Abstract
Cell signaling depends on dynamic protein-protein interaction (PPI) networks, often assembled through modular domains each interacting with multiple peptide motifs. This complexity raises a conceptual challenge, namely to define whether a particular cellular response requires assembly of the complete PPI network of interest, or can be driven by a specific interaction. To address this issue we designed variants of the Grb2 SH2 domain (“pY-clamps”) whose specificity is highly biased toward a single phosphotyrosine (pY)-motif among many potential pYXNX Grb2-binding sites. Surprisingly, directing Grb2 predominantly to a single pY site of the Ptpn11/Shp2 phosphatase, but not other sites tested, was sufficient for differentiation of the essential primitive endoderm lineage from embryonic stem cells. Our data suggest that discrete connections within complex PPI networks can underpin regulation of particular biological events. We propose that this directed wiring approach will be of general utility in functionally annotating specific PPIs.
Intracellular signaling pathways are typically assembled through protein-protein interactions (PPI), and in particular through the ability of protein interaction domains to recognize short peptide motifs, often in a fashion that is regulated by post-translational modification within the motifs (Pawson and Nash, 2003). Such interaction domains frequently bind their targets with relatively low affinity and with only modest selectivity. These characteristics can be rationalized in biological terms, as they allow a single protein containing a given interaction domain to associate with multiple partners in a dynamic fashion, and thereby to achieve the flexibility required for a mammalian cell to respond to varied stimuli. As a consequence, signaling pathways are increasingly viewed as being embedded within multi-protein networks.
This complexity is typified by phosphotyrosine (pY)-based signaling networks, which utilize SH2 domains to engage pY motifs with modest levels of specificity and affinity (Liu et al., 2006; Machida et al., 2007); for example, the Grb2 adaptor binds numerous distinct pYXNX motifs on activated receptor tyrosine kinases and scaffolds through its SH2 domain, and effectors through its flanking SH3 domains (Bisson et al., 2011). Motif-based analyses of a proteome reveals hundreds of potential binding sites for Grb2 SH2 (Obenauer et al., 2003); of these, dozens may be present in a particular cellular context.
During early embryogenesis, Grb2 connects FGF4-dependent pY motif(s) to the Ras guanine nucleotide exchange factors (GEF) Sos1/2 (Findlay et al., 2013; Wilder et al., 1997), resulting in activation of the Ras-Erk MAP kinase pathway and specification of the essential primitive endoderm (PrE) lineage from the epiblast (Chazaud et al., 2006). PrE differentiation is initiated by metastable “priming” of pluripotent mouse embryonic stem cells (mESCs) (Findlay et al., 2013), a state characterized by transcriptional suppression of the pluripotency factor Nanog and elevated expression of the differentiation factor Dnmt3b, followed by full lineage commitment (Figure 1A). During this process, the Grb2 SH2 domain associates with multiple tyrosine-phosphorylated proteins in mESCs (Findlay et al., 2013), but whether all of these interactions or a small subset thereof are required for PrE differentiation is unknown.
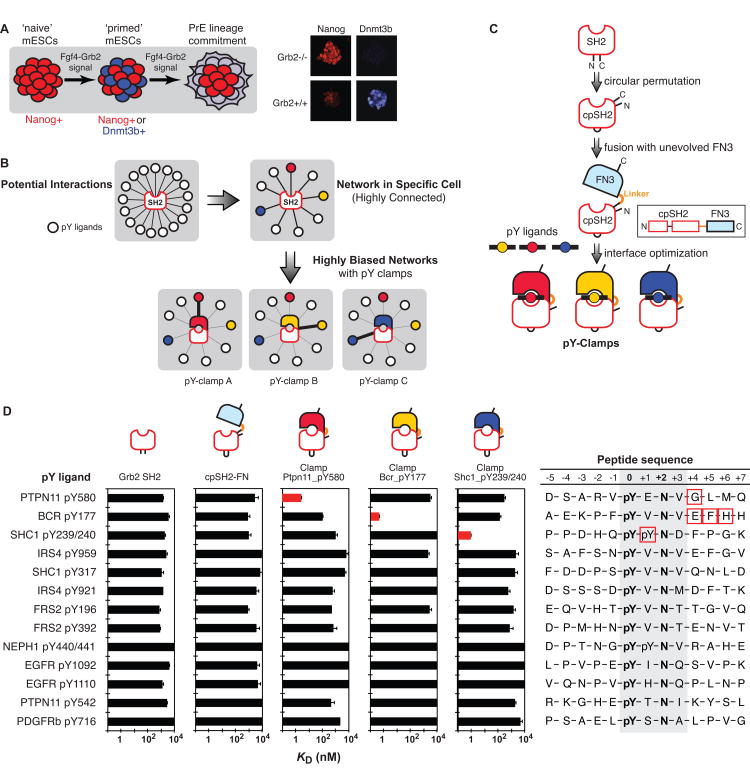
Figure 1. Directed wiring of a biological network using pY clamps.
(A) Model of PrE differentiation from mESCs; upon activation of FGF4-Grb2 signaling, mESCs dynamically segregate into Nanog+ and Dnmt3b+ subpopulations (priming), before proceeding to PrE lineage commitment. Modified from previous data (Findlay et al., 2013). (B) Natural modular interaction domains (e.g. SH2) have modest specificity and bind many ligands (e.g. phosphotyrosine (pY) motifs), leading to a highly connected network. A synthetic binding protein, pY-clamp, binds specifically toward a single pY ligand. By replacing a natural SH2 domain with a pY-clamp, one can dramatically bias a single interaction over the others, thereby allowing directed wiring of the network. (C) An outline of pY-clamp design strategy. The Grb2 SH2 domain is circularly permutated (cpSH2) and fused to the N-terminus of FN3 with a linker. A combinatorial library of FN3 loop residues is constructed, from which pY-clamps with improved affinity and specificity to a pY ligand are identified. See also Figure S1. (D) Binding properties of pY-clamps. KD values of the selected pY-clamps, cpSH2 as well as wild-type Grb2 SH2 to 13 different pY sites measured by the yeast surface display and plotted on the logarithmic scale. Data for the target pY sites are in red. The errors indicated are the standard deviations from curve fitting of the 1:1 binding model. The residues in the red boxes indicate key positions for pY-clamp interaction. See also Figure S2 and Table S1.
This fundamental question in signaling network is extremely difficult to address using conventional approaches. Genetic knockout or knockdown can eliminate a single protein, and the function of the targeted protein is inferred typically based on loss of function. These approaches eliminate an entire protein, often affecting multiple domains and binding motifs simultaneously. Mutation of pY-motifs is an effective method, but our goal here would require many such mutations in different proteins, a technically challenging task. A recent study by Kaneko et al. (Kaneko et al., 2012) generated higher affinity variants, or “superbinders”, of SH2 domains and analyzed the consequences of their introduction to cells. These superbinders act as broad-specificity inhibitors for disrupting binding of any proteins to many pY sites, and thus, while powerful, this technology does not address whether a subset of PPIs within a network are required for a given biological process.
To explore the functional importance of individual PPIs during PrE differentiation, we developed a new approach termed “directed network wiring” (Figure 1B). By employing protein design technologies, we strongly bias the specificity of a modular interaction domain toward a single target site. The full-length, chimeric protein containing such a variant is then introduced to cells that lack the wild-type protein, and the phenotype of the engineered cells is investigated. To test this directed wiring strategy we developed synthetic versions of the Grb2 SH2 domain that bind selectively discrete peptide motifs (dubbed pY-clamps). By applying this technology to an embryonic stem cell system, we were able to connect functional Grb2 signaling complexes to specific pY-motifs in vivo, and demonstrate that the engagement of Grb2 SH2 predominantly with a single pY-motif in Ptpn11 is sufficient to initiate mESC fate change and drive PrE lineage specification. Furthermore, we confirm that this key functional connection is robustly regulated by an FGF4 signal, further defining a role for Ptpn11 in PrE cell fate choice. Our data indicate that individual PPIs within a network play distinct functional roles, and suggest that assembly of complex PPI networks may facilitate the regulation of multiple cellular processes through a single signaling hub.
Results and Discussion
Generation of ligand-specific pY-clamps from the Grb2 SH2 domain
In order to identify key functional Grb2 SH2 domain interactions during embryonic stem cell differentiation, we used affinity clamping technology (Huang et al., 2008; Koide and Huang, 2013) to engineer synthetic protein modules with exquisite binding specificity. Affinity clamping involves attachment of an evolvable fibronectin type III domain (FN3) (Koide et al., 1998; Koide et al., 2012) to a peptide-binding module (in this case a circularly permuted Grb2 SH2 (cpSH2)) and subsequent alteration of surface residues of the FN3 domain via directed evolution so that a target peptide can be “clamped” in the cleft between the two domains (Figures 1C and S1). Not surprisingly, circular permutation reduced the affinity of Grb2 SH2 to most of the pY-motifs tested (Figure 1D). We constructed a large combinatorial library in which the FN3 loops were diversified (Figure S1), and used yeast surface display to generate three pY-clamps, each of which is specific for a distinct pYXNX Grb2 SH2 ligand motif (Figures 1D and S2). The pY-clamps exhibited strong, pY-dependent binding to their target motifs, with KD values in the single or sub-nM range and with minimal cross-reactivity. For example, a pY-clamp directed to the Ptpn11 pY580 motif, termed clamp(Ptpn11_pY580), had a KD value of 2.5 nM for Ptpn11_pY580, but much lower affinity for closely related targets (Figures 1D and S2A and Table S1). These pY peptide-binding properties were recapitulated using purified clamp proteins (Figure S2), confirming the successful generation of pY-clamps that engage three known pY ligands of the Grb2 SH2 domain with high specificity.
Mechanisms of pY-clamp ligand selectivity
To understand the molecular mechanisms underlying highly specific recognition of pY ligand peptides by pY clamps, we first identified important residues for pY-clamp binding within target peptides by alanine-scanning mutagenesis (Figure 2A and Table S2). As expected, position 0, +2 and +3 within the pYXNX recognition motif are critical for binding of all the pY clamps. Clamp(Ptpn11_pY580) and Clamp(Bcr_pY177) extended the recognition motif to the +6 position; consistent with this, Clamp(Bcr_pY177) bound poorly to an IRS4 pY959 peptide that differs from the Bcr pY177 motif only at the +6 position of the seven-residue segment (Figure 1D). In contrast, Clamp(Shc1_pY239/240) did not extend the recognition motif beyond the +3 residue, but showed a strong dependence on the unique +1 pY residue (Figure 2A, right). Thus, the pY-clamps achieved their high specificity by recognizing unique signatures of their target motifs either within or beyond the core pYXNX sequence.
Figure 2. Mechanisms of pY clamp ligand recognition and specificity.
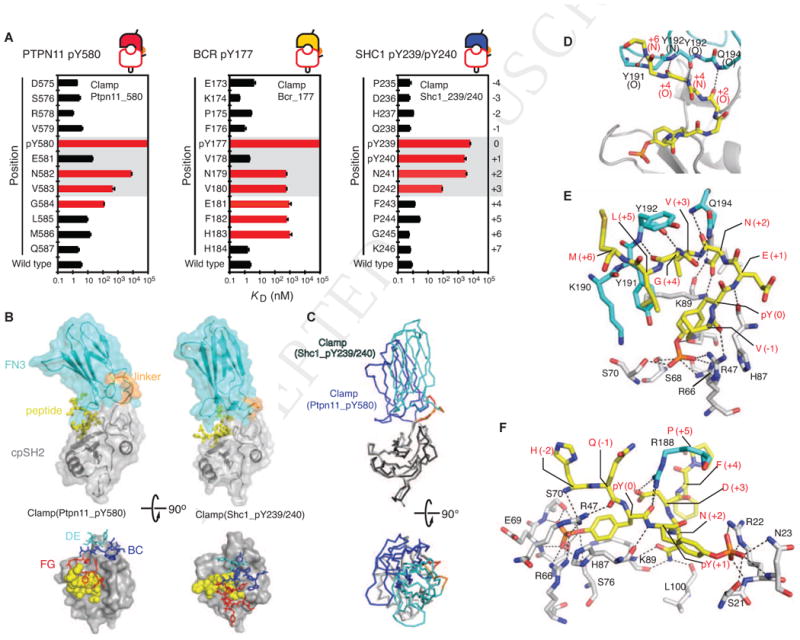
(A) Alanine-scanning analyses of pY peptides. The KD values of the indicated pY clamps for alanine mutants of their cognate peptides, measured by yeast surface display, are shown in the logarithmic scale. The error bars indicate the standard deviations from curve fitting of the 1:1 binding model. See also Table S2. (B) The crystal structures of clamp(Ptpn11_pY580) and clamp(Shc1_pY239/240) in complex with their respective target peptides (top). The cpSH2, FN3 and linker portions of the pY-clamp and the peptide are shown in different colors. The locations of the FN3 loops on the cpSH2 (gray surface) and pY-peptide (yellow spheres) are shown (bottom). (C) Superposition of the two pY-clamps using the cpSH2 domain, showing the different relative orientation of the FN3 enhancer domain. (D) The main-chain interactions of the pY peptide with the FN3 FG loop of clamp(Ptpn11_pY580). The main chain of the residues in the peptide and the FG loop are shown as sticks, hydrogen bonds as dashed lines. The carbon atoms of the FN3 portions are in cyan and those of the peptide in yellow. (E) Polar interactions (dashed lines) in the clamp-peptide interface. The amino acid residues of the cpSH2 and FN3 portions are shown with carbon atoms in gray and cyan, respectively, and labeled in black. The peptide residues are shown with carbon atoms in yellow and labeled in red. The pY residue is engaged completely by cpSH2, whereas Gly(+4) forms extensive interactions with FN3 residues. (F) The interaction between clamp(Shc1_pY239/240) and the SHC1 pY239/240 peptide. See also Table S3.
The crystal structures of clamp(Ptpn11_pY580) and clamp(Shc1_pY239/240) in complex with their target pY peptides revealed that the ligands indeed bind in the cleft between the two domains (Figures 2B and 2C; Table S3). Both pY-clamps substantially enlarged the ligand interaction interfaces compared with the SH2 domain alone (∼1.8- and ∼1.4-fold increases for clamp(Ptpn11_pY580) and clamp(Shc1_pY239/240), respectively; Table S3). As expected, the mode of interaction of the pYXNX segment with the SH2 domain resembles previous SH2-pY peptide structures (Figure 2D–2F), where the pY is engaged by the SH2 pY-binding pocket, and peptides formed a β-turn characteristic of Grb2 SH2 ligands (Rahuel et al., 1996). However, other interactions between the peptide and pY-clamp were distinct for these two complexes. For example, the Y191 and Y192 side chains of the FN3 enhancer in clamp(Ptpn11_pY580) pack against V(+3) and G(+4) in the ligand peptide (Figure 2E), explaining the specificity of this clamp for the Ptpn11 pY580 motif, which uniquely amongst the pYXNX motifs tested contains a G(+4) (Figures 1D and 2A). Similarly, the crystal structure of clamp(Shc1_pY239/240) explains the critical roles of pY(+1) and D(+3) for high-affinity interaction (Figure 2F), consistent with 1,000-fold lower affinity to a peptide containing nonphosphorylated Tyr at 240 (Table S2). Therefore, affinity clamping created extensive new interactions between the FN3 domain and the ligand peptide, resulting in high specificity and affinity. Considerable freedom in the relative orientation of the two connected domains and consequently in the placement of the diversified FN3 loops (Figure 2B and 2C) enables pY-clamps to access diverse interaction modes.
Chimeric Grb2 containing pY-clamps selectively engage their intended pY targets in vivo
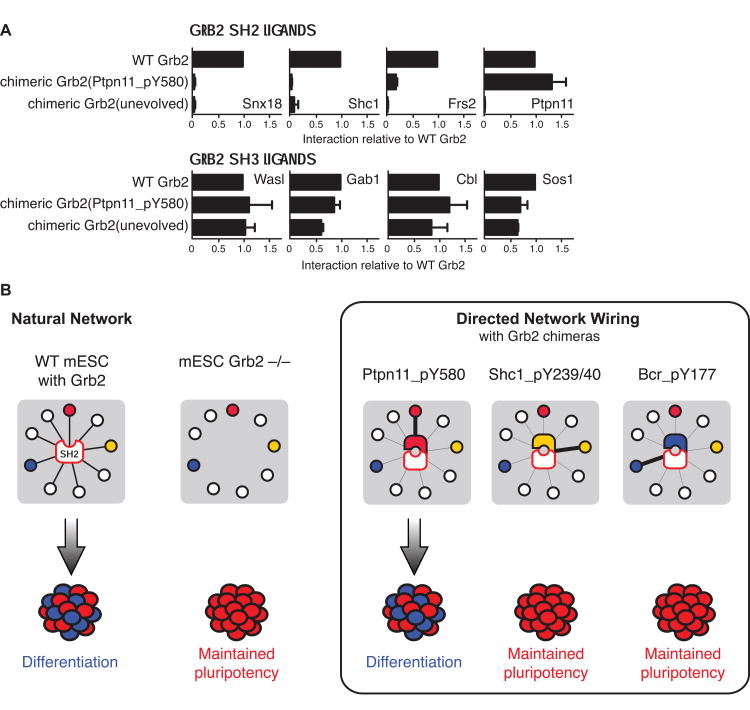
We next examined whether Grb2 SH2 pY-clamps specifically engaged their target ligand in vivo. A chimeric Grb2 protein (chGrb2) in which a pY-clamp replaced the WT SH2 domain (Figure 3A) was introduced into Grb2-/- mESCs. These proteins were expressed at similar levels to endogenous Grb2 and vector-derived WT Grb2 (Figure 3A–3C). Immunoprecipitation followed by immunoblotting with phosphospecific antibodies showed that chGrb2(Ptpn11_pY580), chGrb2(Bcr_pY177) and chGrb2(Shc_pY239/40) bound selectively to the appropriate target motif in endogenous proteins, but not to the other ligands tested (Figure 3B). chGrb2 containing the unevolved FN3 enhancer did not engage these proteins, consistent with the reduced affinity of cpSH2 in vitro (Figure 1D). chGrb2(Ptpn11_pY580) and chGrb2(Bcr_pY177) bound to their target motifs at least as well as WT Grb2. The amount of phosphorylated Shc1 captured by chGrb2(Shc_pY239/240) was much less (Figure 3B), reflecting the fact that this clamp is specific to the doubly phosphorylated form of this motif, a more biologically active but less abundant form than singly phosphorylated Shc1 pY239 (Velazquez et al., 2000; Zheng et al., 2013), and that the antibody used in immunoblotting cross-reacts with additional pY-motifs (see Supplemental Experimental Procedures). We also compared levels of Ptpn11 in the cell lysate to levels found in FLAG IPs of FLAG-tagged WT Grb2 or FLAG-tagged chGrb2(Ptpn11_pY580). Consistent with the improved selectivity and affinity of clamp(Ptpn11_pY580), chGrb2(Ptpn11_pY580) engaged around 38% Ptpn11 in the cell, substantially higher than approximately 20% for WT Grb2 (data not shown). All chGrb2 constructs maintained their ability to interact with Sos1 via the flanking SH3 domains, and can therefore couple functional Grb2 signaling complexes to a specific pY motif (Figure 3B).
Figure 3. Directing Grb2 to Ptpn11 pY580 is sufficient for Grb2 to drive PrE differentiation.
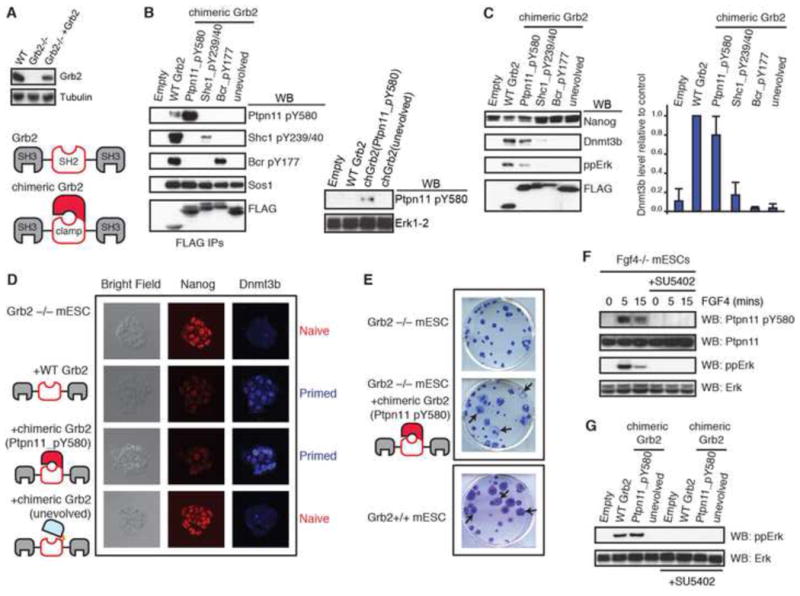
(A) Schematic representation of the domain architecture of wild-type Grb2 and chGrb2 containing a pY-Clamp, and immunoblot showing comparable levels of endogenous and vector-derived Grb2 in mESC. (B) Immunoblot analysis of expression and interaction of FLAG-tagged WT Grb2 or chGrb2 proteins with endogenous Grb2 SH2 ligand motifs Ptpn11 pY580, Shc1 pY239/40, Bcr pY177, and the SH3 ligand Sos1 (left). See also Table S4. See Supplemental Information for technical reasons for the apparently lower engagement of Shc1 pY239/40. An increased level of Ptpn11 pY580 in mESC upon the expression of chGrb2(Ptpn11_pY580) (right). (C) Representative immunoblots of Nanog, Dnmt3b and phospho-Erk levels in Grb2-/- mESCs following expression of WT Grb2 or chGrb2 constructs (left). Quantification of Dnmt3b levels in Grb2-/- mESCs following expression of WT Grb2 or chGrb2 constructs, represented as the average ± standard deviation (n=3; right). (D) Immunostaining of regular Grb2-/- mESC colonies for Nanog and Dnmt3b following expression of WT Grb2 or chGrb2 constructs. (E) PrE differentiation analysis of Grb2-/- mESCs expressing control or chGrb2(Ptpn11_pY580) constructs. The arrows indicate examples of differentiated colonies. Grb2+/+ mESC colony morphology, taken from (Findlay et al., 2013), is given for comparison. (F) Immunoblot analysis of Ptpn11 pY580, total Ptpn11, phospho-Erk and total Erk in Fgf4-/- mESCs. Exogenous Fgf4 (100 ng/ml) was added for the indicated times in the presence or absence of FGFR inhibitor (SU5402, 10 μM). (G) Immunoblot analysis of phospho-Erk and total Erk in Grb2-/- mESCs following expression of WT Grb2 or indicated chGrb2 constructs, and in the presence or absence of FGFR inhibitor (SU5402, 10 μM). See also Figure S3.
The three chGrb2 constructs showed remarkably high selectivity toward their intended target proteins in mESCs, despite pY-clamps exhibiting similar or slightly increased affinity for other pY-motifs compared with WT Grb2 SH2 (Figure 1D and Table S1). These results suggest that pY-ligand engagement by chGrb2 molecules, and probably wild-type Grb2, is determined by relative differences in affinity, rather than the absolute KD values. Slow dissociation rates of the pY-clamps from their target pY-motifs are also likely to contribute to the high selectivity in a cellular context. In addition, the Ptpn11 pY580 site was protected from dephosphorylation in cells expressing chGrb2(Ptpn11_pY580) (Figure 3B), providing further evidence that pY580 is a bona fide target of clamp(Ptpn11_pY580). These observations suggest that different pY-ligands compete for a given SH2 domain in cells, a situation that is not recapitulated in our biochemical experiments measuring binding of SH2 with each ligand separately. Therefore, although the pY-clamps do engage off-targets in vitro, the levels of binding specificity exhibited by these pY-clamps in vivo were sufficiently high to direct chGrb2 molecules predominantly to the desired pY-motifs in cells.
Directing Grb2 SH2 to a pY site in Ptpn11 is sufficient to support FGF4-dependent embryonic stem cell differentiation
We then sought to elucidate the importance of individual interactions within the Grb2 SH2 interaction network during mESC differentiation. Grb2-/- mESCs display a Nanog+ Dnmt3b- ‘naive’ expression signature (Figure 1A), and the metastable ‘primed’ state is reestablished by expression of WT Grb2 (Figure 3C). chGrb2 containing the naïve cpSH2-FN3 (the unevolved clamp) failed to restore mESC priming (Figure 3C), as expected for its reduced affinity for Grb2 SH2 ligands in vitro (Figure 1D) and its inability to bind pY-ligands in mESCs (Figure 3B). Intriguingly, chGrb2(Ptpn11_pY580) activated Erk-MAP kinase and reestablished priming of Grb2-/- mESCs in a manner similar to WT Grb2 (Figure 3C). In contrast, chGrb2(Shc1_pY239/240) or chGrb2(Bcr_pY177) did not restore priming, despite the fact that these clamps engaged their respective ligands in cells. These data suggest that Ptpn11 pY580 may be a functionally important pY-ligand for mESC differentiation.
In order to confirm this finding, we constructed a triple mutant in the pY-binding pocket of cpSH2 (“S88/S90/K109A”) of clamp(Ptpn11_pY580) that had approximately 100-fold weaker affinity to its intended ligand and exhibited much weaker binding to other tested pY-ligands than wild-type Grb2 SH2 (Figure S3A). Remarkably, chGrb2(Ptpn11_pY580) containing these mutations also rescued mESC priming (Figure S3B), suggesting that target specificity, rather than the high affinity of chGrb2(Ptpn11_pY580), is responsible for its biological function. Furthermore, inactivating the ligand-binding properties of either the pY-clamp or the flanking SH3 domains within chGrb2(Ptpn11_pY580) prevented mESC priming (Figure S3B), indicating that recruitment of downstream signaling components, rather than blocking alternative interactions at this pY-motif, is the primary mechanism by which chGrb2(Ptpn11_pY580) functions. Remarkably, Grb2-/- mESCs expressing either WT Grb2 or chGrb2(Ptpn11_pY580), but not Grb2-/- mESCs expressing chGrb2(unevolved), exhibited heterogeneous Nanog and Dnmt3b immunostaining (Figure 3D), similar to that found in the early embryo (Chazaud et al., 2006). Moreover, prolonged expression of chGrb2(Ptpn11_pY580) in Grb2-/- mESCs produced an endodermal-like morphology in a fashion similar to Grb2+/+ mESCs, suggesting that the Grb2 SH2-Ptpn11 pY580 interaction is sufficient for specification of the PrE lineage (Figure 3E).
Autocrine FGF4 signaling is required for mESC priming and PrE differentiation (Feldman et al., 1995). Therefore, we hypothesized that FGF4 may control recruitment of Grb2 to the Ptpn11 pY580 motif in mESCs. Indeed, we confirmed that Ptpn11 Y580 was poorly phosphorylated in Fgf4-/- mESCs, but was induced by FGF4 stimulation in a manner dependent upon FGFR kinase activity (Figure 3F). Furthermore, activation of Erk-MAP kinase by chGrb2(Ptpn11_pY580) or WT Grb2 in Grb2-/- mESCs was also blocked by an FGFR inhibitor, SU5402, indicating that both WT Grb2 and chGrb2(Ptpn11_pY580) require an FGF signal for their biological function (Figure 3G). Finally, we found that FGFR, but not EGFR inhibition, prevents mESC priming (Figure S3C), which, in combination with our previous data showing that only EGFR activity is required for interaction of Grb2 with Shc1 (Findlay et al., 2013), explains the failure of chGrb2(Shc1_pY239/40) to rescue mESC differentiation. Taken together, our results demonstrate that, although the Grb2 SH2 domain is embedded in an extensive network of pYXNX motifs in mESCs, phosphorylation of Ptpn11 Y580 and its engagement by Grb2 is sufficient for FGF-mediated signaling and mESC lineage commitment, whereas other tested pY-Grb2 SH2 interactions are not.
pY-clamp specificity in mESC defined by quantitative mass-spectrometry
Finally, we used immunoprecipitation followed by mass spectroscopy (IP-MS) to investigate chGrb2(Ptpn11_pY580) ligand-specific signaling complexes in mESC (Table S4). Consistent with the analyses using immunoprecipitation followed by immunoblotting above, chGrb2(Ptpn11_pY580) engaged Ptpn11 pY580 and interacted more strongly with Ptpn11 than other Grb2 SH2 ligands, whilst exhibiting a similar interaction profile for SH3 ligands (Table S4). The alanine-scan data (Figure 2A) suggest that clamp(Ptpn11_pY580) can bind tightly to proteins containing the pYXNVG motif, and there are a total of 34 mouse proteins harboring the YXNVG sequence that could produce high-affinity ligands for the clamp upon Tyr phosphorylation. However, we detected none of these proteins except for Ptpn11 in the IP-MS experiments, strongly suggesting that chGrb2(Ptpn11_pY580) did not create a non-natural connection with unintended pY-ligands.
We quantitatively defined the protein interaction network surrounding Grb2 or its variants in mESCs using SWATH IP-MS (Gillet et al., 2012), a technique that can rapidly quantify many individual protein-protein interactions (known Grb2 ligands are defined in (Bisson et al., 2011)). WT Grb2, chGrb2(Ptpn11_pY580) and chGrb2(unevolved) interacted with Sos1, Cbl, Gab1 and Wasl (a representative subset of SH3 ligands) at a similar level (Figure 4A and Figure S3E). In contrast, WT Grb2 bound to the SH2 ligands Ptpn11, Frs2, Shc1 and Snx18, but chGrb2(Ptpn11_pY580) strongly interacted only with Ptpn11 and, to a much lesser extent, Frs2. Clamp(Ptpn11_pY580) has very low affinity for the Grb2 SH2-binding sites of Frs2 (Figure 1D and Table S1) suggesting that chGrb2(Ptpn11_pY580) engages Frs2 indirectly via its interaction with Ptpn11 (Hadari et al., 1998). ChGrb2(unevolved) did not appreciably interact with any of these SH2 ligands (Figure 4A; Table S4). The selective engagement of chGrb2(Ptpn11_pY580) was further corroborated by immunoblotting analysis using an anti-pY antibody, which showed that chGrb2(Ptpn11_pY580) captured much fewer pY-containing proteins than WT Grb2 (Figure S3F). These results are consistent with in vitro binding data (Figure S2A) and those in Figure 3B. Our data therefore indicate that chGrb2(Ptpn11_pY580) connects Grb2 SH3 ligands specifically to the Ptpn11 pY580 motif, and that this interaction is sufficient to drive mESC differentiation (Figure 4B).
Figure 4. Analysis of Ptpn11 pY580 clamp selectivity in vivo using mass-spectrometry.
(A) SWATH-MS based quantification of interaction between chGrb2(Ptpn11_pY580) or chGrb2(unevolved) and a panel of known Grb2 SH2 and SH3 ligands. Data are normalized to WT Grb2, and the relative interaction observed with chGrb2(Ptpn11_pY580) or chGrb2(unevolved) displayed. The average and standard deviation from three replicates are shown. Raw data are provided in Figure S3 and Table S5. (B) Directed network wiring elucidates the role played by individual ligands of the Grb2 SH2 domain during mESC differentiation. The Grb2 SH2 domain complexes with each of Ptpn11 pY580, Shc1 pY239/40 and Bcr pY177 in mESCs, but only Grb2 targeting to Ptpn11 pY580 rescues cell fate change.
In summary, we have developed a protein design strategy for dramatically enhancing the specificity of an SH2 domain toward single pY ligands and employed these synthetic tools to elucidate a functionally important connection within a complex, developmentally critical PPI network (Figure 4B). It is remarkable that highly biasing the Grb2 SH2 domain towards a single pY motif can support a complex biological event such as stem cell differentiation. We therefore propose that complex networks exist to allow related cellular functions to be modulated via a single network hub such as the Grb2 SH2 domain. Whereas the generation of pY-clamps requires substantial protein design expertise, our approach requires only a single cell line for testing different interactions. In theory, one could also address this question by building bypass systems in which two proteins of interest (e.g. Grb2 and Ptpn11 in this case) are directly fused. However, such an approach may be complicated by the necessity to engineer multiple double-knockout cell lines and/or instability and improper regulation of fusion proteins. As genetically encoded in a plasmid, pY-clamps and chimeric proteins harboring them are portable tools for diverse systems. Our success in designing affinity clamps using an SH2 domain here and a PDZ domain previously (Huang et al., 2008) suggests the feasibility of employing affinity clamp technology to diverse modular interaction domains so as to interrogate the biological properties of a wide range of PPI networks, and to construct synthetic pathways and networks with extreme precision.
Experimental Procedures
Full experimental procedures and any associated references are available in the supplemental information. pY-clamps were constructed via the following steps; first, circularly permutated Grb2 SH2 domain (cpSH2) that retained binding of pY-ligands was identified, and two surface mutations, F61Y and F62K were introduced to improve its solubility; second, a yeast-display library was constructed by cloning the genes encoding the cpSH2 domain, a linker and the FN3 domain containing amino acid diversity in the loop regions. pY-peptide ligands were produced as fusion proteins with yeast SUMO and enzymatically phosphorylated. Identities and essentially complete phosphorylation of the peptides were confirmed using mass spectroscopy and chromatography. Sorting of yeast-display libraries was performed using flow cytometry. Individual clones were assessed for their affinity to a panel of pY peptides, and pY-clamps with the highest levels of specificity were produced as isolated proteins and purified for further characterization. Affinity measurements of purified pY-clamps were performed using a Biacore 3000 instrument with a Ni-NTA chip. pY-clamps in complex with their target peptides were crystallized using the hanging-drop vapor-diffusion method, and x-ray diffraction data were collected at the Advanced Photon Source beamlines (Argonne National Laboratories). Data analysis is detailed in the Supplemental Experimental Procedures section.
Grb2-/- (Cheng et al., 1998) and Fgf4-/- (Wilder et al., 1997) mESCs were described previously. Wild-type and chimeric Grb2 constructs were expressed in the pCAGGS vector under the control of a β-actin promoter. Grb2-/- mESCs were transfected using Lipofectamine LTX (Invitrogen), and selected with puromycin for 48 h before analyses. HEK 293T cells were transfected using PEI. For immunoprecipitation, cells were lysed in FLAG IP-MS lysis buffer (Bisson et al., 2011), and triple-FLAG tagged proteins immunoprecipitated with 10 μl of FLAG-M2 Agarose (Sigma-Aldrich). Immunoblotting and mass spectrometry analyses are detailed in the Methods section. SWATH-MS data were acquired on a TripleTOF 5600 instrument (AB SCIEX) in line with a NanoLC-Ultra 1D plus system (Eksigent), with each acquisition recording consecutive high-resolution MS/MS spectra of all precursor ions spanning the mass range of 100-1250 amu. Data were analysed using ProteinPilot, Peakview and MarkerView (AB SCIEX).
Supplementary Material
Highlights.
Defining roles of individual interactions in complex networks is challenging.
Protein design tailors Grb2 SH2 into “pY-clamps” specific for single pY ligands.
Re-wiring the Grb2 PPI network using synthetic Grb2 comprising pY-clamps.
Identified Ptpn11 pY580 as a key Grb2 SH2 ligand in stem cell fate specification.
Acknowledgments
We thank R. J. Hoey for assistance in SPR measurements; S. Dhe-Paganon for reagents. This work was supported by the National Institutes of Health grants R01-GM072688 and R01-GM090324 (SK). Use of the Advanced Photon Source was supported by the U. S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract No. DE-AC02-06CH11357. This paper is dedicated to the memory of Tony Pawson, our beloved colleague and mentor.
Footnotes
Accession numbers: The coordinates and structure factors for clamp(Ptpn11_pY580) and clamp(Shc_pY239/240) have been deposited in the Protein Data Bank under the accession numbers 4JMG and 4JMH, respectively.
Author Contributions: SK and TP conceived the project. NY, JH, WCB, AK and SK developed cpSH2 and pY-clamps. NY biophysically characterized pY-clamps. NY and LS determined the crystal structures. GMF, GG, MSH and TP designed and performed cellular experiments. MT and LT performed mass spectroscopy experiments. NY, GMF, GG, TP and SK wrote the paper with input from the other authors. All authors commented on the paper.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bisson N, James DA, Ivosev G, Tate SA, Bonner R, Taylor L, Pawson T. Selected reaction monitoring mass spectrometry reveals the dynamics of signaling through the GRB2 adaptor. Nat Biotechnol. 2011;29:653–658. doi: 10.1038/nbt.1905. [DOI] [PubMed] [Google Scholar]
- Chazaud C, Yamanaka Y, Pawson T, Rossant J. Early lineage segregation between epiblast and primitive endoderm in mouse blastocysts through the Grb2-MAPK pathway. Dev Cell. 2006;10:615–624. doi: 10.1016/j.devcel.2006.02.020. [DOI] [PubMed] [Google Scholar]
- Cheng AM, Saxton TM, Sakai R, Kulkarni S, Mbamalu G, Vogel W, Tortorice CG, Cardiff RD, Cross JC, Muller WJ, et al. Mammalian Grb2 regulates multiple steps in embryonic development and malignant transformation. Cell. 1998;95:793–803. doi: 10.1016/s0092-8674(00)81702-x. [DOI] [PubMed] [Google Scholar]
- Feldman B, Poueymirou W, Papaioannou VE, DeChiara TM, Goldfarb M. Requirement of FGF-4 for postimplantation mouse development. Science. 1995;267:246–249. doi: 10.1126/science.7809630. [DOI] [PubMed] [Google Scholar]
- Findlay GM, Smith MJ, Lanner F, Hsiung MS, Gish GD, Petsalaki E, Cockburn K, Kaneko T, Huang H, Bagshaw RD, et al. Interaction domains of sos1/grb2 are finely tuned for cooperative control of embryonic stem cell fate. Cell. 2013;152:1008–1020. doi: 10.1016/j.cell.2013.01.056. [DOI] [PubMed] [Google Scholar]
- Gillet LC, Navarro P, Tate S, Rost H, Selevsek N, Reiter L, Bonner R, Aebersold R. Targeted data extraction of the MS/MS spectra generated by data-independent acquisition: a new concept for consistent and accurate proteome analysis. Mol Cell Proteomics. 2012;11:O111 016717. doi: 10.1074/mcp.O111.016717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadari YR, Kouhara H, Lax I, Schlessinger J. Binding of Shp2 tyrosine phosphatase to FRS2 is essential for fibroblast growth factor-induced PC12 cell differentiation. Mol Cell Biol. 1998;18:3966–3973. doi: 10.1128/mcb.18.7.3966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Koide A, Makabe K, Koide S. Design of protein function leaps by directed domain interface evolution. Proc Natl Acad Sci USA. 2008;105:6578–6583. doi: 10.1073/pnas.0801097105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko T, Huang H, Cao X, Li X, Li C, Voss C, Sidhu SS, Li SS. Superbinder SH2 domains act as antagonists of cell signaling. Sci Signal. 2012;5:ra68. doi: 10.1126/scisignal.2003021. [DOI] [PubMed] [Google Scholar]
- Koide A, Bailey CW, Huang X, Koide S. The fibronectin type III domain as a scaffold for novel binding proteins. J Mol Biol. 1998;284:1141–1151. doi: 10.1006/jmbi.1998.2238. [DOI] [PubMed] [Google Scholar]
- Koide A, Wojcik J, Gilbreth RN, Hoey RJ, Koide S. Teaching an Old Scaffold New Tricks: Monobodies Constructed Using Alternative Surfaces of the FN3 Scaffold. J Mol Biol. 2012;415:393–405. doi: 10.1016/j.jmb.2011.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koide S, Huang J. Generation of high-performance binding proteins for Peptide motifs by affinity clamping. Methods Enzymol. 2013;523:285–302. doi: 10.1016/B978-0-12-394292-0.00013-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu BA, Jablonowski K, Raina M, Arce M, Pawson T, Nash PD. The human and mouse complement of SH2 domain proteins-establishing the boundaries of phosphotyrosine signaling. Mol Cell. 2006;22:851–868. doi: 10.1016/j.molcel.2006.06.001. [DOI] [PubMed] [Google Scholar]
- Machida K, Thompson CM, Dierck K, Jablonowski K, Karkkainen S, Liu B, Zhang H, Nash PD, Newman DK, Nollau P, et al. High-throughput phosphotyrosine profiling using SH2 domains. Mol Cell. 2007;26:899–915. doi: 10.1016/j.molcel.2007.05.031. [DOI] [PubMed] [Google Scholar]
- Obenauer JC, Cantley LC, Yaffe MB. Scansite 2.0: Proteome-wide prediction of cell signaling interactions using short sequence motifs. Nucleic Acids Res. 2003;31:3635–3641. doi: 10.1093/nar/gkg584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawson T, Nash P. Assembly of cell regulatory systems through protein interaction domains. Science. 2003;300:445–452. doi: 10.1126/science.1083653. [DOI] [PubMed] [Google Scholar]
- Rahuel J, Gay B, Erdmann D, Strauss A, Garcia-Echeverria C, Furet P, Caravatti G, Fretz H, Schoepfer J, Grutter MG. Structural basis for specificity of Grb2-SH2 revealed by a novel ligand binding mode. Nat Struct Biol. 1996;3:586–589. doi: 10.1038/nsb0796-586. [DOI] [PubMed] [Google Scholar]
- Velazquez L, Gish GD, van Der Geer P, Taylor L, Shulman J, Pawson T. The shc adaptor protein forms interdependent phosphotyrosine-mediated protein complexes in mast cells stimulated with interleukin 3. Blood. 2000;96:132–138. [PubMed] [Google Scholar]
- Wilder PJ, Kelly D, Brigman K, Peterson CL, Nowling T, Gao QS, McComb RD, Capecchi MR, Rizzino A. Inactivation of the FGF-4 gene in embryonic stem cells alters the growth and/or the survival of their early differentiated progeny. Dev Biol. 1997;192:614–629. doi: 10.1006/dbio.1997.8777. [DOI] [PubMed] [Google Scholar]
- Zheng Y, Zhang C, Croucher DR, Soliman MA, St-Denis N, Pasculescu A, Taylor L, Tate SA, Hardy WR, Colwill K, et al. Temporal regulation of EGF signalling networks by the scaffold protein Shc1. Nature. 2013;499:166–171. doi: 10.1038/nature12308. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.