Abstract
Despite current treatment regimens, heart failure remains the leading cause of morbidity and mortality in the developed world due to the limited capacity of adult mammalian ventricular cardiomyocytes to divide and replace ventricular myocardium lost from ischemia-induced infarct1,2. As a result, there is great interest to identify potential cellular sources and strategies to generate new ventricular myocardium3. Past studies have shown that lower vertebrate and early postnatal mammalian ventricular cardiomyocytes can proliferate to help regenerate injured ventricles4–6; however, recent studies have suggested that additional endogenous cellular sources may contribute to this overall ventricular regeneration3. Here, we have developed in the zebrafish a combination of fluorescent reporter transgenes, genetic fate-mapping strategies, and a ventricle-specific genetic ablation system to discover that differentiated atrial cardiomyocytes can transdifferentiate into ventricular cardiomyocytes to contribute to zebrafish cardiac ventricular regeneration. Using in vivo time-lapse and confocal imaging, we monitored the dynamic cellular events during atrial-to-ventricular cardiomyocyte transdifferentiation to define intermediate cardiac reprogramming stages. Importantly, we observed that Notch signaling becomes activated in the atrial endocardium following ventricular ablation, and discovered that inhibiting Notch signaling blocked the atrial-to-ventricular transdifferentiation and cardiac regeneration. Overall, these studies not only provide evidence for the plasticity of cardiac lineages during myocardial injury, but more importantly reveal an abundant new potential cardiac resident cellular source for cardiac ventricular regeneration.
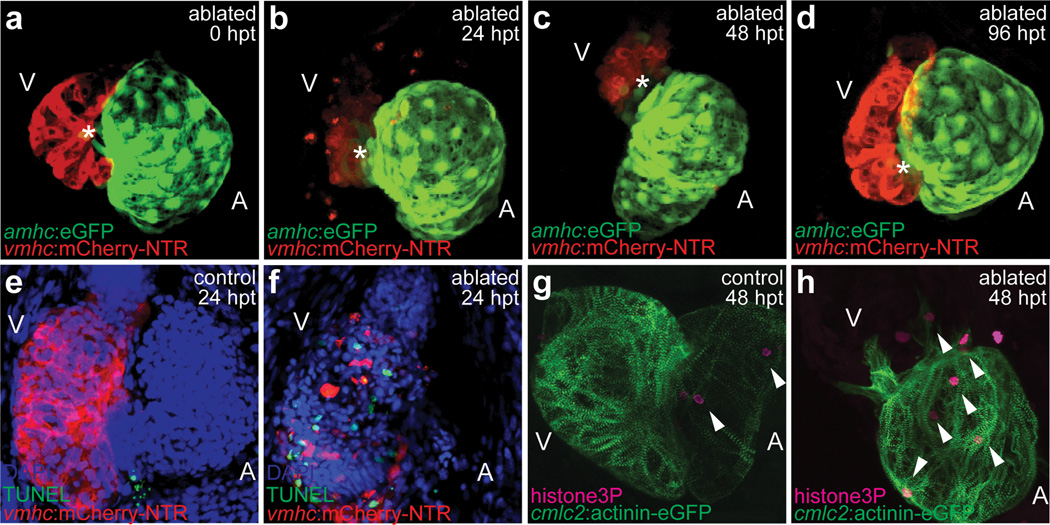
Atrial cardiomyocytes appear to have the remarkable capacity to divide in many adult vertebrate hearts including mammals during cardiac ventricular injury7,8; however, it remains unclear as to the role of these proliferating atrial cardiomyocytes. To address this issue, we have generated in the zebrafish, a genetic cardiac ventricle-specific nitroreductase (NTR)-mediated ablation system9, Tg(vmhc:mCherry-NTR)s957, using the ventricular myosin heavy chain (vmhc) promoter10. Thus, this ablation system permits the targeted destruction of ventricular cardiomyocytes after metronidazole (MTZ) treatment (Fig. 1). Furthermore, to track both atrial and ventricular cardiomyocytes during ventricular injury, we have fused the mCherry fluorescent protein to NTR to label ventricular cardiomyocytes in red (vCherry) as well as used the atrial myosin heavy chain (amhc) promoter to create the Tg(amhc:eGFP)s958 transgenic line, which marks atrial cardiomyocytes in green (aGFP). As a result, we observed that the expression of mCherry and GFP in these reporter lines were restricted to ventricular and atrial cardiomyocytes, respectively throughout development (Supplementary Fig. 1a–d). In order to visually monitor in vivo the dynamic cellular events that transpire throughout ventricular injury and regeneration within the same animal, the ventricles of Tg(vmhc:mCherry-NTR);Tg(amhc:eGFP) hearts were ablated at 3–4 days post-fertilization (dpf), an age when the zebrafish heart has completed cardiac looping and cardiac chamber cardiomyocytes have fully differentiated11, but the zebrafish remains optically clear. As a result, 4 dpf ablated vmhc:mCherry-NTR ventricles displayed significant reduction in vCherry fluorescence by 24 hours post MTZ treatment (hpt) (Fig. 1b, Supplementary Fig. 2a). This rapid decrease in vCherry fluorescence was accompanied by a reduction in ventricular size (Fig. 1b, Supplementary Fig. 2b) and function (fractional area change decreased from 42% to 16% at 24 hpt/N=5, p=0.004; Supplementary videos 1, 2; Supplementary Fig. 3a–c), leading to pericardial edema and overall decreased blood circulation. TUNEL cell death assays revealed that this decrease in ventricular function and size was due to an increase in ventricular cardiomyocyte death (Fig. 1f). Conversely, we observed that the atria significantly enlarged (Supplementary Fig. 2b) and maintained normal contractile function (Supplementary Fig. 3d) during this reduction in ventricular size and function.
Figure 1. Genetic ventricular-specific cardiomyocyte ablation results in ventricular cardiomyocyte death and subsequent regeneration.
(a–d) Metronidazole (MTZ)-ablated Tg(vmhc:mCherry-NTR;amhc:eGFP) ventricular cardiomyocytes at (a) 0, (b) 24, (c) 48, (d) 96 hours post-treatment (hpt). Red - ventricular cardiomyocytes; Green - atrial cardiomyocytes; Asterisks - AVC. (e, f) TUNEL staining (green) in 24 hpt Tg(vmhc:mCherry-NTR) (e) control and (f) ablated ventricles (red) at 4 dpf. Blue - DAPI. (g, h) Anti-phospho-histone H3 staining (magenta) in 48 hpt Tg(vmhc:mCherry-NTR;cmlc2:actinin-eGFP) (g) control and (h) ablated hearts (green) at 5 dpf. Arrowheads - anti-phospho-histone H3 positive cells. A - atrium; V - ventricle.
By 48 hpt, we observed in these injured ventricles an accretion of new vCherry+ cardiomyocytes adjacent to the atrioventricular canal (AVC) (Fig. 1c, asterisk), which continued to expand across the ventricle over the next 48 hours to restore lost ventricular myocardium nearest the AVC by 96 hpt (Fig. 1d, asterisk). Subsequently, ventricular cardiac function recovered in these injured hearts to nearly that of age-matched controls by 96 hpt (fractional area change of ablated and control ventricle was 38% and 39%, respectively; N=5, p=0.49; Supplementary Fig. 3c), allowing for survival of these ventricle-ablated fish into adulthood. Anti-phospho-histone H3 immunostaining revealed that ventricular injury resulted in not only ventricular but also atrial cardiomyocyte proliferation (Fig. 1g, h; Supplementary Fig. 4), as observed in other cardiac injury and regeneration models4,6,12. Although this cardiomyocyte cell division peaked by 48 hpt in both cardiac chambers (Supplementary Fig. 4d, e), there was a particularly significant atrial cardiomyocyte proliferative increase that remained elevated up to 84 hpt.
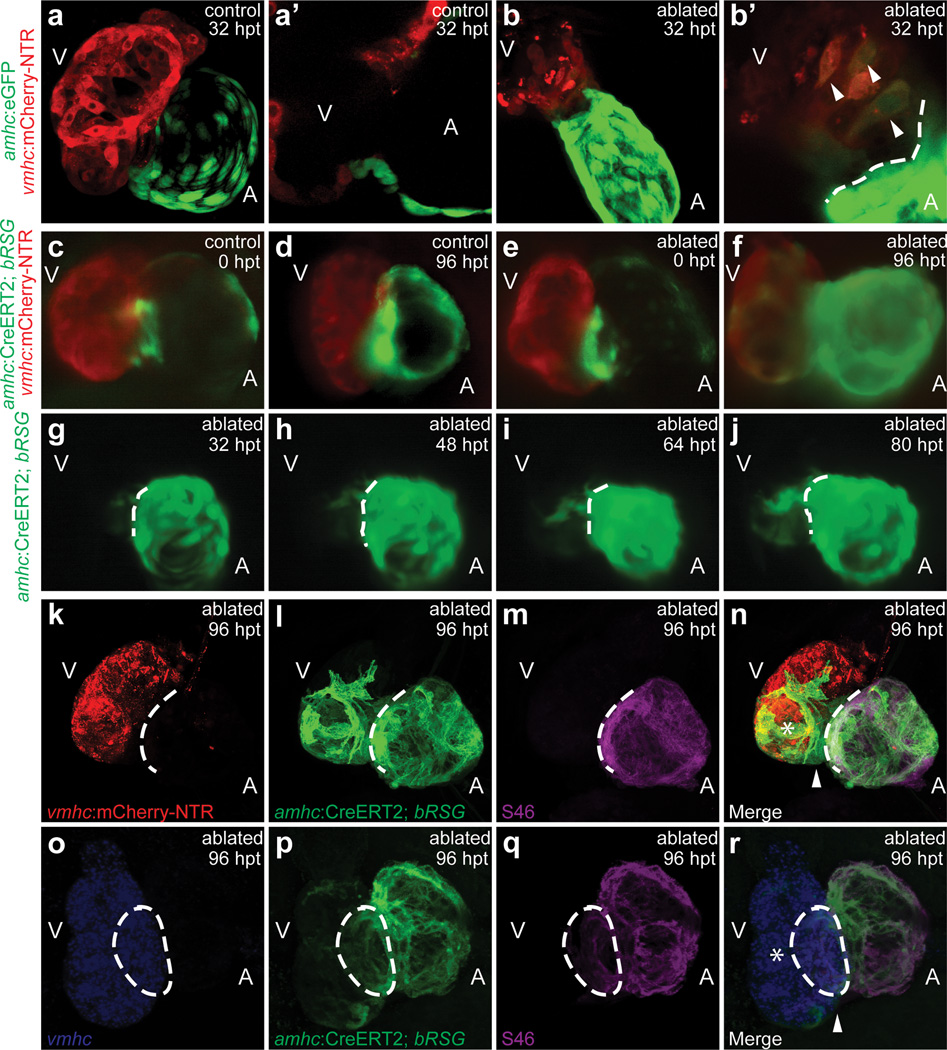
During the ventricular recovery, we observed the presence of low expressing aGFP cardiomyocytes (aGFPlo) within the injured ventricle near the AVC between 24–48 hpt (Fig. 1b, c, 2b), whereas aGFP cardiomyocytes were never observed in the ventricle of control hearts (Fig. 2a). Optical sections of the AVC region revealed that these aGFPlo cardiomyocytes co-expressed the vCherry marker in the ablated ventricles (Fig. 2b’, arrowheads), but were not detected in unablated ventricles (Fig. 2a’). To further confirm these findings, we examined the gene expression of amhc and vmhc in the hearts of these ablated ventricles and observed that amhc was expressed throughout the atrium and frequently in the ventricle at the AVC by 24–48 hpt, where we observed the double labeled aGFPlo;vCherry cardiomyocytes; whereas, vmhc was expressed throughout the ventricle but rarely in the atrium at 12–24 hpt only (Supplementary Fig. 5). Thus, these findings suggest that during injury, preexisting atrial cardiomyocytes may migrate to the ventricle to transdifferentiate into ventricular cardiomyocytes, or alternatively, proliferating dedifferentiated ventricular cardiomyocytes may need to activate an atrial program prior to fully differentiating into ventricular cardiomyocytes. Given that we still observe some proliferating pre-existing vCherry cardiomyocytes that never express the aGFP marker, we hypothesize that the atrial-to-ventricular cardiomyocyte transdifferentiation model may be more plausible.
Figure 2. In vivo cardiac reprogramming contributes to zebrafish ventricular regeneration.
(a, b) 32 hpt Tg(vmhc:mCherry-NTR;amhc:eGFP) (a) control and (b) ablated hearts. (a’, b’) AVC section of a and b. Arrowheads - mCherry+/GFP+ cardiomyocytes. (c–j) 4-HT-treated Tg(vmhc:mCherry-NTR;amhc:CreERT2;β-act2:RSG) zebrafish in (c, d) control or (e–j) ablated hearts at (c, e) 0, (g) 32, (h) 48, (i) 64, (j) 80, (d, f) 96 hpt. (g–j) 32–80 hpt GFP time-lapse imaging. (k–r) 4-HT-treated Tg(vmhc:mCherry-NTR;amhc:CreERT2;β-act2:RSG) ablated hearts at 96 hpt. (k) red - vCherry+; (l, p) green - c-aGFP+; (m, q) magenta - S46/anti-Amhc; (n, r) merge; and (o) blue - vmhc RNA. Asterisk - (n) vCherry+/c-aGFP+/S46−, (r) vmhc+/c-aGFP+/S46− ventricular cardiomyocytes. Arrowhead - (n) vCherry−/c-aGFP+/S46−, (r) vmhc−/c-aGFP+/S46− ventricular cardiomyocytes. A - atrium; V - ventricle. Dashed lines - atrioventricular boundary.
To further investigate this transdifferentiation model, we performed genetic lineage tracing of atrial cardiomyocytes in the Tg(vmhc:mCherry-NTR) ventricle ablation model using an atrial cardiomyocyte specific tamoxifen-inducible Cre transgenic line, Tg(amhc:CreERT2)sd20, and the indicator transgenic line, Tg(β-actin2:loxP-DsRed-STOP-loxP-eGFP)s928 (β-act2:RSG), which can effectively label most cells, including cardiomyocytes and endocardial cells, in GFP after tissue-specific Cre-mediated excision of the floxed dsRed cassette5,13. Treating the Tg(vmhc:mCherry-NTR;amhc:CreERT2;β-act2:RSG) fish with 4-hydroxytamoxifen (4-HT) at 3 dpf for 4–6 hours, effectively GFP labeled all atrial cardiomyocytes by cre recombination (c-aGFP+) without ectopic GFP-labeling of ventricular cardiomyocytes by 5 dpf (Fig. 2c, e; Supplementary Fig. 6a–c); whereas vehicle control treatment resulted in no cre-mediated GFP labeling of any cells (Supplementary Fig. 6d). We subsequently MTZ-ablated the ventricles of these c-aGFP-labeled hearts at 5 dpf (Fig. 2e). In the ablated group, these c-aGFP+ cardiomyocytes at 96 hpt and at 12 months were present in not only the atrium but also in the ventricle where many of them also co-expressed vCherry (n = 73/76; Fig. 2f, Supplementary Fig. 7i, j, 8b, c). However, in the control non-ablated group at 96 hpt and at 12 months, c-aGFP+ cardiomyocytes were only observed in the atrium but never in the ventricle (n = 0/54; Fig. 2d; Supplementary Fig. 6c, 8a). Using time-lapse imaging within the same ventricle-ablated Tg(vmhc:mCherry-NTR;amhc:CreERT2;β-act2:RSG) hearts, we observed the gradual and contiguous extension of these c-aGFP+ cardiomyocytes at the AVC into the ventricle where they began to restore ventricular contractile function nearest the AVC and also to exhibit vCherry expression, suggesting that they were transdifferentiating from atrial to ventricular cardiomyocytes (Fig. 2g–j, Supplementary Videos 3–5). Electrophysiologic intracellular recordings revealed that these c-aGFP+/vCherry+ cardiomyocytes within the regenerating ventricle exhibited electrical attributes similar to endogenous c-aGFP−/vCherry+ ventricular cardiomyocytes and distinct from atrial cardiomyocytes (i.e. c-aGFP+/vCherry−) (Supplementary Fig. 9). Furthermore, in situ analysis showed that the ventricular specific markers, irx1a and vmhc, were expressed in ventricular cardiomyocytes at the AVC where c-aGFP+ ventricular cardiomyocytes reside (Supplementary Fig. 10). Moreover, we also observed that the atrial chamber c-aGFP+ fluorescence intensity and size in hearts with ablated ventricles appeared to be significantly greater than that of control hearts (compare Fig. 2f to d, Supplementary Fig. 7k, l), an observation consistent with the increased atrial size, fluorescence intensity, and cardiomyocyte proliferation observed after ventricular injury (Supplementary Fig. 2a, b, 4d). Finally, in adult zebrafish ventricles ablated at 4 months, we discovered that the contribution of transdifferentiated atrial cardiomyocytes to the regenerating ventricle was diminished (Supplementary Fig. 11), suggesting that this cardiac transdifferentation process may be age-dependent.
To investigate whether these atrial-derived ventricular cardiomyocytes maintained the atrial program, we examined Amhc protein expression by S46 immunostaining in the ventricle-ablated Tg(vmhc:mCherry-NTR;amhc:CreERT2;β-act2:RSG) hearts, and observed that c-aGFP+ atrial cardiomyocytes continued to express Amhc in the atrium (Fig. 2m, n), but not in the ventricle. However, prior to their subsequent differentiation into ventricular cardiomyocytes (c-aGFP+, Amhc/S46−, vCherry+, Fig. 2n, Supplementary Fig. 7d, d’ asterisk), we discovered that c-aGFP+ cardiomyocytes may first exist in an intermediate dedifferentiation stage (c-aGFP+, Amhc/S46−, vCherry−, Fig. 2n, Supplementary Fig. 7d, d’ arrowheads), where they no longer maintain the atrial program (Amhc/S46−) but have not yet activated the ventricular program (vCherry−). Consistent with these findings, vmhc fluorescent in situ hybridization also revealed a population of c-aGFP+ cardiomyocytes devoid of vmhc expression within the recovering ventricle (Fig. 2r, Supplementary Fig. 7h, h’ arrowheads). Because of this possible dedifferentiation of atrial cardiomyocytes, we also examined the expression of cardiac progenitor markers in the ventricle-ablated hearts. Although we did not detect Isl1 in the injured hearts (data not shown), we did observe the re-expression of gata4, hand2, nkx2.5, tbx5 and tbx20 (Supplementary Fig. 12) as well as increased expression of Mef2 in the atrium, ventricle, and outflow tract of the recovering ventricle-ablated hearts (Supplementary Fig. 13). Furthermore, Mef2 immunofluorescence revealed not only that many of the intermediate dedifferentiated c-aGFP+ cardiomyocytes within the recovering ventricle (i.e. c-aGFP+/vCherry−) expressed Mef2 (Supplementary Fig. 13p’, arrowheads), but also that the new Mef2+ cells at the outflow tract did not express myosin heavy chain (Supplementary Fig. 13e–h), suggesting that these cells may be cardiac progenitor cells, which have not yet differentiated into mature cardiomyocytes. Overall, these data suggest that atrial cardiomyocytes may acquire cardiac progenitor cell attributes to become ventricular cardiomyocytes and moreover that the zebrafish second heart field residing near the outflow tract14,15 may also contribute ventricular cardiomyocytes to recover the distal portion of the injured ventricle.
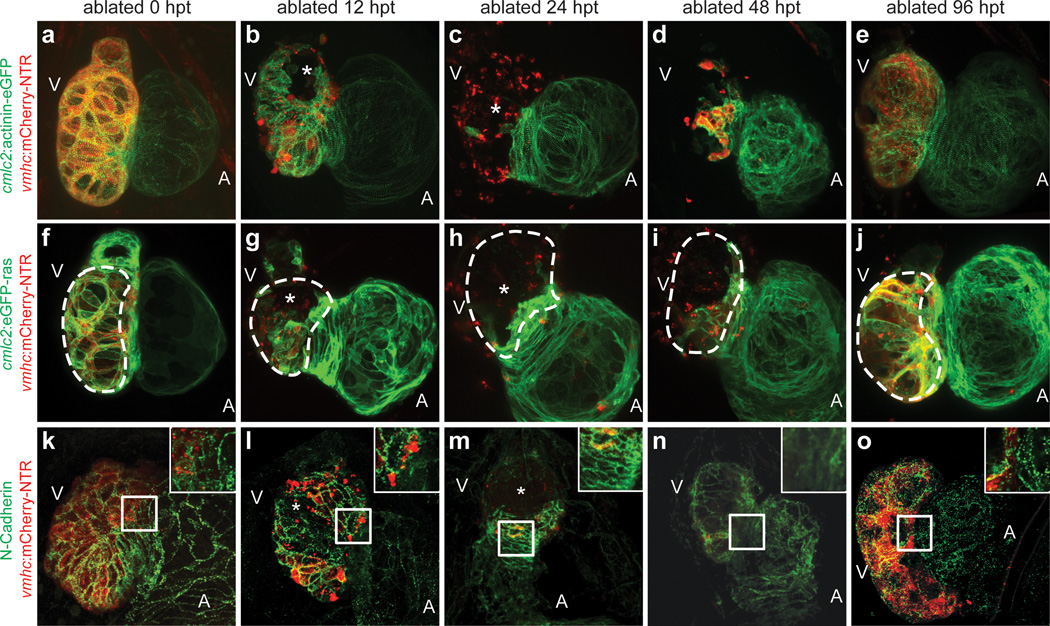
To further elucidate the cellular events that take place during ventricular injury and recovery including the atrial-to-ventricular cardiac lineage switch, we analyzed the cardiomyocyte structural and morphologic changes after ventricular ablation. To this end, we crossed the Tg(vmhc:mCherry-NTR) fish to the reporter lines, Tg(cmlc2:actinin-eGFP)sd10 or Tg(cmlc2:eGFP-ras)s883, which labels sarcomeric structures16 or outlines cardiomyocytes17, respectively (Fig. 3a–j, Supplementary Fig. 14), as well as performed N-cadherin immunostaining to examine cardiomyocyte cell-cell junctions (Fig. 3k–o). During ventricular injury, damaged ventricular cardiomyocytes lost their rod-like shape18, particularly at the outer curvature (Fig. 3g, h, asterisks), and as reported4,5, exhibited disorganized or complete loss of sarcomeres and cell junctions between 12–24 hpt (Fig. 3b, c, l, m, asterisks), when ventricular function has reached its nadir. In contrast, during ventricular recovery (36–48 hpt), we observed the reorganization of not only ventricular but also atrial cardiomyocyte sarcomeres, morphologies, and adhesion junctions, (Fig. 3d, i, n, Supplementary Fig. 14e). In particular, atrial cardiomyocytes at the AVC became more circular (Supplementary Fig. 14b, e) and lost N-cadherin localization at their cell junctions when they began to extend into the ventricle and differentiate into ventricular cardiomyocytes (Fig. 3n). Moreover, we also observed that these migrating atrial cardiomyocytes at the AVC initially displayed disorganized sarcomeric structures (Supplementary Fig. 14d asterisk), but after populating the ventricle, exhibited enriched actinin-eGFP at the cell periphery/cortex (Fig. 3d, Supplementary Fig. 14d - box, d’ - arrow) and frequently short bundles of actinin-eGFP labeled z-line structures (Fig. 3d, Supplementary Fig. 14d’, arrowhead), closely resembling de novo sarcomere assembly16. By 96 hpt when the ventricle has functionally recovered, the ventricular cardiomyocytes derived from reprogrammed atrial cardiomyocytes exhibited not only organized sarcomeres that align with other ventricular cardiomyocytes but also localization of N-cadherin at the cell junctions (Fig. 3e, o). Overall, these results reveal that atrial cardiomyocytes undergo activated cellular reprogramming following ventricular myocardial injury in order to initiate cellular events that permit their proliferation, migration, and transdifferentiation to facilitate ventricular regeneration.
Figure 3. Dynamic cellular remodeling occurs during zebrafish cardiac regeneration.
MTZ-treated (a–e) Tg(vmhc:mCherry-NTR;cmlc2:actinin-eGFP); (f–j) Tg(vmhc:mCherry-NTR;cmlc2:eGFP-ras); and (k–o) N-cadherin immunostained Tg(vmhc:mCherry-NTR) zebrafish. (a, f, k) 0 hpt; (b, g, l) 12 hpt; (c, h, m) 24 hpt; (d, i, n) 48 hpt; (e, j, o) 96 hpt. Asterisks - ventricular cardiomyocyte injury and death due to ablation. Dashed lines - outer curvature ventricular cardiomyocytes. Inset - enlargement of boxed area in k-o at AVC. A - atrium; V - ventricle.
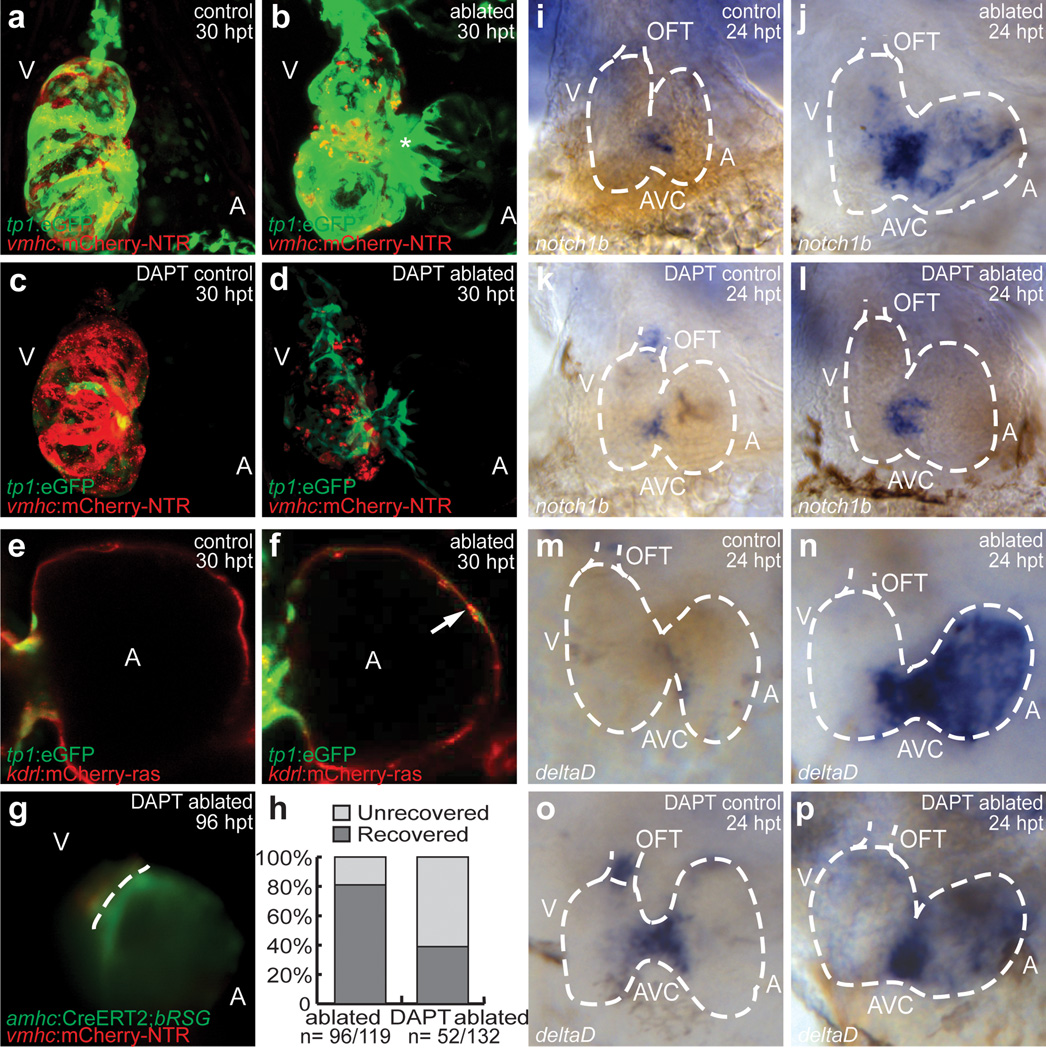
Because previous studies have shown that endocardial activation may be essential to regulate cardiac regeneration19, we examined Raldh2 expression in ventricle ablated Tg(vmhc:mCherry-NTR) fish. Although Raldh2 is expressed weakly in control treated hearts (Supplementary Fig. 15a, c, g, h), its expression is induced throughout the endocardium of the ventricle-ablated hearts by 24–48 hpt (Supplementary Fig. 15b, d, k, l). Using the Tg(tp1:eGFP)um14 Notch reporter line20, we also observed that Notch signaling (i.e. tp1:eGFP expression) was specifically activated in atrial endocardial cells of Tg(vmhc:mCherry-NTR) MTZ-ablated hearts, as detected by the overlapping expression between tp1:eGFP and kdrl:mCherry (Tg(kdrl:ras-mCherry)s896), which labels endothelial/endocardial cell membranes in red21 (compare Fig. 4a to b, 4e to f). Whole mount in situ hybridization of Notch signaling components revealed that notch1b and deltaD, which are normally present in the AVC and OFT of uninjured hearts (Fig. 4i, m), exhibited increased AVC expression as well as activated atrial expression in ventricular-ablated hearts (Fig. 4j, n); however, deltaC was absent (Supplementary Fig. 16). Because tp1:eGFP expression was strong in the ventricle (Fig. 4a, b), but notch1b and deltaD expression was not, we performed gfp RNA in situ analysis to confirm this tp1:eGFP ventricular fluorescence and discovered that gfp RNA expression was weak to negligible in both uninjured and injured ventricles (Supplementary Fig. 17a, b), suggesting that the relatively robust tp1:eGFP ventricular fluorescence may be due to eGFP protein perdurance. Finally, we investigated whether these Notch-activated atrial endocardial cells genetically marked by Tg(kdrl:Cre)s898;Tg(β-act2:RSG)13 could reprogram into cardiomyocytes to contribute to the regeneration of the ventricle-injured heart, but found no evidence for such an event (data not shown). Conversely, we did not observe any of the genetically marked atrial cardiomyocytes (c-aGFP+) becoming endocardial cells (Supplementary Fig. 15i–l).
Figure 4. Notch signaling is required for zebrafish cardiac regeneration.
(a–d) 30 hpt Tg(vmhc:mCherry-NTR;tp1:eGFP) (a) control; (b) ablated; (c) DAPT-treated control; (d) DAPT-treated ablated hearts. (e, f) Atrium of 30 hpt Tg(vmhc:mCherry-NTR;tp1:eGFP;kdrl:mCherry) (e) control and (f) ablated hearts. Arrow - Endocardial tp1:eGFP expression. (g) DAPT-treated 96 hpt Tg(vmhc:mCherry-NTR;amhc:CreERT2;β-act2:RSG) ablated ventricles. Dashed line - atrioventricular boundary. (h) Recovery of 96 hpt control and DAPT-treated Tg(vmhc:mCherry-NTR;amhc:CreERT2;β-act2:RSG) ablated fish. (i–l) notch1b and (m–p) deltaD in situ hybridization on (i, k, m, o) control or (j, l, n, p) ablated Tg(vmhc:mCherry-NTR) hearts (dashed lines) at 24 hpt. (k, l, o, p) DAPT-treated. A - atrium; AVC - atrioventricular canal; OFT - outflow tract; V - ventricle.
Thus, to further investigate the significance of this injury-induced atrial Notch activation, we treated ventricle-ablated hearts with the γ-secretase inhibitor, DAPT. As a result, DAPT treatment significantly decreased the ability of these hearts to restore their ventricular morphology and function after injury (control 81%, N=96/119 vs. DAPT-treated 39%, N=52/132; Fig. 4h), which may be due to overall decreased atrial and ventricular cardiomyocyte proliferation (Supplementary Fig. 18). To determine how DAPT impairs this recovery process, we examined the expression of tp1:eGFP as well as the migration of genetically labeled atrial cardiomyocytes in DAPT-treated ventricle-injured hearts. We observed not only decreased tp1:eGFP expression in the atrium (Fig. 4d) but also a reduction in the migration of c-aGFP+ atrial cardiomyocytes into the injured-ventricle (Fig. 4g). Consistent with these findings, DAPT also inhibited notch1b, deltaD, and tp1:egfp expression in the atrium of ventricle-injured hearts (Fig. 4l, p, Supplementary Fig. 17d), but interestingly increased notch1b and deltaD, in the OFT of uninjured hearts (Fig. 4k, o). Thus, these data suggest that atrial endocardial Notch activation may be crucial to non-cell-autonomously regulate ventricle regeneration through the modulation of atrial-to-ventricular transdifferentiation.
Overall, these results reveal that in vivo reprogramming of differentiated atrial cardiomyocytes can be induced by cardiac ventricular injury through Notch-mediated signaling and proceeds in distinct intermediate stages (Supplementary Fig. 19) in order to provide a novel endogenous cellular source for cardiac ventricular regeneration in zebrafish. Similar to recent cardiac-injury studies in zebrafish and mouse neonatal hearts, this process may be mediated through the activation of Raldh2 and Notch pathways19,22, which may lead to sarcomeric reorganization4,5, cardiomyocyte migration4,5, and the re-expression of key early cardiac transcriptional regulators, such as Gata4, Hand2, Mef2, Nkx2.5, Tbx5, and Tbx205,6,23. Although Raldh2 was expressed throughout the endocardium of the ventricle-injured heart as previously described6,19, Notch signaling was activated primarily in the atrial endocardium. As a result, we speculate that Notch-activated atrial endocardium may influence the atrial myocardium, just as Notch signaling in the ventricular endocardium influences ventricular trabeculation24. Given that previous studies have shown that specific congenital heart diseases can arise from aberrant Notch signaling25, our studies also raise the possibility that abnormal reprogramming of cardiac lineages may be partially responsible for these heart defects.
Though it remains to be elucidated whether mammalian atrial cardiomyocytes may also have a comparable transdifferentiation capacity, recent studies have indicated that mammalian cardiomyocyte progenitor cells (CPMCs) appear to be enriched in the atrium26–28 suggesting that the zebrafish transdifferentiating atrial cardiomyocytes may be analogous to the atrial-resident adult CPMCs. Alternatively, mammalian atrial cardiomyocytes could reprogram into ventricular cardiomyocytes through the reactivation of early cardiac transcriptional factors, which has been recently reported to reprogram cardiac fibroblasts into ventricular cardiomyocytes in mouse hearts29,30. Thus, future studies in mammalian ventricular injury models6 using similar atrial genetic lineage tracing strategies as presented here, are warranted to further investigate the potential of this cardiac reprogramming process as an endogenous cellular regenerative therapy in human heart failure patients. Such a cardiac regenerative strategy could overcome current obstacles facing the translation of stem cell therapies for heart failure patients, including the differentiation of autologous cardiac progenitor cells into ventricular cardiomyocytes as well as the delivery and integration of these differentiated ventricular cardiomyocytes into the patient’s ventricular myocardium.
Methods Summary
Chamber-specific ablation and reporter lines were generated using the standard I-SceI meganuclease transgenesis technique (details in Methods). To perform ventricular cardiomyocyte ablation, Tg(vmhc:mCherry-NTR) zebrafish were treated with 5 mM MTZ as previously described9. For lineage tracing experiments, Tg(vmhc:mCherry-NTR;amhc:CreERT2;β-act2:RSG) zebrafish were treated with 10 µM 4-hydroxytamoxifen as previously described5. For Notch inhibition studies, zebrafish were treated with 100 µM DAPT. Live imaging, heart contraction, immunofluorescence, and whole mount in situ hybridization were performed as described in Methods.
Methods
Zebrafish husbandry and generation of transgenic fish lines
Zebrafish were raised under standard laboratory conditions at 28°C. We used the following transgenic lines: Tg(β-actin2:loxP-DsRed-STOP-loxP-eGFP)s928 - Tg(β-act2:RSG)5, Tg(cmlc2:actinin-eGFP)sd10 (16), Tg(cmlc2:eGFP-ras)s883(17), Tg(tp1:eGFP)um14(20)s, Tg(kdrl:ras-mCherry)s896(21) and Tg(kdrl:Cre)s898(13). The Tg(tp1:eGFP)um14 notch reporter line used for these studies was previously confirmed for reporting notch activity in the heart as well as other tissues using the mindbomb mutant25,31,32. The construct to create Tg(vmhc:mCherry-NTR)s957 was generated by cloning a 2 kb vmhc promoter10 into the pBSKi vector33, and the mCherry-nitroreductase fusion gene was then cloned downstream of the vmhc promoter. The construct to create Tg(amhc:eGFP)s958 was generated by cloning a 982 bp amhc promoter (forward primer: 5’-gctaaagtggcagtgtgccg and reverse primer: 5’-cgtgaatatggttttcaggag) into the pBSKi vector, and GFP was cloned downstream of the amhc promoter. The construct to create Tg(amhc:CreERT2)sd20 was generated by cloning a 4.5 kb amhc promoter (forward primer: 5’-cctggaactcaacaatgctc and reverse primer: 5’-gatctggatctctcctcagttg) into the pBSKi vector, and CreERT234 was cloned downstream of the amhc promoter. We injected 200 pg of each linearized DNA into one-cell-stage embryos and selected individual transgenic carrier adults by screening for fluorescent progeny.
MTZ treatment
Tg(vmhc:mCherry-NTR) larval and adult zebrafish were treated with 5 or 10 mM MTZ in egg or zebrafish system water for four hours and two days, respectively at 28°C in the dark as previously described35. As controls, age-matched Tg(vmhc:mCherry-NTR) siblings were incubated in 0.2% DMSO in egg or zebrafish system water. Treated zebrafish were washed with several changes of fresh egg or zebrafish system water at the end of ablation and then allowed to continue to grow in fresh egg or zebrafish system water.
TUNEL
Using the in situ cell death detection kit, fluorescein from Roche (#11684795910), fish hearts were examined for cell death by TUNEL staining. Specifically, zebrafish at the indicated stages were fixed with 4% PFA, permeabilized with PBS/0.5% TritonX-100 and then incubated in TUNEL staining solution at 37°C for 2 hours.
Immunofluorescence
Immunofluorescence staining was performed as previously described36. The primary antibodies used in this study include: anti-GFP (chicken; Aves Labs), anti-Isl11 (mouse; Developmental Studies Hybridoma Bank), anti-Mef2/C-2115 (rabbit; Santa Cruz Biotechnology), MF20/anti-MHC37 (mouse; Developmental Studies Hybridoma Bank), anti-N-cadherin38 (rabbit; courtesy of Dr Q. Liu), anti-phospho-histone H3 (rabbit; Upstate), anti-Raldh219 (rabbit; Abmart), and S46/anti-Amhc37 (mouse; Developmental Studies Hybridoma Bank). For the vmhc fluorescent in situ hybridization studies, GFP antibody was used to detect the GFP genetically labeled atrial cardiomyocytes. Raldh2 antibody was used to examine endocardial and epicardial activation during cardiac injury19. The secondary antibodies used in this study include: Alexa Fluor 488 goat anti-mouse IgG, Alexa Fluor 488 goat anti-rabbit IgG, Alexa Fluor 594 goat anti-mouse IgG, Alexa Fluor 633 goat anti-mouse IgG, and Alexa Fluor 647 goat anti-rabbit IgG from Invitrogen, and Anti-chicken IgY-FITC from Sigma. Fluorescent images were obtained using a Leica SP5 or Nikon C2 confocal microscope.
in situ hybridization
Whole mount in situ hybridization was performed as previously described18, using the following probes: amhc, deltaC (courtesy of Dr J. Lenis), deltaD (courtesy of Dr W. Clements), gata4, gfp, hand2, irx1a, nkx2.5, notch1b (courtesy of Dr M. Lardelli), tbx5, tbx20, and vmhc. vmhc fluorescent in situ hybridization37 was performed as previously described39 using single digoxygenin-labeled riboprobe and detected by TSA Plus Cy5 Solution (Perkin Elmer).
Cardiac contractile analysis
Live zebrafish were embedded in 1% low melting agarose in a glass bottom culture dish (MatTek), and the heart contraction was recorded by a high speed EMCCD camera (Hamamatsu). The heart function was analyzed as previously described40. The fractional area change was calculated as FC = (End diastolic area – End systolic area)/End diastolic area * 100%.
Intracellular action potential recording
Adult zebrafish hearts were mounted in a chamber containing Tyrode’s solution of the following composition (mM): NaCl 150, KCl 5.4, MgSO4 1.5, NaH2PO4 0.4, CaCl2 2, Glucose 10, HEPES 10. The pH of the solution was adjusted to 7.4 with NaOH. Glass pipettes with tip resistance 30–40 MΩ were filled with 3M KCl solutions. The hearts were allowed to equilibrate for 10 min before spontaneous intracellular action potentials were recorded using an Axopatch 200B amplifier and pClamp10.3 software (Molecular Devices, LLC). Electrophysiologic intracellular recordings were performed on transdifferentiated ventricular cardiomyocytes (c-aGFP+/vCherry+), endogenous ventricular cardiomyocytes (c-aGFP−/vCherry+) and atrial cardiomyocytes (c-aGFP+/vCherry−). All experiments were performed at room temperature (20–22°C). N=5 hearts.
Tamoxifen treatment and lineage tracing
Tg(vmhc:mCherry-NTR;amhc:CreERT2; β-act2:RSG) zebrafish were treated with 10 µM 4-Hydroxytamoxifen (4-HT) solution (Sigma) or 0.1% ethanol alone (control) at 72 hpf for 6 h at 28°C and then washed with fresh egg water several times. The ventricles of hearts with GFP genetically labeled atrial cardiomyocytes were then ablated at 96–120 hpf (n = 76) or at 4 months (n = 23). Live zebrafish were embedded in 0.5% low melting agarose in a glass bottom culture dish as previously described41 for time lapse imaging using a Leica M205FA stereomicroscope with a DFC310FX camera. The hearts were then collected at indicated time points post treatment, cryo-sectioned and examined under a Nikon C2 confocal microscope.
Cryosection and histology staining
Adult zebrafish hearts were fixed in 4% PFA for two hours at room temperature and equilibrated through 15% and then 30% sucrose in PBS for several hours until they sank down to the bottom. Hearts were embedded in O.C.T. compound (Tissue-Tek, 4583) and frozen in liquid nitrogen. 10 µm cryosections were prepared on a Leica CM3050 S cryostat. Masson’s trichrome staining was performed using the Sigma HT15 kit.
DAPT treatment
Zebrafish were incubated in egg water with 100 µM DAPT (Sigma) or 0.1% DMSO alone (control) right after ablation until the zebrafish were fixed or examined at indicated stages. Treated zebrafish were washed several times in egg water after DAPT or DMSO treatment.
Quantification and statistical analysis
To measure cardiac chamber size, Tg(vmhc:mCherry-NTR) hearts were visualized by bright field microscopy at the indicated conditions, and the cardiac chamber size was calculated by measuring the surface area of outlined chambers using Leica LAS AF software (N=10). To determine average fluorescence intensity for each chamber, Tg(vmhc:mCherry-NTR; cmlc2:actinin-eGFP) hearts were visualized by confocal microscopy at the indicated stages, and average fluorescence intensity was calculated by measuring the fluorescence intensity of outlined maximal projections of cardiac chambers using Nikon NIS Elements software (N=5). Cardiomyocyte surface area and circularity were measured by outlining single cardiomyocytes on confocal maximal projection images of Tg(vmhc:mCherry-NTR; cmlc2:eGFP-ras) hearts using the Nikon NIS Elements software (N=15, 5 cells per heart, 3 hearts) as previously described18. Circularity is calculated as Circularity = 4 *π* Area / Perimeters2. To determine the percentage of the Tg(vmhc:mCherry-NTR;amhc:CreERT2;β-act2:RSG) ventricles that is GFP positive, the measured GFP positive surface area of the cardiac ventricles as detected by confocal microscopy was divided by the measured total surface area of the cardiac ventricles as detected by brightfield microscopy. To determine the surface area and average fluorescence intensity of the Tg(vmhc:mCherry-NTR;amhc:CreERT2;β-act2:RSG) atria, the atrial chamber surface area and fluorescence were measured in the bright field and GFP channel using the Nikon NIS Elements software (N=5). p values were obtained by unpaired two-tailed Student's t-test.
Supplementary Material
Acknowledgements
We thank N. Tedeschi, L. Pandolfo, and A. Ayala for fish care; O. Huang, J. Kim, T. Kuo, and J. Sun for experimental assistance; S. Tu and other lab members for comments on the manuscript; Q. Liu for anti-N-cadherin antibody; M. Lardelli, J. Lenis, and W. Clements for plasmids; and N. Lawson for the Notch reporter line. This work was supported in part by grants from the American Heart Association to D.Y. (0940041N), R.Z. (11POST7090024) and H.Y. (12POST12050080); the Packard Foundation and the NIH (HL54737) to D.Y.R.S.; NIH/NHLBI to J.C.; and the NIH (OD007464, HL104239) to N.C.C.
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Author Contributions: N.C.C. initiated the project when he was in D.Y.R.S.’ lab by generating and validating some of the cardiac chamber specific transgenic lines. Additional experimental design was done with the help of R.Z. and D.Y. R.Z., P.H., K.O., D.L., G.K., and N.C.C. conducted experiments. R.Z., H.Y., and N.C.C. generated and characterized transgenic lines for lineage tracing. K.O. and J.C. helped with cardiac function analysis. Y.F.L. and D.Y. provided key reagents. R.Z., P.H., D.L., D.Y. and N.C.C. prepared the manuscript. All authors commented on the manuscript.
Author Information: Reprints and permissions information is available at www.nature.com/reprints. The authors declare no competing financial interests.
Reference
- 1.Bergmann O, et al. Evidence for cardiomyocyte renewal in humans. Science. 2009;324:98–102. doi: 10.1126/science.1164680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Senyo SE, et al. Mammalian heart renewal by pre-existing cardiomyocytes. Nature. 2013;493:433–436. doi: 10.1038/nature11682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Laflamme MA, Murry CE. Heart regeneration. Nature. 2011;473:326–335. doi: 10.1038/nature10147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jopling C, et al. Zebrafish heart regeneration occurs by cardiomyocyte dedifferentiation and proliferation. Nature. 2010;464:606–609. doi: 10.1038/nature08899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kikuchi K, et al. Primary contribution to zebrafish heart regeneration by gata4(+) cardiomyocytes. Nature. 2010;464:601–605. doi: 10.1038/nature08804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Porrello ER, et al. Transient regenerative potential of the neonatal mouse heart. Science. 2011;331:1078–1080. doi: 10.1126/science.1200708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McDonnell TJ, Oberpriller JO. The response of the atrium to direct mechanical wounding in the adult heart of the newt, Notophthalmus viridescens. An electron-microscopic and autoradiographic study. Cell Tissue Res. 1984;235:583–592. doi: 10.1007/BF00226956. [DOI] [PubMed] [Google Scholar]
- 8.Oberpriller JO, Oberpriller JC, Aafedt BC. Changes in binucleation and cellular dimensions of rat left atrial myocytes after induced left ventricular infarction. Am J Anat. 1987;179:285–290. doi: 10.1002/aja.1001790310. [DOI] [PubMed] [Google Scholar]
- 9.Curado S, et al. Conditional targeted cell ablation in zebrafish: a new tool for regeneration studies. Dev Dyn. 2007;236:1025–1035. doi: 10.1002/dvdy.21100. [DOI] [PubMed] [Google Scholar]
- 10.Zhang R, Xu X. Transient and transgenic analysis of the zebrafish ventricular myosin heavy chain (vmhc) promoter: an inhibitory mechanism of ventricle-specific gene expression. Dev Dyn. 2009;238:1564–1573. doi: 10.1002/dvdy.21929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Pater E, et al. Distinct phases of cardiomyocyte differentiation regulate growth of the zebrafish heart. Development. 2009;136:1633–1641. doi: 10.1242/dev.030924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Poss KD, Wilson LG, Keating MT. Heart regeneration in zebrafish. Science. 2002;298:2188–2190. doi: 10.1126/science.1077857. [DOI] [PubMed] [Google Scholar]
- 13.Bertrand JY, et al. Haematopoietic stem cells derive directly from aortic endothelium during development. Nature. 2010;464:108–111. doi: 10.1038/nature08738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhou Y, et al. Latent TGF-beta binding protein 3 identifies a second heart field in zebrafish. Nature. 2011;474:645–648. doi: 10.1038/nature10094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hinits Y, et al. Zebrafish Mef2ca and Mef2cb are essential for both first and second heart field cardiomyocyte differentiation. Dev Biol. 2012;369:199–210. doi: 10.1016/j.ydbio.2012.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lin YF, Swinburne I, Yelon D. Multiple influences of blood flow on cardiomyocyte hypertrophy in the embryonic zebrafish heart. Dev Biol. 2012;362:242–253. doi: 10.1016/j.ydbio.2011.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.D'Amico L, Scott IC, Jungblut B, Stainier DY. A mutation in zebrafish hmgcr1b reveals a role for isoprenoids in vertebrate heart-tube formation. Curr Biol. 2007;17:252–259. doi: 10.1016/j.cub.2006.12.023. [DOI] [PubMed] [Google Scholar]
- 18.Chi NC, et al. Genetic and physiologic dissection of the vertebrate cardiac conduction system. PLoS Biol. 2008;6:e109. doi: 10.1371/journal.pbio.0060109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kikuchi K, et al. Retinoic acid production by endocardium and epicardium is an injury response essential for zebrafish heart regeneration. Dev Cell. 2011;20:397–404. doi: 10.1016/j.devcel.2011.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Parsons MJ, et al. Notch-responsive cells initiate the secondary transition in larval zebrafish pancreas. Mech Dev. 2009;126:898–912. doi: 10.1016/j.mod.2009.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chi NC, et al. Foxn4 directly regulates tbx2b expression and atrioventricular canal formation. Genes Dev. 2008;22:734–739. doi: 10.1101/gad.1629408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Raya A, et al. Activation of Notch signaling pathway precedes heart regeneration in zebrafish. Proc Natl Acad Sci U S A. 2003;100(Suppl 1):11889–11895. doi: 10.1073/pnas.1834204100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lepilina A, et al. A dynamic epicardial injury response supports progenitor cell activity during zebrafish heart regeneration. Cell. 2006;127:607–619. doi: 10.1016/j.cell.2006.08.052. [DOI] [PubMed] [Google Scholar]
- 24.Grego-Bessa J, et al. Notch signaling is essential for ventricular chamber development. Dev Cell. 2007;12:415–429. doi: 10.1016/j.devcel.2006.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Luxan G, et al. Mutations in the NOTCH pathway regulator MIB1 cause left ventricular noncompaction cardiomyopathy. Nat Med. 2013;19:193–201. doi: 10.1038/nm.3046. [DOI] [PubMed] [Google Scholar]
- 26.Bu L, et al. Human ISL1 heart progenitors generate diverse multipotent cardiovascular cell lineages. Nature. 2009;460:113–117. doi: 10.1038/nature08191. [DOI] [PubMed] [Google Scholar]
- 27.Laugwitz KL, et al. Postnatal isl1+ cardioblasts enter fully differentiated cardiomyocyte lineages. Nature. 2005;433:647–653. doi: 10.1038/nature03215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Genead R, et al. Early first trimester human embryonic cardiac Islet-1 progenitor cells and cardiomyocytes: Immunohistochemical and electrophysiological characterization. Stem Cell Res. 2010;4:69–76. doi: 10.1016/j.scr.2009.10.001. [DOI] [PubMed] [Google Scholar]
- 29.Song K, et al. Heart repair by reprogramming non-myocytes with cardiac transcription factors. Nature. 2012;485:599–604. doi: 10.1038/nature11139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Qian L, et al. In vivo reprogramming of murine cardiac fibroblasts into induced cardiomyocytes. Nature. 2012 doi: 10.1038/nature11044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ninov N, Borius M, Stainier DY. Different levels of Notch signaling regulate quiescence, renewal and differentiation in pancreatic endocrine progenitors. Development. 2012;139:1557–1567. doi: 10.1242/dev.076000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang Y, Rovira M, Yusuff S, Parsons MJ. Genetic inducible fate mapping in larval zebrafish reveals origins of adult insulin-producing beta-cells. Development. 2011;138:609–617. doi: 10.1242/dev.059097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thermes V, et al. I-SceI meganuclease mediates highly efficient transgenesis in fish. Mech Dev. 2002;118:91–98. doi: 10.1016/s0925-4773(02)00218-6. [DOI] [PubMed] [Google Scholar]
- 34.Feil R, et al. Ligand-activated site-specific recombination in mice. Proc Natl Acad Sci U S A. 1996;93:10887–10890. doi: 10.1073/pnas.93.20.10887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Curado S, Stainier DY, Anderson RM. Nitroreductase-mediated cell/tissue ablation in zebrafish: a spatially and temporally controlled ablation method with applications in developmental and regeneration studies. Nat Protoc. 2008;3:948–954. doi: 10.1038/nprot.2008.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huang W, Zhang R, Xu X. Myofibrillogenesis in the developing zebrafish heart: A functional study of tnnt2. Dev Biol. 2009;331:237–249. doi: 10.1016/j.ydbio.2009.04.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stainier DY, Fishman MC. Patterning the zebrafish heart tube: acquisition of anteroposterior polarity. Dev Biol. 1992;153:91–101. doi: 10.1016/0012-1606(92)90094-w. [DOI] [PubMed] [Google Scholar]
- 38.Kerstetter AE, Azodi E, Marrs JA, Liu Q. Cadherin-2 function in the cranial ganglia and lateral line system of developing zebrafish. Dev Dyn. 2004;230:137–143. doi: 10.1002/dvdy.20021. [DOI] [PubMed] [Google Scholar]
- 39.Brend T, Holley SA. Zebrafish whole mount high-resolution double fluorescent in situ hybridization. J Vis Exp. 2009 doi: 10.3791/1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fink M, et al. A new method for detection and quantification of heartbeat parameters in Drosophila, zebrafish, and embryonic mouse hearts. Biotechniques. 2009;46:101–113. doi: 10.2144/000113078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dong Z, Wagle M, Guo S. Time-lapse live imaging of clonally related neural progenitor cells in the developing zebrafish forebrain. J Vis Exp. 2011 doi: 10.3791/2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.