 (391) |
S/p HSCT |
| 9 | Female | 55 | Unknown; followed out of state | T-B+NK+ Typical SCID CD3 (45), CD4 (32), CD8 (25), CD19 (509), CD16CD56 (414) |
S/p HSCTat 3.5 months of age; h/o autoimmune hemolytic anemia at 11 months of age |
| 10 | Male | Undetectable | IL2RG mutation | T-B+NK- Typical SCID CD3 (<20), CD4 (<20), CD8 (<20), CD19 (498), CD16CD56 (47) |
S/p HSCT at 2 months of age with cord blood, patient currently doing well |
A specimen was submitted for a patient presenting in the PICU who was born 5 months before newborn screening for SCID started in NYS. The patient was diagnosed with SCID based on clinical symptoms. TRECs were undetectable. Two mutations were identified in JAK3. Data for this baby is not included in this report
(flow cytometry) is provided in cells/µL; reference values are not indicated since the flow analysis was performed in multiple testing laboratories
Syndrome with T-Cell Impairment
Most of the infants identified with a syndrome with T-cell impairment had chromosome disorders with multi-system involvement. The most common syndrome with T-cell impairment identified by the TREC assay was 22q11 deletion syndrome (Table V). This diagnosis was established either clinically by phenotype or by a molecular cytogenetic study. Approximately 1 in 27,000 newborns were referred for immunologic evaluation based on low TREC copy numbers and also had 22q11 deletion syndrome. Congenital heart defects were present in ten and two had an absent kidney. One patient did not have major defects associated with 22q11 deletion syndrome at birth, had mild T-cell lymphopenia and was not diagnosed with DiGeorge syndrome until 10 months of age. Two patients with the DiGeorge phenotype did not carry the chromosome 22q11 deletion. One was identified with a DiGeorge anomaly (absent thymus) secondary to diabetic embryopathy, which has been reported in other patients [22]. The second case was found to carry a deletion at the second DiGeorge syndrome locus (DGS2): 46,XY,del(10)(p13) and had clinical findings of imperforate anus, bilateral deafness, hypothyroidism, hypocalcemia and patent ductus arteriosus.
Other chromosome disorders identified in our cohort include one infant with 17q12 duplication syndrome (arr 17q12 (34,611,377–36,248,889x3)), four with Down syndrome, one with trisomy 18, one with chromosome 6p deletion syndrome and one with a ring chromosome 17. Two of the patients with Down syndrome are included in Table VII describing patients with secondary T-cell lymphopenia due to severe T-cell lymphopenia related to other factors (Table VII, Case 52, 56). One infant was found to have severe immune deficiency and CHAR GE syndrome (confirmed by a mutation detected in CHD7).
Table VII.
Patients with secondary T-cell lymphopenia; N=18
| Case number | Gender | TREC average on initial specimen (TRECs/µL) | Description |
|---|---|---|---|
| 40 | Male | 98 | Congenital diaphragmatic hernia, mild hypocalcemia, congenital heart disease, corticosteroid therapy in NICU, familial macrocephaly |
| 41 | Female | 1,670 pre-surgery; 116 post-surgery | Surgical thymectomy during congenital cardiac defect repair |
| 42 | Female | 769 pre-surgery; 95 post-surgery | Hypoplastic left heart, thymectomy |
| 43 | Female | Undetectable | Dysmorphic features, non-immune hydrops, VSD, micrognathia, cleft palate, baby expired |
| 44 | Male | 28 | Previous diagnosis of leukemia, patient receiving chemotherapy, screened at 6 months of age |
| 45 | Male | 101 | Hypoplastic left heart |
| 46 | Male | 102 | TGA and right congenital diaphragmatic hernia, low TREC likely due to third spacing, Negative FISH for 22q11.2 |
| 47 | Male | 6 | Gastroschisis |
| 48 | Female | 58 | Complex congenital heart defect and chylothorax |
| 49 | Male | 181 | Dandy Walker malformation, heart defect, cleft palate |
| 50 | Male | 63 | Hypoplastic left heart |
| 51 | Male | Undetectable | Gastroschisis |
| 52 | Male | 31 | Trisomy 21 with non-immune hydrops; loss of lymphocytes in severe effusions, bone marrow biopsy showed normal lymphocyte counts and subsets |
| 53 | Female | Undetectable | Non-immune hydrops, dysmorphic features, VSD, micrognathia, cleft palate |
| 54 | Male | 96 | Gastroschisis and congenital diaphragmatic hernia |
| 55 | Male | Undetectable | Omphalocele |
| 56 | Male | 1,642 pre-surgery; 46 post-surgery | Trisomy 21 with surgical thymectomy during cardiac defect repair |
| 57 | Male | 78 | Elevated 3-methylglutaric acid and 3-methylglutaconic acid on urine organic acid, possible mitochondrial disease, expired |
Idiopathic T-Cell Lymphopenia
Nearly one-third (30.9 %) of newborns with a clinically significant diagnosis after screening positive in the TREC assay were classified as idiopathic T-cell lymphopenia. To our knowledge, a definition of idiopathic T-cell lymphopenia (ITCL) in the context of newborn screening does not exist in the literature currently. We define ITCL as infants without SCID, a birth defect or another syndrome that required ongoing monitoring or treatment for a deficiency of T-cells. The TREC copy numbers, T-cell counts and clinical treatment of these 30 infants were variable. Six were administered antibiotic prophylaxis (20 %). Interestingly, a pair of brothers had undetectable TRECs and both were administered antibiotics. The flow cytometry abnormalities resolved in 11 patients (37.9 %) at an average age of 11.9 months (Table VI; age available in eight cases). These infants likely have transient lymphopenia of infancy, previously unidentified, that is the corollary of the well known condition, transient hypogammaglobulinemia of infancy. Eighteen patients still exhibit T-cell lymphopenia and are being monitored.
Table VI.
Patients with resolved idiopathic T-cell lymphopenia; N=11
| Patient number |
Gender | TREC average on initial specimen (TRECs/µL) |
Immunologic findingsa | Treatment | Age resolved (months) |
Comments |
|---|---|---|---|---|---|---|
| 29 | Male | Undetectable | 2 months of age: CD3 (989), CD4 (640), CD8 (271) 12.5 months of age: CD3 (2,091), CD4 (1,211), CD8 (796) |
None | 12.5 | Normal response to vaccines, Negative gene testing (IL2RG, JAK3, RAG1, RAG2, IL7R, ADA, CD3D, CD3E, DCLERE1C) |
| 30 | Male | Undetectable | 2 months of age: CD3 (470), CD4 (323), CD8 (135) 12.5 months of age: CD3 (1,348), CD4 (813), CD8 (487) |
None | 12.5 | Normal response to vaccines, Negative gene testing (IL2RG, JAK3, RAG1, RAG2, IL7R, ADA, CD3D, CD3E, DCLERE1C) |
| 31 | Female | 75 | 2 months of age: ALC 1.750 (3.4–7.6); CD3 of 228 (2,500–5,500) 9.9 months of age: ALC 4.420 (3.4–9.0); CD3 2829 (1,900–5,900) |
Prophylaxis and isolation | 9.9 | |
| 32 | Male | 79 | 2 weeks of age: CD3 (1,166), CD4 (717), CD8 (434), follow-up data not available (verbal communication that all values in normal range) | None | Not reported | |
| 33 | Female | 107 | 13 days of age: CD3 (1,407), CD4 (686), CD8 (780), follow-up data not available (verbal communication that all values in normal range) | Bactrim prophylaxis | 9 | Received rotavirus at 2 months had diarrhea, otherwise healthy |
| 34 | Male | Undetectable | T-B-NK+ on first assessment (changed to T-B+NK+ for all subsequent phenotyping). Initial flow at 2 weeks of age abnormal: low CD3 (258), CD4 (94), CD8 (140), CD19 (166) | PJP prophylaxis | 9 | Negative testing for ADA, IL7R, JAK3, IL2RG, CD3E/D, Artemis, RAG1/ 2. Now well, off PJP prophylaxis. Plan to cautiously administer MMR/Varivax after 1 year of age. |
| 35 | Female | 156 | T-B+NK+, initial flow at 1 month of age abnormal: low CD3 (741), CD4 (613), CD8 (117), follow-up data not available (verbal communication that all values in normal range) | PJP prophylaxis | 12 | Normal genetic testing of ADA, IL7R, IL2RG, CD3D/E. Now well, off PJP prophylaxis. Received MMR and Varivax without problem. |
| 36 | Male | 79 | 6 months of age: CD3 (2,359), CD4 (1,342), CD8 (874), follow-up data not available (verbal communication that all values in normal range) | None | Not reported | |
| 37 | Male | 118 | 7 weeks of age: CD3 (2,237), CD4 (1,593), CD8 (515), follow-up data not available (verbal communication that all values in normal range) | None | 22 | |
| 38 | Male | 98 | 1 month age: CD3 (1,912), CD4 (1,211), CD8 (677), follow-up data not available (verbal communication that all values in normal range) | None | Not reported | |
| 39 | Female | 29 | 1 month of age: CD3 (1,125), CD4 (769), CD8 (314), follow-up data not available (verbal communication that all values in normal range) | None | 8 |
(Flow cytometry) is provided in cells/µL
Secondary T-Cell Lymphopenia
The majority of infants with secondary T-cell lymphopenia had major birth defects including hypoplastic left heart syndrome, congenital diaphragmatic hernia and gastroschisis (Table VII). TRECs in these infants varied from undetectable to 181.
Premature Infants
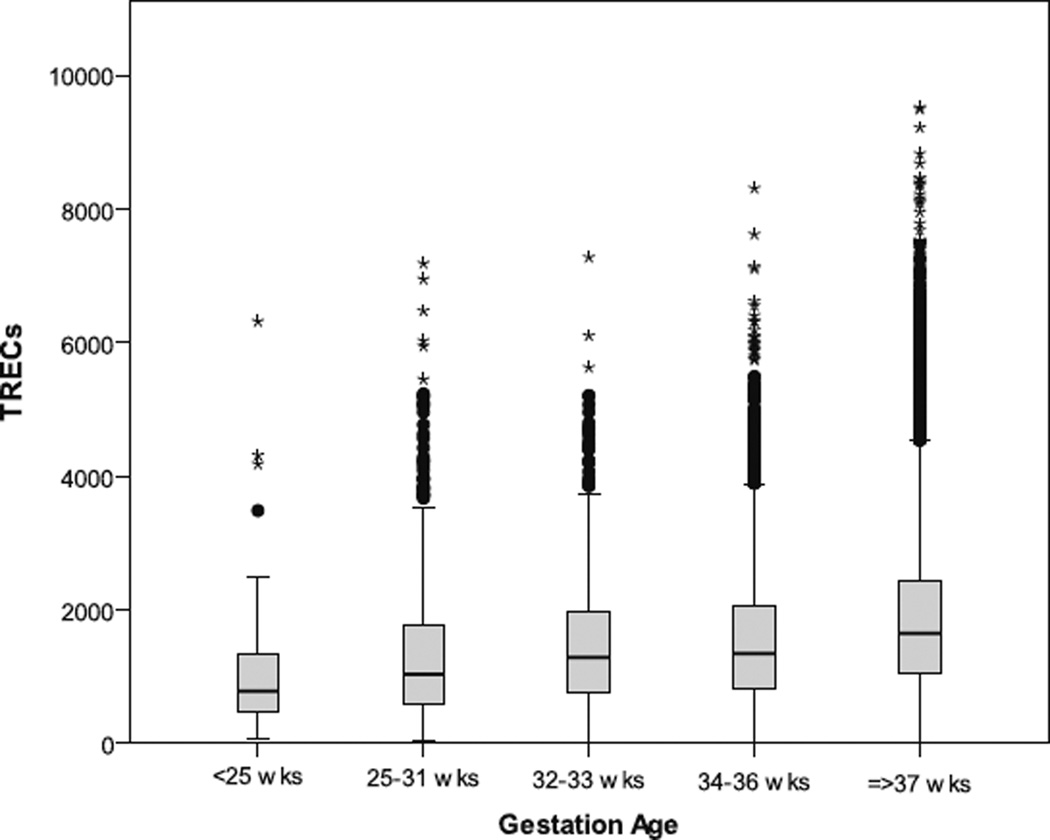
TREC averages varied by gestational age. The mean TREC level in the overall premature population was 1,521 (95%CI= 1,494–1,548; Fig. 2), and there was a trend towards fewer TRECs in babies born at earlier gestational ages. One premature infant with undetectable TRECs and multiple congenital anomalies was also diagnosed with SCID, demonstrating the importance of immediately referring all infants with undetectable TRECs, regardless of gestational age.
Fig. 2.
TREC data from premature infants in groups of gestational age on the x-axis (<25, 25–31, 32–33, 34–36, ≥37 weeks). Data shown for 3 month dataset. Mean TRECs increased with increasing gestation age: 1,072 (<25), 1,319 (25–31), 1,508 (32–33), 1,570 (34–36), 1,862 (≥37 weeks). The difference in mean TRECs in term and pre-term infants was significant (p<0.001, t-test). Minimum, first quartile, median, third quartile and maximum are depicted in the figure. Outliers are shown as dots, and extreme outliers are shown as stars. Created using SPSS v.17.0
Infants with Undetectable TRECs and Normal Flow Cytometry
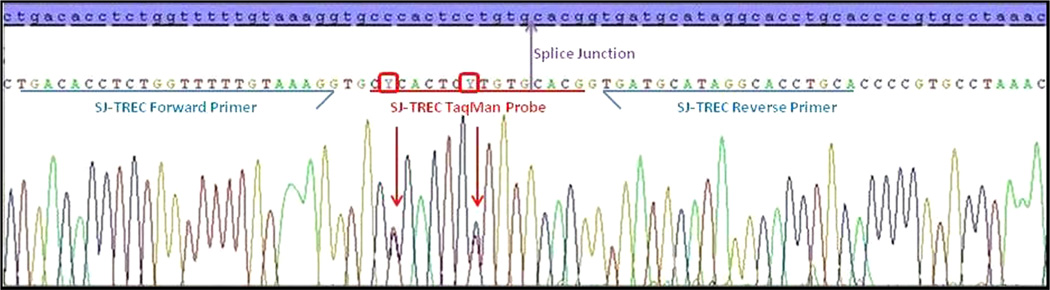
Among cases with undetectable TRECs on the initial screen identified as false positives by flow cytometry (i.e., referred but no disease), TRECs were detectable on a second sample. The only exception was a single infant who was referred for a clinical evaluation based on undetectable TRECs. Laboratory studies including CBC, flow cytometry, memory cells and mitogens were normal. TRECs were undetectable on a repeat newborn screen. Failure of the probe to detect its targeted region was suspected. The patient’s TRECs were sequenced and two SNPs were detected in the sequence targeted by the TREC probe (Fig. 3). Both SNPs in the TREC probe binding site are reported in dbSNP. The first, rs76132819 (NT_026437.12:g.3944295C>T) has a reported minor allele frequency (MAF) of 8.3 % and 15.3 % among 120 CEU and 118YRI chromosomes, respectively [23]. The MAF for the second SNP, rs79211180 (NT_026437.12:g.3944301C>T), is reported to be 0.8 % in 118 YRI chromosomes. Given the complete failure of TRECs to amplify, the SNPs are likely on opposite chromosomes.
Fig. 3.
Sequence analysis of δRec-ψJα Signal Joint region from an infant with normal flow cytometry and undetectable TREC on newborn screen specimens. The reference sequence is highlighted in blue. The forward and reverse primers (blue arrows) flank the SJ-TREC Taqman Probe (underlined in red), which spans across the splice junction (purple arrow). Two SNPs, both C>T changes at the second and eighth base pair within the FAM probe, are indicated by red arrows. Y is the IUPAC code for a C/T heterozygote
Discussion
New York State implemented SCID NBS in 2010, screening 485,912 newborns in 2 years. According to discussions with regional pediatric medical centers, every infant born in NYS with SCID during this time has been identified by newborn screening. Previous estimates of the incidence of SCID were 1 in 40,000 to 1 in 100,000 [2, 10]. Newborn screening data reported by California and Wisconsin suggests an incidence of 1 in 40,000 to 1 in 70,000. California reported an incidence of 1 in 66,250 infants requiring hematopoietic stem cell transplant, thymus transplant or gene therapy [7]. Wisconsin reports an incidence 1 in 41,539 infants with SCID or severe T-cell lymphopenia [16]. Based on newborn screening in more than 450,000 infants, it appears that the incidence of typical and leaky SCID in NYS is closer to between 1 in 50,000 and 1 in 60,000 and is consistent with the incidence observed by other states screening for SCID.
The NYS algorithm was modified twice in the first year of screening to increase the PPV while still keeping the risk of missing an infant with leaky SCID low. To date we are not aware of any missed cases of leaky SCID. Our goal is to improve the sensitivity and specificity of our screen to maximize the PPV as we expand our understanding of TREC point estimates and distributions in the true positive, false positive and normal populations. To remain conservative, we created a borderline category in lieu of a lower cut-off. We believe many false positive screens are likely due to either poor specimen quality, biological variation in the normal range of TRECs in newborns (our data shows that TRECs vary by gestational age, gender and race/ethnicity), or the presence of SNPs in the T-cell receptor region of genomic DNA. These data demonstrated that TRECs are lower in infants reported as Black, which is likely contributing to the higher referral rate in Black infants in NY. It is possible that lower TRECs are a consequence of white blood cell counts, which are known to be 10–20 % lower in African Americans compared to European Americans, partly due to the common variant rs2814778 in the DARC gene that confers selective advantage against malaria in the African American population [24–26]. Further studies are needed to determine why TRECs are lower in males, which probably contributes to the observed skewed referral ratio. The screening algorithm may be refined further as additional data to support or refute these hypotheses are collected and analyzed.
Low TREC values detected by screening in premature infants were first reported by Wisconsin and later by Massachusetts [12, 14]. While, there is a report in the literature that addresses diagnostic challenges in premature infants, we are not aware of published normal reference ranges for TRECs in a premature population [27]. Our data also supports an association of TRECs with gestational age and we have observed that TRECs typically ‘normalize’ (i.e. return to the mean) at an equivalent of 37 weeks gestation. We agree with Drs. Ward and Baptist [27] and many states like ours who implemented the use of repeat specimens to enhance specificity [21]. Our case of a premature infant with undetectable TRECs and typical SCID confirmed for us an approach developed independently by early screening states to ensure that all infants with undetectable TRECs are referred for a diagnostic evaluation regardless of gestational age [21]. Other states planning to screen or currently screening for SCID might consider this in their algorithm development.
In addition to premature infants, we identified one baby with undetectable TRECs, yet a normal diagnostic evaluation. Further investigation demonstrated this infant had two SNPs in the TREC region used for our probe, resulting in complete failure of amplification. This possibility should be considered for cases with a normal diagnostic evaluation and repeated undetectable TRECs on the newborn screen, although this is a rare occurrence in NYS infants (1 of 485,912). It is also possible that infants referred with low TRECs or infants in the lower end of the normal distribution carry a SNP(s) in the TREC primer or probe binding sites. This possibility is being investigated in a subset of referred infants (of varying race/ethnicities) with normal CBC and flow cytometry results.
One consideration for Programs is the extent babies are detected with diagnoses other than typical or leaky SCID. For example, in NYS, the TREC assay identified more infants with DiGeorge syndrome than typical SCID. The incidence of DiGeorge syndrome is estimated at 1 in 4,000 [28, 29]. In NYS, we can infer that the TREC assay identified approximately 14.8 % of individuals with DiGeorge syndrome. Previous estimates suggest that 80 % of individuals with DiGeorge syndrome have some immune system involvement [28, 29]. Therefore, we conclude that the TREC assay will not identify all infants with immune involvement related to DiGeorge syndrome. Conversely, patients with TRECs in the low normal to normal range demonstrating any phenotype associated with DiGeorge syndrome should be evaluated for immune dysfunction. Our assay identified patients with DiGeorge syndrome and severe immune involvement, which occurs in less than 1.5%of newborns [28, 29]. Identifying this group is important for medical management [29]. The importance of early identification of the group with milder involvement is unknown, and assessment of long-term outcomes may be helpful in the ongoing discussion about newborn screening for 22q11 deletions [30].
Another non-SCID syndrome detected in NYS was Down syndrome (n=4 newborns). Precocious aging is a feature of Down syndrome, involving many organ systems, including the thymus [31, 32]. Two studies in children with Down syndrome noted that TRECs were decreased compared to a control population, and there was a strong negative correlation with age. Thus the reduced TREC numbers in individuals with Down syndrome may be age dependent.
In order to understand the underlying causes and possible outcomes in infants with low TRECs and ITCL identified in the newborn period, long-term follow-up is required. Additional molecular testing, including sequencing of known immunodeficiency genes and/or whole exome sequencing, may help elucidate the cause of immunodeficiency in patients with idiopathic T-cell lymphopenia [33]. Interestingly, one infant diagnosed with idiopathic T-cell lymphopenia and an average of 75 TRECs had 228 CD3 cells. The clinician chose the “wait and watch” approach described by Verbsky et al., and the patient was placed in isolation, received antibiotic prophylaxis and was not given live vaccines, but HSCT was not done [15].By 9 months of age, the CD3 count normalized to 2,829. This case demonstrates the diagnostic challenges facing clinicians in states that are screening for SCID and the need for long-term follow-up. Currently, in NYS, short-term follow-up concludes when a diagnosis is obtained. Results of additional testing and treatment are considered long-term follow-up. Long-term follow-up data for SCID are not actively being collected in NYS at the present time, however, there are plans to implement long-term follow-up in the future, in accordance with national efforts.
Conclusions
Newborn screening for severe T-cell deficiencies in NYS identified ten infants with SCID. Nine of the ten patients (90 %) received HSCT or enzyme replacement therapy and are doing well demonstrating that SCID NBS is beneficial. Additional data and follow-up may allow for further adjustments and improvements to the screening algorithm, leading to an increased PPV. It appears that a very conservative cut-off could be used to detect typical SCID. However, lowering the cutoff would reduce the number of patients identified with non-SCID disorders that benefit from early or pre-symptomatic identification, requiring medical management. Long-term follow-up is necessary to determine the benefits of identifying individuals with idiopathic T-cell lymphopenia, syndromes with T-cell impairment and secondary T-cell lymphopenia. Because only one patient with leaky SCID was identified, more data are needed to determine whether further algorithm adjustments would impact detection of this group. National collaboration is also essential to obtain enough data to learn about the long-term impact of SCID NBS, and outcomes in patients with and without typical SCID. Comprehensive data collection using a common national dataset is important. The role of groups such as the Newborn Screening Translational Research Network will be invaluable to assess laboratory practice, diagnosis and short- and long-term follow-up.
Acknowledgments
Funding was provided by the Eunice Kennedy Shriver Institute for Child Health and Human Development, the Jeffrey Modell Foundation and the New York State Department of Health. Victoria Popson was responsible for most of the follow-up at the Newborn Screening Program.
Appendix
Contributor Information
Beth H. Vogel, Email: bmh06@health.state.ny.us, Newborn Screening Program, Wadsworth Center, New York State, Department of Health, PO Box 509, Albany, NY 12201-0509, USA.
Vincent Bonagura, Steven and Alexandra Cohen Children’s Medical Center of NY, 865 Northern Boulevard, Room 101, Great Neck, NY 11021, USA.
Geoffrey A. Weinberg, University of Rochester School of Medicine and Dentistry, 601 Elmwood Avenue, Box 690, Rochester, NY 14642, USA
Mark Ballow, Women and Children’s Hospital of Buffalo, 219 Bryant Street, Buffalo, NY 14222, USA.
Jason Isabelle, Newborn Screening Program, Wadsworth Center, New York State, Department of Health, PO Box 509, Albany, NY 12201-0509, USA.
Lisa DiAntonio, Newborn Screening Program, Wadsworth Center, New York State, Department of Health, PO Box 509, Albany, NY 12201-0509, USA.
April Parker, Newborn Screening Program, Wadsworth Center, New York State, Department of Health, PO Box 509, Albany, NY 12201-0509, USA.
Allison Young, Newborn Screening Program, Wadsworth Center, New York State, Department of Health, PO Box 509, Albany, NY 12201-0509, USA.
Charlotte Cunningham-Rundles, Mount Sinai Medical Center, 1425 Madison Avenue, Room 1120, New York, NY 10029, USA.
Chin-To Fong, University of Rochester School of Medicine and Dentistry, 601 Elmwood Avenue, Box 690, Rochester, NY 14642, USA.
Jocelyn Celestin, Asthma and Allergy Center of Albany Medical Center, 176 Washington Ave Extension, Suite 102, Albany, NY 12203, USA.
Heather Lehman, Women and Children’s Hospital of Buffalo, 219 Bryant Street, Buffalo, NY 14222, USA.
Arye Rubinstein, Division of Allergy and Immunology, Montefiore Medical Park, 1525 Blondell Avenue, Room 100, Bronx, NY 10461, USA.
Subhadra Siegel, New York Medical College, Westchester Medical Center, Munger Pavilion Room 106, Valhalla, NY, USA.
Leonard Weiner, Department of Pediatrics, State University of New York Upstate Medical University, 750 East Adams Street, Room 5400, Syracuse, NY 13210, USA.
Carlos Saavedra-Matiz, Newborn Screening Program, Wadsworth Center, New York State, Department of Health, PO Box 509, Albany, NY 12201-0509, USA.
Denise M. Kay, Newborn Screening Program, Wadsworth Center, New York State, Department of Health, PO Box 509, Albany, NY 12201-0509, USA
Michele Caggana, Newborn Screening Program, Wadsworth Center, New York State, Department of Health, PO Box 509, Albany, NY 12201-0509, USA.
References
- 1.Buckley RH. The long quest for neonatal screening for severe combined immunodeficiency. J Allergy Clin Immunol. 2012;129(3):597–604. doi: 10.1016/j.jaci.2011.12.964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adeli MM, Buckley RH. Why newborn screening for severe combined immunodeficiency is essential: a case report. Pediatrics. 2010;126(2):e465–e469. doi: 10.1542/peds.2009-3659. [DOI] [PubMed] [Google Scholar]
- 3.Puck JM. Laboratory technology for population-based screening for severe combined immunodeficiency in neonates: the winner is T-cell receptor excision circles. J Allergy Clin Immunol. 2012;129(3):607–616. doi: 10.1016/j.jaci.2012.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chan A, Scalchunes C, Boyle M, Puck JM. Early vs. delayed diagnosis of severe combined immunodeficiency: a family perspective survey. Clin Immunol (Orlando, FL) 2011;138(1):3–8. doi: 10.1016/j.clim.2010.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gennery AR, Slatter MA, Grandin L, Taupin P, Cant AJ, Veys P, et al. Transplantation of hematopoietic stem cells and long-term survival for primary immunodeficiencies in Europe: entering a new century, do we do better? J Allergy Clin Immunol. 2010;126(3):602, 10.e1–11.e1. doi: 10.1016/j.jaci.2010.06.015. [DOI] [PubMed] [Google Scholar]
- 6.Baker MW, Grossman WJ, Laessig RH, Hoffman GL, Brokopp CD, Kurtycz DF, et al. Development of a routine newborn screening protocol for severe combined immunodeficiency. J Allergy Clin Immunol. 2009;124(3):522–527. doi: 10.1016/j.jaci.2009.04.007. [DOI] [PubMed] [Google Scholar]
- 7.Kwan A, Church JA, Cowan MJ, Agarwal R, Kapoor N, Kohn DB, et al. Newborn screening for severe combined immunodeficiency and T-cell lymphopenia in California: results of the first 2 years. J Allergy Clin Immunol. 2013;132(1):140–147. doi: 10.1016/j.jaci.2013.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Buckley RH. Transplantation of hematopoietic stem cells in human severe combined immunodeficiency: longterm outcomes. Immunol Res. 2011;49(1–3):25–43. doi: 10.1007/s12026-010-8191-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Puck JM. Population-based newborn screening for severe combined immunodeficiency: steps toward implementation. J Allergy Clin Immunol. 2007;120(4):760–768. doi: 10.1016/j.jaci.2007.08.043. [DOI] [PubMed] [Google Scholar]
- 10.Chan K, Puck JM. Development of population-based newborn screening for severe combined immunodeficiency. J Allergy Clin Immunol. 2005;115(2):391–398. doi: 10.1016/j.jaci.2004.10.012. [DOI] [PubMed] [Google Scholar]
- 11.Comeau AM, Hale JE, Pai S-Y, Bonilla FA, Notarangelo LD, Pasternack MS, et al. Guidelines for implementation of population-based newborn screening for severe combined immunodeficiency. J Inherit Metab Dis. 2010;33(Suppl 2):S273–S281. doi: 10.1007/s10545-010-9103-9. [DOI] [PubMed] [Google Scholar]
- 12.Thompson JG, Wilkey JF, Baptiste JC, Navas JS, Pai S-Y, Pass KA. Development of a high throughput multiplexed TREC qPCR assay with internal controls for detection of severe combined immunodeficiency in population-based newborn screening. Clin Chem. 2010;56(9):1466–1474. doi: 10.1373/clinchem.2010.144915. [DOI] [PubMed] [Google Scholar]
- 13.Routes JM, Grossman WJ, Verbsky J, Laessig RH, Hoffman GL, Brokopp CD, et al. Statewide newborn screening for severe T-cell lymphopenia. J Allergy Clin Immunol. 2009;302(22):2465–2470. doi: 10.1001/jama.2009.1806. [DOI] [PubMed] [Google Scholar]
- 14.Baker MW, Laessig RH, Katcher ML, Routes JM, Grossman WJ, Verbsky J, et al. Implementing routine testing for severe combined immunodeficiency within Wisconsin’s newborn screening program. Public Health Rep (Washington, DC: 1974) 2010;125(Suppl 2):88–95. doi: 10.1177/00333549101250S211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Verbsky J, Thakar M, Routes J. The Wisconsin approach to newborn screening for severe combined immunodeficiency. J Allergy Clin Immunol. 2012;129(3):622–627. doi: 10.1016/j.jaci.2011.12.004. [DOI] [PubMed] [Google Scholar]
- 16.Verbsky JW, Baker MW, Grossman WJ, Hintermeyer M, Dasu T, Bonacci B, et al. Newborn screening for severe combined immunodeficiency; the Wisconsin experience (2008–2011) J Clin Immunol. 2012;32(1):82–88. doi: 10.1007/s10875-011-9609-4. [DOI] [PubMed] [Google Scholar]
- 17.Borte S, Wang N, Oskarsdóttir S, Von Döbeln U, Hammarström L. Newborn screening for primary immunodeficiencies: beyond SCID and XLA. Ann N YAcad Sci. 2011;1246:118–130. doi: 10.1111/j.1749-6632.2011.06350.x. [DOI] [PubMed] [Google Scholar]
- 18.Saavedra-Matiz CA, Isabelle JT, Biski CK, Duva SJ, Sweeney ML, Parker AL, et al. Cost-effective and scalable DNA extraction method from dried blood spots. Clin Chem. 2013 doi: 10.1373/clinchem.2012.198945. [DOI] [PubMed] [Google Scholar]
- 19.Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, et al. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem. 2009;55(4):611–622. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- 20.No Title [Internet] Available from: https://www.nbstrn.org/sites/default/files/SCID%20National%20Monthly%20March%202012.pdf. [Google Scholar]
- 21.Hannon WH, Abraham RS, Kobrynski L, Vogt RF, Adair O, Constantino A, et al. NBS06-A Newborn blood spot screening for severe combined immunodeficiency by measurement of T-cell receptor excision circles; approved guideline. Clinical Standards and Laboratory Institute. 2013:1–92. [Google Scholar]
- 22.Rope AF, Cragun DL, Saal HM, Hopkin RJ. DiGeorge anomaly in the absence of chromosome 22q11.2 deletion. J Pediatr. 2009;155(4):560–565. doi: 10.1016/j.jpeds.2009.04.010. [DOI] [PubMed] [Google Scholar]
- 23.1000 Genomes Project Consortium. Abecasis GR, Auton A, Brooks LD, DePristo MA, Durbin RM, et al. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491(7422):56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lim E-M, Cembrowski G, Cembrowski M, Clarke G. Race-specific WBC and neutrophil count reference intervals. Int J Lab Hematol. 2010;32(6 Pt 2):590–597. doi: 10.1111/j.1751-553X.2010.01223.x. [DOI] [PubMed] [Google Scholar]
- 25.Tollerud DJ, Brown LM, Blattner WA, Mann DL, Pankiw-Trost L, Hoover RN. Tcell subsets in healthy black smokers and nonsmokers. Evidence for ethnic group as an important response modifier. Am Rev Respir Dis. 1991;144(3 Pt 1):612–616. doi: 10.1164/ajrccm/144.3_Pt_1.612. [DOI] [PubMed] [Google Scholar]
- 26.Reiner AP, Lettre G, Nalls MA, Ganesh SK, Mathias R, Austin MA, et al. Genome-wide association study of white blood cell count in 16, 388 African Americans: the Continental Origins and Genetic Epidemiology Network (COGENT) . PLoS Genet. 2011;7(6):e1002108. doi: 10.1371/journal.pgen.1002108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ward CE, Baptist AP. Challenges of newborn severe combined immunodeficiency screening among premature infants. Pediatrics. 2013;131(4):e1298–e1302. doi: 10.1542/peds.2012-1921. [DOI] [PubMed] [Google Scholar]
- 28.Lima K, Abrahamsen TG, Foelling I, Natvig S, Ryder LP, Olaussen RW. Low thymic output in the 22q11.2 deletion syndrome measured by CCR9+CD45RA+T cell counts and T cell receptor rearrangement excision circles. Clin Exp Immunol. 2010;161(1):98–107. doi: 10.1111/j.1365-2249.2010.04152.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McDonald-McGinn DM, Sullivan KE. Chromosome 22q11.2 deletion syndrome (DiGeorge syndrome/velocardiofacial syndrome) . Medicine (Baltimore) 2011;90(1):1–18. doi: 10.1097/MD.0b013e3182060469. [DOI] [PubMed] [Google Scholar]
- 30.Bales AM, Zaleski CA, McPherson EW. Newborn screening programs: should 22q11 deletion syndrome be added? Genet Med. 2010;12(3):135–144. doi: 10.1097/GIM.0b013e3181cdeb9a. [DOI] [PubMed] [Google Scholar]
- 31.Prada N, Nasi M, Troiano L, Roat E, Pinti M, Nemes E, et al. Direct analysis of thymic function in children with Down’s syndrome. Immun Ageing. 2005;2(1):4. doi: 10.1186/1742-4933-2-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Roat E, Prada N, Lugli E, Nasi M, Ferraresi R, Troiano L, et al. Homeostatic cytokines and expansion of regulatory T cells accompany thymic impairment in children with Down syndrome. Rejuvenation Res. 2008;11(3):573–583. doi: 10.1089/rej.2007.0648. [DOI] [PubMed] [Google Scholar]
- 33.Mallott J, Kwan A, Church J, Gonzalez-Espinosa D, Lorey F, Tang LF, et al. Newborn screening for SCID identifies patients with ataxia telangiectasia. J Clin Immunol. 2013;33(3):540–549. doi: 10.1007/s10875-012-9846-1. [DOI] [PMC free article] [PubMed] [Google Scholar]