Abstract
Development of SAR in an aryl ether series of mGlu5 NAMs leading to the identification of pyrazine analog VU0431316 is described in this Letter. VU0431316 is a potent and selective non-competitive antagonist of mGlu5 that binds at a known allosteric binding site. VU0431316 demonstrates an attractive DMPK profile, including moderate clearance and good bioavailability in rats. Intraperitoneal (IP) dosing of VU0431316 in a mouse marble burying model of anxiety, an assay known to be sensitive to mGlu5 antagonists and other anxiolytics, produced dose proportional effects.
The metabotropic glutamate receptors (mGlus) comprise a family of eight related G-protein-coupled receptors (GPCRs) wherein each receptor acts through binding glutamate, the major excitatory transmitter in the mammalian central nervous system (CNS). In these seven transmembrane spanning (7TM) receptors, the orthosteric binding sites are located in the extracellular domain while known allosteric binding sites are contained in the transmembrane domain.1 Design of highly selective orthosteric ligands has continually proven difficult due to the extensive homology of the binding sites across the mGlu family. In many instances, the development of allosteric modulators of mGlus has been established as a viable solution to enhancing selectivity among family members.2 Among the individual mGlus investigated as potential drug targets, a substantial portion of that attention has been devoted to the design of small molecule negative allosteric modulators (NAMs), or non-competitive antagonists, of mGlu5.3
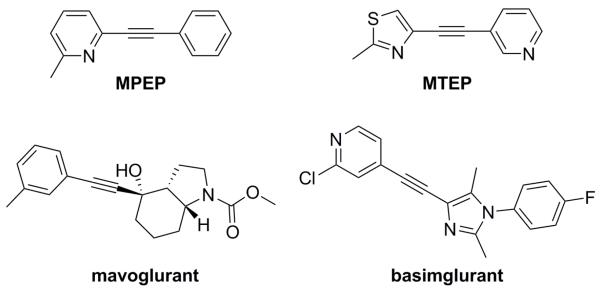
The vast majority of the preclinical behavioral work with mGlu5 NAMs has been conducted using one of two structurally related disubstituted alkyne tool compounds, MPEP4 and MTEP5 (Fig. 1). Efficacy has been reported with these compounds across a number of different disease models. Examples include pain,6 anxiety,7 gastroesophageal reflux disease (GERD),8 Parkinson’s disease levodopa induced dyskinesia (PD-LID),9 fragile X syndrome (FXS),10 and other autism spectrum disorders.11 Furthermore, both MPEP and MTEP have been used effectively in various animal models of addictive behavior with well-known drugs of abuse, such as cocaine,12 nicotine,12g,13 methamphetamine,14 morphine,15 and ethanol.16
Figure 1.
mGlu5 NAM tool and advanced clinical compounds
Currently, several mGlu5 NAMs have progressed to human trials, and results from studies in patients with GERD,17 FXS,18 and PD-LID19 have been encouraging. Though structural diversity among mGlu5 NAMs in the literature has expanded considerably in recent years, the majority of clinical compounds have been from the disubstituted alkyne structure class.3a Furthermore, the most advanced clinical compounds, mavoglurant (AFQ056) and basimglurant (RG7090, RO4917523), each contain the alkyne moiety (Fig. 1).20 Recently, concerns that such alkyne compounds might be prone to metabolic activation and resultant toxicities have proven warranted, at least in one instance. Pfizer has now disclosed their observation of biliary epithelial hyperplasia in non-human primate regulatory toxicology studies with the disubstituted alkyne compound known as GRN-529. Glutathione conjugation at the alkyne moiety was believed to be related to these adverse findings.21
Our mGlu5 NAM program has long been centered on the identification and optimization of compounds from chemotypes that do not contain a disubstituted alkyne motif. The majority of this effort has been spent on the optimization of hits identified from a functional cell-based high-throughput screen (HTS) of a collection of 160,000 compounds;22 however, rational design approaches23 and a virtual screening approach also produced new non-alkyne based mGlu5 NAM tool compounds.24 We recently reported on a lead optimization effort based around hit compound 1 from our functional HTS (Fig. 2).22a This particular optimization effort, based on 1, culminated in discovery of the in vivo tool compound VU0409106.
Figure 2.
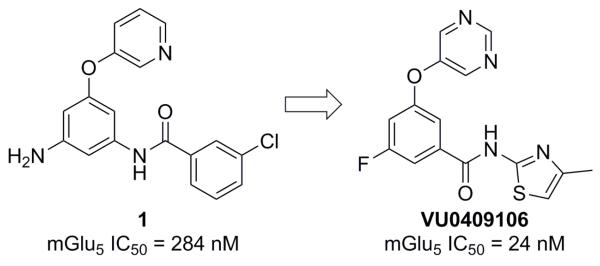
HTS hit 1 and mGlu5 NAM in vivo tool VU0409106
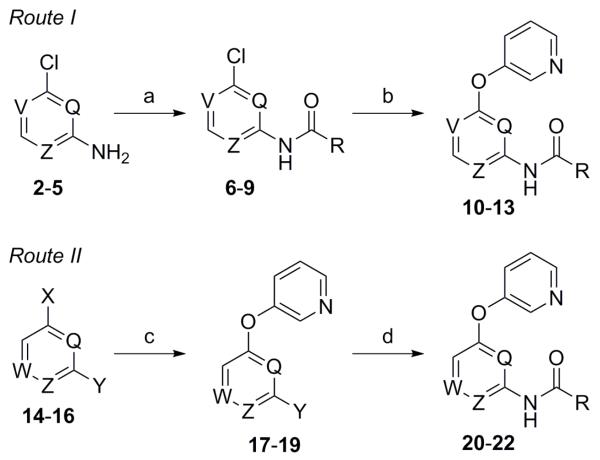
Concomitant to the recently described work that led to the discovery of VU0409106, we were also pursuing additional analogs of 1. Reasoning that a potential route of metabolism for analogs of 1 might include amide bond cleavage, we immediately sought to identify compounds that would not produce electron rich anilines should the amide bond indeed be cleaved in vivo. In the case of VU0409106 and associated analogs, we achieved this goal by reversing the orientation of the amide bond; however, the work described herein centers on the replacement of the phenyl core with heteroaryl rings. Preparation of the initial heteroaryl ether analogs of 1 was executed according to one of the two general methods outlined here (Scheme 1).25
Scheme 1. Reagents and conditions: (a) For 2 (Z = N; V = Q = CH), 3 (V = N; Q = Z = CH), 4 (Q = N; V = Z = CH), and 5 (V = Z = N; Q = CH); RCO2H (R = 3-chlorophenyl), EDC, DMAP, CH2Cl2, (83–94%); (b) 3-hydroxypyridine, CuI, Cs2CO3, Me2NCH2CO2H·HCl (35–55%); (c) For 14 (W = N; Q = Z = CH; X = F; Y = Br), 15 (Q = W = N; Z = CH; X = Y = Cl), and 16 (Q = Z = N; W = CH; X = Y = Cl); 3-hydroxypyridine, K2CO3, DMF, microwave, 150 °C (60–93%); (d) RCONH2 (R = 3-chlorophenyl), NaOtBu, Pd(OAc)2, Xantphos, PhMe, 100 °C (50-78%).
Certain pyridine (10-12) and pyrimidine (13) analogs were prepared by first coupling the commercial amines 2-5 with 3-chlorobenzoic acid to afford the corresponding amides 6-9 (Route I). Reaction with 3-hydroxypyridine in the presence of copper (I) iodide and dimethylglycine afforded the desired compounds 10-13. Alternatively, pyridine 20, pyrazine 21, and pyrimidine 22 were prepared via a route relying on initial installation of the aryl ether (Route II). Reaction of the commercial monomers 14-16 with 3-hydroxypyridine in a microwave assisted nucleophilic aromatic substitution (SNAr) reaction provided heteroaryl halide intermediates 17-19. The final analogs were prepared directly through a Buchwald-Hartwig coupling with 3-chlorobenzamide in moderate to high yields.26
Evaluation of these initial analogs against mGlu5 yielded clear SAR (Table 1). Our functional assay measures the ability of the compound to block the mobilization of calcium induced by an EC80 concentration of glutamate in HEK293A cells expressing rat mGlu5.27 Among the pyridine analogs, compounds 12 and 20 were superior to compounds 10 and 11. In fact both 12 and 20 exhibited potency at a level near hit 1. Pyrimidine analogs 13 and 22 were weak antagonists; however, pyrazine 21 exhibited the best potency in this set of analogs. Having established the pyrazine core as a favorable group for further SAR development, lead optimization continued in that area.
Table 1. Core SAR.
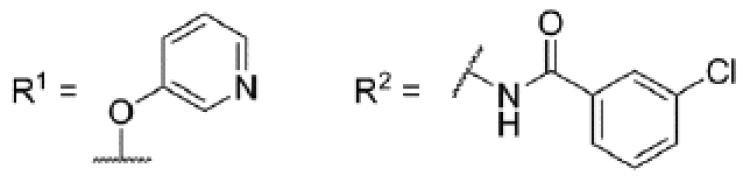
| ||||
|---|---|---|---|---|
|
| ||||
| Cpd | Core | mGlu5 pIC50a (± SEM) |
mGlu5 IC50 (nM) |
% Glu Maxa,b(± SEM) |
| 1 |
|
6.55 ± 0.19 | 284 | 1.3 ± 0.2 |
| 10 |
|
6.00 ± 0.21 | 994 | 1.8 ± 0.7 |
| 11 |
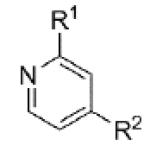
|
5.43 ± 0.26 | 3700 | −4.9 ± 4.7 |
| 12 |
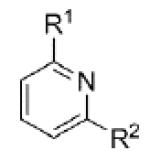
|
6.44 ± 0.19 | 359 | 2.0 ± 0.7 |
| 13 |
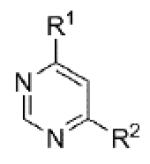
|
5.53 ± 0.23 | 2930 | 1.1 ± 0.5 |
| 20 |
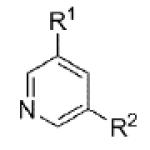
|
6.39 ± 0.07 | 403 | 1.1 ± 0.4 |
| 21 |
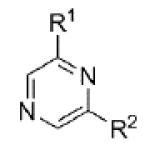
|
6.89 ± 0.04 | 129 | 1.0 ± 0.3 |
| 22 |
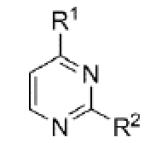
|
<5.0c | >10,000 | 44 ± 8 |
Calcium mobilization mGlu5 assay; values are average of n ≥ 3
Amplitude of response in the presence of 30 μM test compound as a percentage of maximal response (100 μM glutamate); average of n ≥ 3
Concentration response curve (CRC) does not plateau
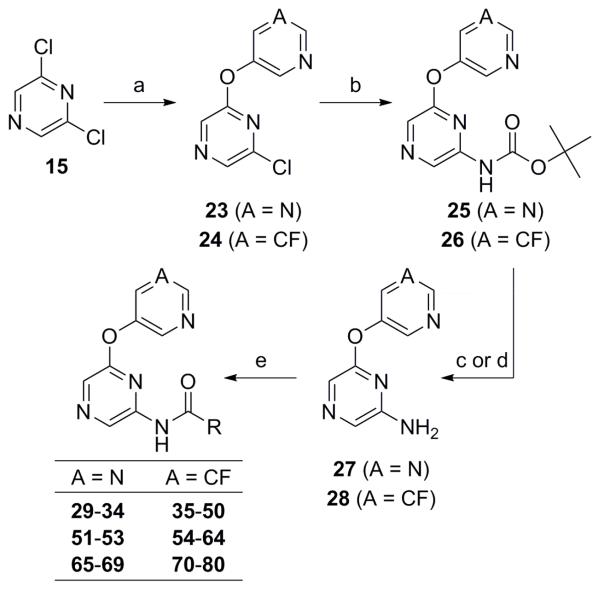
Ongoing research has identified the 5-fluoropyridin-3-yl and pyrimidin-5-yl ethers as optimal groups in the northern portion of the chemotype.22a Thus, much of the SAR was developed in the context of one or both of these moieties. Compounds containing the pyrimidine moiety are less lipophilic than their 5-fluoropyridine counterparts,28 a feature that can often provide advantages with respect to drug-like properties. Though the synthesis outlined in Scheme I (Route II) was utilized to prepare some new pyrazine analogs, a new synthetic route allowing for the preparation of a broader diversity of amides was utilized in most cases (Scheme 2).29 This route also begins with a similar SNAr reaction, providing ethers 23-24. A Buchwald-Hartwig coupling with t-butyl carbamate was employed to afford intermediates 25-26.30 Cleavage of the protecting group was carried out under acidic conditions to yield amines 27-28. Conversion to the desired amide products was accomplished using standard coupling conditions.
Scheme 2. Reagents and conditions: (a) 3-fluoro-5-hydroxypyridine or 5-hydroxypyrimidine, K2CO3, DMF, microwave, 120 °C (51% for 23; 62% for 24); (b) H2NCO tBu, NaO2 tBu, Pd2(dba)3·CHCl3, tBuXPhos, PhMe (46% for 25; 64% for 26); (c) For 25 27, TFA, CH2Cl2 (96%); (d) For 26 28, 4N HCl in dioxane (100%); (e) RCO2H, EDC, DMAP, CH2Cl2 or RCO2H, HATU, DIEA, CH2Cl2, DMF or RCO2H, POCl3, pyridine or RCOCl, DMAP, CH2Cl2 (30–80%).
Evaluation of various substituted benzamides revealed some additional potent compounds (Table 2). As anticipated, both the 5-fluoropyridin-3-yl (35) and pyrimidin-5-yl (29) ethers proved competent replacements for the simple pyridine-3-yl (21) ether. Furthermore, the importance of the 3-chloro substituent on the benzamide was established through preparation of unsubstituted analogs 30 and 36. Many additional 3-substituted analogs were prepared and tested (31-34 and 36-41); however, only the 3-methyl analogs 32 and 38 demonstrated potency comparable to 29 and 35. 3-Cyanobenzamides 33 and 39 were approximately six fold less potent than 3-chlorobenzamide comparators 29 and 35. Additional monosubstituted benzamides demonstrated moderate to weak antagonist activity. Several disubstituted benzamides were also evaluated (42-50), with only 2-fluoro-5-chlorobenzamide 42 demonstrating an IC50 less than one micromolar.
Table 2. Benzamide SAR.
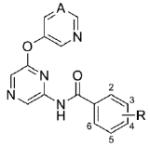
|
|||||
|---|---|---|---|---|---|
|
| |||||
| Cpd | A | R | mGlu5pIC50a (± SEM) |
mGlu5 IC50 (nM) |
% Glu Maxa,b (± SEM) |
| 29 | N | 3-Cl | 6.94 ± 0.10 | 116 | 1.2 ± 0.0 |
| 30 | N | H | <5.0c | >10,000 | 12 ± 11 |
| 31 | N | 3-F | 5.29 ± 0.17 | 5140 | 1.5 ± 2.7 |
| 32 | N | 3-Me | 6.67 ± 0.16 | 212 | 1.5 ± 0.5 |
| 33 | N | 3-CN | 6.15 ± 0.17 | 709 | 1.5 ± 0.2 |
| 34 | N | 3-OMe | 5.39 ± 0.15 | 4110 | −4.3 ± 2.7 |
| 35 | CF | 3-Cl | 6.93 ± 0.10 | 118 | 2.0 ± 0.3 |
| 36 | CF | H | 5.06 ± 0.09 | 8650 | −9.4 ± 2.3 |
| 37 | CF | 3-F | 5.42 ± 0.09 | 3820 | −0.4 ± 1.7 |
| 38 | CF | 3-Me | 6.70 ± 0.18 | 199 | 2.1 ± 0.4 |
| 39 | CF | 3-CN | 6.18 ± 0.16 | 662 | 1.2 ± 0.3 |
| 40 | CF | 3-OMe | 5.36 ± 0.17 | 4400 | −5.6 ± 5.1 |
| 41 | CF | 3-NMe2 | <5.0c | >10,000 | 52 ± 11 |
| 42 | CF | 2-F, 3-Cl | <4.5 | >30,000 | — |
| 43 | CF | 3-Cl, 4-F | 5.34 ± 0.15 | 4590 | −2.8 ± 6.2 |
| 44 | CF | 3-Cl, 5-F | <5.0c | >10,000 | 48 ± 10 |
| 45 | CF | 2-F, 5-Cl | 6.05 ± 0.13 | 894 | 1.0 ± 0.8 |
| 46 | CF | 3,5-di-F | <4.5 | >30,000 | — |
| 47 | CF | 3,5-di-Cl | <4.5 | >30,000 | — |
| 48 | CF | 3,5-di-Me | 5.18 ± 0.18 | 6600 | 6.6 ± 2.0 |
| 49 | CF | 3,5-di-OMe | <4.5 | >30,000 | — |
| 50 | CF | 3-CN, 5-F | <4.5 | >30,000 | — |
Calcium mobilization mGlu5 assay; values are average of n ≥ 3
Amplitude of response in the presence of 30 μM test compound as a percentage of maximal response (100 μM glutamate); average of n ≥ 3
CRC does not plateau
Turning our attention from benzamide analogs to analogs with a heteroaryl amide moiety identified a mixture of weak antagonists and compounds that were inactive up to the top concentration tested of 30 μM (Table 3). Recognizing the importance of substitution that was observed in the case of the benzamide analogs, particularly at the 3-position, we decided to prepare some additional analogs of the weak antagonists identified from this initial set (Table 4). Methyl substitution resulted in modest potency enhancement in the case of thiophene 70 relative to 55. More dramatic potency improvement was noted with picolinamides 67 and 75 when compared to 59. Preparation of additional 4-substituted picolinamides (68, 69, and 76) identified 4-chloropicolinamide 68 (VU0431316) as the most potent mGlu5 NAM in this series.
Table 3. Heteroaryl Amide SAR.
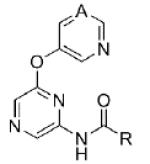
|
|||||
|---|---|---|---|---|---|
|
| |||||
| Cpd | A | R | mGlu5 pIC50a (± SEM) |
mGlu5 IC50 (nM) |
% Glu Maxa,b (± SEM) |
| 51 | N |
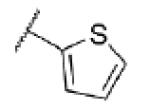
|
<5.0c | >10,000 | 20d |
| 52 | N |
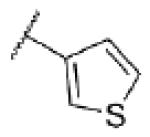
|
<5.0c | >10,000 | 15 ± 8 |
| 53 | N |
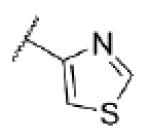
|
<5.0c | >10,000 | 35 ± 6 |
| 54 | CF |
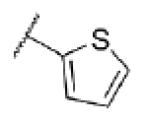
|
5.2 ± 0.11 | 6370 | 7.8 ± 2.8 |
| 55 | CF |
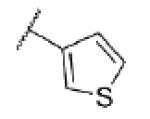
|
5.5 ± 0.15 | 3450 | 0.1 ± 2.4 |
| 56 | CF |
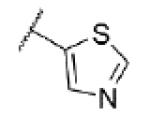
|
<4.5 | >30,000 | — |
| 57 | CF |
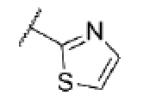
|
<5.0c | >10,000 | 40 ± 9 |
| 58 | CF |
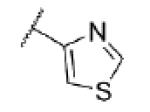
|
5.07 ± 0.05 | 8430 | 22 ± 11 |
| 59 | CF |
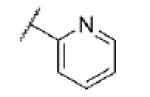
|
<5.0c | >10,000 | 40 ± 15 |
| 60 | CF |
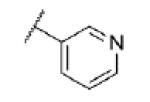
|
<4.5 | >30,000 | — |
| 61 | CF |
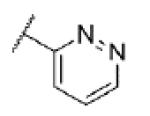
|
<4.5 | >30,000 | — |
| 62 | CF |
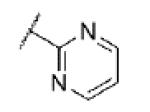
|
<4.5 | >30,000 | — |
| 63 | CF |
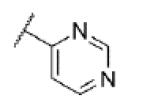
|
<4.5 | >30,000 | — |
| 64 | CF |
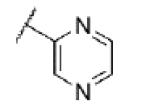
|
<4.5 | >30,000 | — |
Calcium mobilization mGlu5 assay; values are average of n ≥ 3
Amplitude of response in the presence of 30 μM test compound as a percentage of maximal response (100 μM glutamate); average of n ≥ 3
CRC does not plateau
average of n = 2
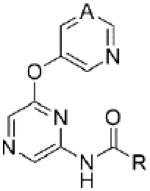
Table 4. Substituted Heteroaryl Amide SAR.
|
|||||
|---|---|---|---|---|---|
|
| |||||
| Cpd | A | R | mGlu5 pIC50a (± SEM) |
mGlu5 IC50 (nM) |
% Glu Maxa,b (± SEM) |
| 65 | N |
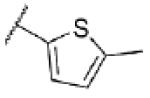
|
<4.5 | >30,000 | — |
| 66 | N |
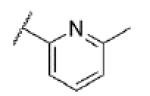
|
<4.5 | >30,000 | — |
| 67 | N |
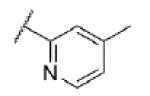
|
6.72 ± 0.22 | 193 | 2.1 ± 0.8 |
| 68 | N |
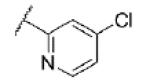
|
7.20 ± 0.06 | 62.4 | 1.6 ± 0.1 |
| 69 | N |
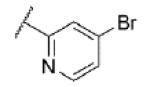
|
7.00 ± 0.29 | 100 | 2.3 ± 0.6 |
| 70 | CF |
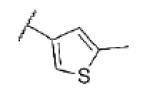
|
6.00 ± 0.04 | 990 | 1.1 ± 0.75 |
| 71 | CF |
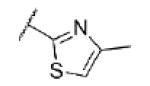
|
<5.0c | >10,000 | 20 ± 10 |
| 72 | CF |
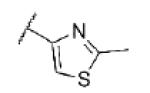
|
5.33 ± 0.17 | 4640 | 19 ± 6 |
| 73 | CF |
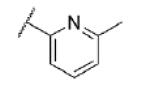
|
<4.5 | >30,000 | — |
| 74 | CF |
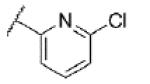
|
<4.5 | >30,000 | — |
| 75 | CF |
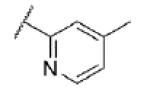
|
6.51 ± 0.15 | 311 | 1.7 ± 0.4 |
| 76 | CF |
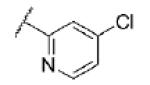
|
6.95 ± 0.29 | 111 | 2.0 ± 0.3 |
Calcium mobilization mGlu5 assay; values are average of n ≥ 3
Amplitude of response in the presence of 30 μM test compound as a percentage of maximal response (100 μM glutamate); average of n ≥ 3
CRC does not plateau
Further in vitro characterization of 68 (VU0431316) was subsequently initiated. Competitive displacement of the established radioligand [3H]3-methoxy-5-(pyridin-2-ylethynyl)pyridine31 confirmed the interaction of the compound with a known mGlu5 allosteric binding site (mGlu5 Ki = 37 nM (n=1)).32 Evaluation of 68 (VU0431316) in cell based functional assays against the other seven mGlus showed no detectable activity at 10μM.33 Additionally, the functional activity of 68 (VU0431316) at human mGlu5 was determined and was essentially identical to that at the rat receptor (human mGlu5 IC50 = 85 nM (n=1)).34 Finally, 68 (VU0431316) was tested in a commercially available radioligand binding assay panel of 68 clinically relevant GPCRs, ion channels, kinases, and transporters,35 and no significant responses were found at 10 μM compound.36
The pharmacological profile of 68 (VU0431316) warranted further evaluation of its drug-likeness and potential utility as an in vivo tool (Table 5). Evaluation of the compound’s propensity to bind to plasma proteins revealed similar results across multiple species. Nonspecific binding to mouse brain homogenates was also evaluated to enable the estimation of the fraction unbound in the CNS.37 Bidirectional permeability was assessed in both Madin Darby canine kidney (MDCK) and human intestinal epithelial (Caco-2) cells, and permeability was high with no evidence of efflux.38 Determination of the cytochrome P450 (CYP) inhibition profile of 68 (VU0431316) indicated potent inhibition of CYP1A2 with no inhibition of other isoforms up to the top concentration tested of 30 μM.39 As was the case with the related tool compound VU0409106, the common pyrimidine ether moiety resulted in a major non-P450 mediated metabolic pathway for 68 (VU0431316).40
Table 5. In vitro DMPK Profile for 68 (VU0431316).
| Protein Binding (Fu)a,b |
MDCK-WT Permeability |
||||
|---|---|---|---|---|---|
| rat PPB | 0.179 | Papp (A-B) | 50.4 × 10−6 cm/sec | ||
| cynomolgus PPB | 0.167 | Papp (B-A) | 45.0 × 10−6 cm/sec | ||
| human PPB | 0.120 | efflux ratio | 0.89 | ||
|
|
|||||
| mouse BHB | 0.054 | ||||
|
|
|
||||
| Caco-2 Permeability |
|||||
| CYP Inhibition IC50 (μM)c |
Papp (A-B) | 66.8 × 10−6 cm/sec | |||
| 3A4 | 2D6 | 2C9 | 1A2 | Papp (B-A) | 71.4 × 10−6 cm/sec |
| >30 | >30 | >30 | 0.6 | efflux ratio | 1.07 |
Fu = fraction unbound
PPB = plasma protein binding; BHB = brain homogenate binding
Assayed in pooled HLM in the presence of NADPH
The in vitro DMPK profile of 68 (VU0431316) was deemed supportive of in vivo evaluation (Table 6). Pharmacokinetic parameters were calculated from an IV dosing of 68 (VU0431316) to male Sprague-Dawley rats; hepatic clearance was moderate with an approximate three hour half-life. Bioavailability from a single oral (PO) dose was also encouraging, approaching fifty percent. A satellite tissue distribution study was conducted one hour after a 10 mg/kg PO dose. Seventy percent of the compound was detected in plasma relative to the hepatic portal vein, indicative of a low first-pass effect and consistent with the previously observed clearance. CNS penetration was also excellent with a brain to plasma ratio (B/P) of 1.6.41
Table 6. Rat PK Results for 68 (VU0431316).
| IVa,b | POa,c | ||
|---|---|---|---|
| half-life | 193 min | plasma Cmax | 1.12 μM |
| clearance | 30.2 mL/min/kg | plasma Tmax | 240 min |
| VSS | 2.7 L/kg | AUC | 8.37 μM·h |
| AUC | 1.73 μM·h | F | 48% |
Male Sprague-Dawley rats (n=2 per time point)
1 mg/kg; vehicle = 10% ethanol, 90% PEG400
10 mg/kg; vehicle = 10% Tween 80 in 0.5% MC
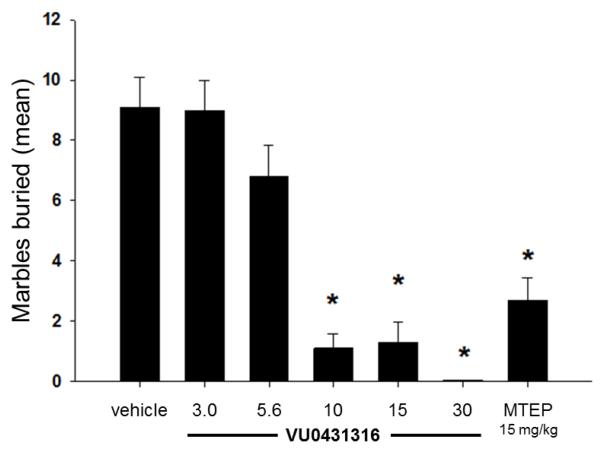
It is recognized that naïve mice will bury foreign objects such as glass marbles in deep bedding. Pretreating such mice with low doses of anxiolytic benzodiazepines will consistently inhibit this behavior.42 Additionally, the well-known and thoroughly characterized mGlu5 NAMs MPEP and fenobam each inhibit marble burying in this model.7a,d Furthermore, we have used this model successfully to evaluate several of our own novel mGlu5 NAM tool compounds.22a, 23b, 24 More recent reports have raised the possibility that marble burying reflects a repetitive and perseverative behavior as opposed to novelty-induced anxiety.43 Still, given the convenience and reliability of the marble burying assay, it has served as the frontline behavioral assay for our mGlu5 NAM discovery program.44 Thus, a dose response study with 68 (VU0431316) using a 15 minute pretreatment time following intraperitoneal (IP) administration was carried out in this model (Fig. 3).45 Statistically significant inhibition of marble burying was noted at all doses greater than or equal to 10 mg/kg. A satellite tissue distribution experiment in mice at the top dose of 30 mg/kg (IP) showed considerable concentrations of compound in the brain.46 Using the results from this study to extrapolate exposures at 10 mg/kg along with the aforementioned brain homogenate binding data indicates that unbound compound concentrations in the brain were likely near the functional IC50 at the minimum effective dose.
Figure 3.
Dose dependent inhibition of marble burying with 68 (VU0431316); n = 7-8 per dose; *, P < 0.05 vs. vehicle control group, Dunnett’s test. Bars denote marbles buried. Vehicle = 10% Tween 80
In conclusion, a second in vivo tool compound has been developed from HTS hit 1. VU0431316 offers an advantage to VU0409106 in that it is orally bioavailable in rats. VU0431316 is a potent and selective mGlu5 NAM binding to a known allosteric site. CNS exposure in both mice and rats is quite good and efficacy has been established in a proven anxiolytic model. A host of additional behavioral models associated with mGlu5 are either planned or already underway with VU0431316 and will be the subject of future communications.
Acknowledgements
We thank NIDA (R01 DA023947) and Seaside Therapeutics (VUMC33842) for their support of our programs in the development of non-competitive antagonist of mGlu5. We also thank Tammy S. Santomango for technical contributions with the protein binding assays.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and notes
- 1.(a) Schoepp DD, Jane DE, Monn JA. Neuropharmacology. 1999;38:1431. doi: 10.1016/s0028-3908(99)00092-1. [DOI] [PubMed] [Google Scholar]; (b) Conn PJ, Pin J-P. Annu. Rev. Pharmacol. Toxicol. 1997;37:205. doi: 10.1146/annurev.pharmtox.37.1.205. [DOI] [PubMed] [Google Scholar]
- 2.(a) Melancon BJ, Hopkins CR, Wood MR, Emmitte KA, Niswender CM, Christopoulos A, Conn PJ, Lindsley CW. J. Med. Chem. 2012;55:1445. doi: 10.1021/jm201139r. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Conn PJ, Christopolous A, Lindsley CW. Nat. Rev. Drug Discovery. 2009;8:41. doi: 10.1038/nrd2760. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Bridges TM, Lindsley CW. ACS Chem. Biol. 2008;3:530. doi: 10.1021/cb800116f. [DOI] [PubMed] [Google Scholar]; (d) Ritzén A, Mathiesen JM, Thomsen C. Basic Clin. Pharmacol. Toxicol. 2005;97:202. doi: 10.1111/j.1742-7843.2005.pto_156.x. [DOI] [PubMed] [Google Scholar]; (e) Kew JNC. Pharmacol. Ther. 2004;104:233. doi: 10.1016/j.pharmthera.2004.08.010. [DOI] [PubMed] [Google Scholar]
- 3.(a) Emmitte KA. Expert Opin. Ther. Pat. 2013;23:393. doi: 10.1517/13543776.2013.760544. [DOI] [PubMed] [Google Scholar]; (b) Emmitte KA. ACS Chem. Neurosci. 2011;2:411. doi: 10.1021/cn2000266. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Rocher JP, Bonnet B, Boléa C, Lütjens R, Le Poul E, Poli S, Epping-Jordan M, Bessis A-S, Ludwig B, Mutel V. Curr. Top. Med. Chem. 2011;11:680. doi: 10.2174/1568026611109060680. [DOI] [PubMed] [Google Scholar]; (d) Lindsley CW, Emmitte KA. Curr. Opin. Drug Discov. Dev. 2009;12:446. [PubMed] [Google Scholar]; (e) Gasparini F, Bilbe G, Gomez-Mancilla B, Spooren W. Curr. Opin. Drug Discov. Dev. 2008;11:655. [PubMed] [Google Scholar]
- 4.Gasparini F, Lingenhöhl K, Stoehr N, Flor PJ, Heinrich M, Vranesic I, Biollaz M, Allgeier H, Heckendorn R, Urwyler S, Varney MA, Johnson EC, Hess SD, Rao SP, Sacaan AI, Santori EM, Veliocelebi G, Kuhn R. Neuropharmacology. 1999;38:1493. doi: 10.1016/s0028-3908(99)00082-9. [DOI] [PubMed] [Google Scholar]
- 5.Cosford ND, Tehrani L, Roppe J, Schweiger E, Smith ND, Anderson J, Bristow L, Brodkin J, Jiang X, McDonald I, Rao S, Washburn M, Varney MA. J. Med. Chem. 2003;46:204. doi: 10.1021/jm025570j. [DOI] [PubMed] [Google Scholar]
- 6.Zhu CZ, Wilson SG, Mikusa JP, Wismer CT, Gauvin DM, Lynch JJ, Wade CL, Decker MW, Honore P. Eur. J. Pharmacol. 2004;506:107. doi: 10.1016/j.ejphar.2004.11.005. [DOI] [PubMed] [Google Scholar]
- 7.(a) Nicolas LB, Kolb Y, Prinssen EPM. Eur. J. Pharmacol. 2006;547:106. doi: 10.1016/j.ejphar.2006.07.015. [DOI] [PubMed] [Google Scholar]; (b) Pietraszek M, Sukhanov I, Maciejak P, Szyndler J, Gravius A, Wislowska A, Plaznik A, Bespalov AY, Danysz W. Eur. J. Pharmacol. 2005;514:25. doi: 10.1016/j.ejphar.2005.03.028. [DOI] [PubMed] [Google Scholar]; (c) Busse CS, Brodkin J, Tattersall D, Anderson JJ, Warren N, Tehrani L, Bristow LJ, Varney MA, Cosford NDP. Neuropsychopharmacology. 2004;29:1971. doi: 10.1038/sj.npp.1300540. [DOI] [PubMed] [Google Scholar]; (d) Klodzinska A, Tatarczynska E, Chojnacka-Wójcik E, Nowak G, Cosford NDP, Pilc A. Neuropharmacology. 2004;47:342. doi: 10.1016/j.neuropharm.2004.04.013. [DOI] [PubMed] [Google Scholar]; (e) Spooren WPJM, Vassout A, Neijt HC, Kuhn R, Gasparini F, Roux S, Porsolt RD, Gentsch C. J. Pharmacol. Exp. Ther. 2000;295:1267. [PubMed] [Google Scholar]
- 8.(a) Jensen J, Lehmann A, Uvebrant A, Carlsson A, Jerndal G, Nilsson K, Frisby C, Blackshaw LA, Mattsson JP. Eur. J. Pharmacol. 2005;519:154. doi: 10.1016/j.ejphar.2005.07.007. [DOI] [PubMed] [Google Scholar]; (b) Frisby CL, Mattsson JP, Jensen JM, Lehmann A, Dent J, Blackshaw LA. Gastroenterology. 2005;129:995. doi: 10.1053/j.gastro.2005.06.069. [DOI] [PubMed] [Google Scholar]
- 9.Morin N, Grégoire L, Gomez-Mancilla B, Gasparini F, Di Paolo T. Neuropharmacology. 2010;58:981. doi: 10.1016/j.neuropharm.2009.12.024. [DOI] [PubMed] [Google Scholar]
- 10.(a) de Vrij FMS, Levenga J, van der Linde HC, Koekkoek SK, De Zeeuw CI, Nelson DL, Oostra BA, Willemsen R. Neurobiol. Dis. 2008;31:127. doi: 10.1016/j.nbd.2008.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Yan QJ, Rammal M, Tranfaglia M, Bauchwitz RP. Neuropharmacology. 2005;49:1053. doi: 10.1016/j.neuropharm.2005.06.004. [DOI] [PubMed] [Google Scholar]
- 11.Silverman JL, Tolu SS, Barkan CL, Crawley JN. Neuropsychopharmacology. 2010;35:976. doi: 10.1038/npp.2009.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.(a) Martin-Fardon R, Baptista MAS, Dayas CV, Weiss F. J. Pharmacol. Exp. Ther. 2009;329:1084. doi: 10.1124/jpet.109.151357. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Kumaresan V, Yuan M, Yee J, Famous KR, Anderson SM, Schmidt HD, Pierce RC. Behav. Brain Res. 2009;202:238. doi: 10.1016/j.bbr.2009.03.039. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Platt DM, Rowlett JK, Spealman RD. Psychopharmacology. 2008;200:167. doi: 10.1007/s00213-008-1191-y. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Bäckstrom P, Hyytiä P. Neuropsychopharmacology. 2006;31:778. doi: 10.1038/sj.npp.1300845. [DOI] [PubMed] [Google Scholar]; (e) Lee B, Platt DM, Rowlett JK, Adewale AS, Spealman RD. J. Pharmacol. Exp. Ther. 2005;312:1232. doi: 10.1124/jpet.104.078733. [DOI] [PubMed] [Google Scholar]; (f) Kenny PJ, Boutrel B, Gasparini F, Koob GF, Markou A. Psychopharmacology. 2005;179:247. doi: 10.1007/s00213-004-2069-2. [DOI] [PubMed] [Google Scholar]; (g) Tessari M, Pilla M, Andreoli M, Hutcheson DM, Heidbreder CA. Eur. J. Pharmacol. 2004;499:121. doi: 10.1016/j.ejphar.2004.07.056. [DOI] [PubMed] [Google Scholar]
- 13.Tronci V, Vronskaya S, Montgomery N, Mura D, Balfour DJK. Psychopharmacology. 2010;211:33. doi: 10.1007/s00213-010-1868-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gass JT, Osborne MPH, Watson NL, Brown JL, Olive MF. Neuropsychopharmacology. 2009;34:820. doi: 10.1038/npp.2008.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kotlinska J, Bochenski M. Eur. J. Pharmacol. 2007;558:113. doi: 10.1016/j.ejphar.2006.11.067. [DOI] [PubMed] [Google Scholar]
- 16.(a) Adams CL, Short JL, Lawrence AJ. Br. J. Pharmacol. 2010;159:534. doi: 10.1111/j.1476-5381.2009.00562.x. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Besheer J, Grondin JJM, Salling MC, Spanos M, Stevenson RA, Hodge CW. J. Neurosci. 2009;29:9582. doi: 10.1523/JNEUROSCI.2366-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Gass JT, Olive MF. Psychopharmacology. 2009;204:587. doi: 10.1007/s00213-009-1490-y. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Schroeder JP, Spanos M, Stevenson JR, Besheer J, Salling M, Hodge CW. Neuropharmacology. 2008;55:546. doi: 10.1016/j.neuropharm.2008.06.057. [DOI] [PMC free article] [PubMed] [Google Scholar]; (e) Lominac KD, Kapasova Z, Hannun RA, Patterson C, Middaugh LD, Szumlinski KK. Drug Alcohol Depend. 2006;85:142. doi: 10.1016/j.drugalcdep.2006.04.003. [DOI] [PubMed] [Google Scholar]
- 17.(a) Rohof WO, Lei A, Hirsch DP, Ny L, Astrand M, Hansen MB, Boeckxstaens GE. Aliment. Pharmacol. Ther. 2012;35:1231. doi: 10.1111/j.1365-2036.2012.05081.x. [DOI] [PubMed] [Google Scholar]; (b) Keywood C, Wakefield M, Tack J. Gut. 2009;58:1192. doi: 10.1136/gut.2008.162040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.(a) Jacquemont S, Curie A, des Portes V, Torrioli MG, Berry-Kravis E, Hagerman RJ, Ramos FJ, Cornish K, He Y, Paulding C, Neri G, Chen F, Hadjikhani N, Martinet D, Meyer J, Beckmann JS, Delange K, Brun A, Bussy G, Gasparini F, Hilse T, Floesser A, Branson J, Bilbe G, Johns D, Gomez-Mancilla B. Sci. Transl. Med. 2011;3:64ra1. doi: 10.1126/scitranslmed.3001708. [DOI] [PubMed] [Google Scholar]; (b) Berry-Kravis EM, Hessl D, Coffey S, Hervey C, Schneider A, Yuhas J, Hutchinson J, Snape M, Tranfaglia M, Nguyen DV, Hagerman R. J. Med. Genet. 2009;46:266. doi: 10.1136/jmg.2008.063701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.(a) Tison F, Durif F, Corvol JC, Eggert K, Trenkwalder C, Lew M, Isaacson S, Keywood C, Rascol O. Neurology. 2013;80:S23.004. Meeting Abstracts 1. [Google Scholar]; (b) Berg D, Godau J, Trenkwalder C, Eggert K, Csoti I, Storch A, Huber H, Morelli-Canelo M, Stamelou M, Ries V, Wolz M, Schneider C, Di Paolo T, Gasparini F, Hariry S, Vandemeulebroecke M, Abi-Saab W, Cooke K, Johns D, Gomez-Mancilla B. Movement Disorders. 2011;26:1243. doi: 10.1002/mds.23616. [DOI] [PubMed] [Google Scholar]
- 20.Searching at www.clinicaltrials.gov on 24 March 2014 found 20 studies when searching mavoglurant or AFQ056 and 12 studies when searching RO4917523.
- 21.Zhang L, Balan G, Barreiro G, Boscoe BP, Chenard LK, Cianfrogna J, Claffey MM, Chen L, Coffman KJ, Drozda SE, Dunetz JR, Fonseca KR, Galatsis P, Grimwood S, Lazzaro JT, Mancuso JY, Miller EL, Reese MR, Rogers BN, Sakurada I, Skaddan M, Smith DL, Stepan AF, Trapa P, Tuttle JB, Verhoest PR, Walker DP, Wright AS, Zaleska MM, Zasadny K, Shaffer CL. J. Med. Chem. 2014;57:861. doi: 10.1021/jm401622k. [DOI] [PubMed] [Google Scholar]
- 22.(a) Felts AS, Rodriguez AL, Morrison RD, Venable DF, Manka JT, Bates BS, Blobaum AL, Byers FW, Daniels JS, Niswender CM, Jones CK, Conn PJ, Lindsley CW, Emmitte KA. Bioorg. Med. Chem. Lett. 2013;23:5779. doi: 10.1016/j.bmcl.2013.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Amato RJ, Felts AS, Rodriguez AL, Venable DF, Morrison RD, Byers FW, Daniels JS, Niswender CM, Conn PJ, Lindsley CW, Jones CK, Emmitte KA. ACS Chem. Neurosci. 2013;4:1217. doi: 10.1021/cn400070k. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Felts AS, Saleh SA, Le U, Rodriguez AL, Weaver CD, Conn PJ, Lindsley CW, Emmitte KA. Bioorg. Med. Chem. Lett. 2009;19:6623. doi: 10.1016/j.bmcl.2009.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Zhou Y, Rodriguez AL, Williams R, Weaver CD, Conn PJ, Lindsley CW. Bioorg. Med. Chem. Lett. 2009;19:6502. doi: 10.1016/j.bmcl.2009.10.059. [DOI] [PMC free article] [PubMed] [Google Scholar]; (e) Rodriguez AL, Williams R, Zhou Y, Lindsley SR, Le U, Grier MD, Weaver CD, Conn PJ, Lindsley CW. Bioorg. Med. Chem. Lett. 2009;19:3209. doi: 10.1016/j.bmcl.2009.04.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.(a) Lamb JP, Engers DW, Niswender CM, Rodriguez AL, Venable DF, Conn PJ, Lindsley CW. Bioorg. Med. Chem. Lett. 2011;21:2711. doi: 10.1016/j.bmcl.2010.11.119. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Lindsley CW, Bates BS, Menon UN, Jadhav SB, Kane AS, Jones CK, Rodriguez AL, Conn PJ, Olsen CM, Winder DG, Emmitte KA. ACS Chem. Neurosci. 2011;2:471. doi: 10.1021/cn100099n. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Felts AS, Lindsley SR, Lamb JP, Rodriguez AL, Menon UN, Jadhav S, Jones CK, Conn PJ, Lindsley CW, Emmitte KA. Bioorg. Med. Chem. Lett. 2010;20:4390. doi: 10.1016/j.bmcl.2010.06.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mueller R, Dawson ES, Meiler J, Rodriguez AL, Chauder BA, Bates BS, Felts AS, Lamb JP, Menon UN, Jadhav SB, Kane AS, Jones CK, Gregory KJ, Niswender CM, Conn PJ, Olsen CM, Winder DG, Emmitte KA, Lindsley CW. Chem. Med. Chem. 2012;7:406. doi: 10.1002/cmdc.201100510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Conn PJ, Lindsley CW, Emmitte KA, Weaver CD, Rodriguez AL, Felts AS, Jones CK, Bates BS. PCT Int. Patent Appl. Detailed synthetic procedures for the preparation of analogs and characterization are described in. WO 2011/035174. [Google Scholar]
- 26.Yin J, Buchwald SL. Org. Lett. 2000;2:1101. doi: 10.1021/ol005654r. [DOI] [PubMed] [Google Scholar]
- 27.HEK293A cells expressing rat mGlu5 were cultured and plated. The cells were loaded with a Ca2+ sensitive fluorescent dye and the plates were washed and placed in the Functional Drug Screening System (Hamamatsu). Compounds (10 mM in DMSO) were serially diluted 1:3 into 10 point CRC (30 μM to 1 nM final) and transferred to daughter plates. Compounds were diluted into assay buffer and applied to cells 3 seconds after baseline readings were taken. Cells were incubated with the test compounds for 140 seconds and then stimulated with an EC20 concentration of glutamate; 60 seconds later an EC80 concentration of agonist was added and readings taken for an additional 40 seconds. Allosteric modulation by the compounds was measured by comparing the amplitude of the responses at the time of glutamate addition plus and minus test compound. For a more detailed description of the assay, see reference 22b.
- 28.According to the cLogP calculator developed by ADRIANA.Code (www.molecular-networks.com) 5-fluoropyridin-3-yl ether compounds are ~1.2 units more lipophilic than their pyrimidin-5-yl ether counterparts in this chemotype (e.g. 76 cLogP = 3.72; 68 cLogP = 2.51).
- 29.The synthesis of VU0431316 (68) is representative: (i) 5-((6-chloropyrazin-2-yl)oxy)pyrimidine (23). A mixture of 2,6-dichloropyrazine (1.00 g, 6.71 mmol, 1.00 eq), pyrimidin-5-ol (645 mg, 6.71 mmol, 1.00 eq), and potassium carbonate (1.39 g, 10.1 mmol, 1.50 eq) in DMF (20 mL) was heated via microwave irradiation at 120 °C for 10 min. The reaction was cooled and diluted with EtOAc and washed with H2O (3×) and brine (1×). The organic layer was dried (MgSO4), filtered, and concentrated in vacuo. Purification by flash chromatography on silica gel afforded 710 mg (51%) of the title compound. 1H NMR (400 MHz, DMSO-d6) 9.15 (s, 1H), 8.93 (s, 2H), 8.73 (s, 1H), 8.62 (s, 1H); ES-MS [M+H]+: 209.1. (ii) tert-butyl (6-(pyrimidin-5-yloxy)pyrazin-2-yl)carbamate (25). Compound 23 (760 mg, 3.64 mmol, 1.00 eq), tert-butyl carbamate (117 mg, 4.37 mmol, 1.20 eq), Pd2(dba)3·CHCl3 (113 mg, 0.110 mmol, 0.030 eq), NaOtBu (490 mg, 5.10 mmol, 1.40 eq), and tBuXPhos (164 mg, 0.330 mmol, 0.090 eq) were stirred in toluene (14 mL) at rt overnight. The mixture was filtered through celite and washed with 5% MeOH in CH2Cl2. The filtrate was collected and concentrated in vacuo. Purification by flash chromatography on silica gel afforded 480 mg (46%) of the title compound. 1H NMR (400 MHz, DMSO-d6) 10.14 (s, 1H), 9.09 (s, 1H), 8.91 (s, 2H), 8.82 (s, 1H), 8.25 (s, 1H), 1.45 (s, 9H); ES-MS [M+H]+: 290.1. (iii) 6-(pyrimidin-5-yloxy)pyrazin-2-amine (27). Compound 25 (480 mg, 1.66 mmol, 1.00 eq) was stirred in 4:1 CH2Cl2:TFA (2 mL) at rt overnight. The reaction mixture was concentrated in in vacuo and diluted EtOAc. The organic layer was washed with saturated NaHCO3 (3×) and brine (1×), dried (MgSO4), filtered and concentrated in vacuo to give 300 mg (96%) of the title compound. 1H NMR (400 MHz, DMSO-d6) 9.04 (s, 1H), 8.79 (s, 2H), 7.65 (s, 1H), 7.60 (s, 1H), 6.64 (s, 2H); ES-MS [M+H]+: 190.1. (iv) 4-chloro-N-(6-(pyrimidin-5-yloxy)pyrazin-2-yl)picolinamide (68, VU0431316). Compound 27 (150 mg, 0.790 mmol, 1.00 eq) and 4-chloropicolinic acid (125 mg, 0.790 mmol, 1.00 eq) were dissolved in pyridine (10 mL) under argon and cooled to −15 °C with stirring. POCl3 (0.080 mL, 0.870 mmol, 1.10 eq) was added dropwise. The resulting reaction mixture was stirred at −15 °C for 30 min. The reaction was then warmed to rt and quenched with ice water. The mixture was extracted with CH2Cl2. The organics were washed with water (3×) and brine (1×). The organic layer was dried (MgSO4), filtered, and concentrated in vacuo. The resulting residue was purified by flash chromatography on silica gel to provide 125 mg (48%) of the desired product as a white solid. 1H NMR (400 MHz, DMSO-d6) 10.64 (s, 1H), 9.24 (s, 1H), 9.14 (s, 1H), 8.99 (s, 2H), 8.71 (d, J = 5.3 Hz 1H) 8.50 (s, 1H), 8.19 (d, J = 1.8 Hz, 1H), 7.89 (dd, J = 5.3, 2.0 Hz, 1H); ES-MS [M+H]+: 329.1.
- 30.Bhagwanth S, Waterson AG, Adjabeng GM, Hornberger KR. J. Org. Chem. 2009;74:4634. doi: 10.1021/jo9004537. [DOI] [PubMed] [Google Scholar]
- 31.Cosford NDP, Roppe J, Tehrani L, Schweiger EJ, Seiders TJ, Chaudary A, Rao S, Varney MA. Bioorg. Med. Chem. Lett. 2003;13:351. doi: 10.1016/s0960-894x(02)00997-6. [DOI] [PubMed] [Google Scholar]
- 32.For a detailed description of the mGlu5 radioligand binding assay see reference 23b.
- 33.mGlu selectivity assays are described in Noetzel MJ, Rook JM, Vinson PN, Cho H, Days E, Zhou Y, Rodriguez AL, Lavreysen H, Stauffer SR, Niswender CM, Xiang Z, Daniels JS, Lindsley CW, Weaver CD, Conn PJ. Mol. Pharmacol. 2012;81:120. doi: 10.1124/mol.111.075184.
- 34.Analogous to reference 27 with the exception that HEK293A cells expressing human mGlu5 were used.
- 35.LeadProfilingScreen®, Eurofins Panlabs, Inc. (http://www.eurofinspanlabs.com)
- 36.Significant responses are defined as those that inhibited more than 50% of radioligand binding. In the case of VU0431316, no inhibition greater than 23% was observed.
- 37.Binding to plasma and brain homogenates were measured using equilibrium dialysis according to methods similar to those described in Kalvass JC, Maurer TS. Biopharm. Drug Dispos. 2002;23:327. doi: 10.1002/bdd.325.
- 38.Bidirectional permeability was carried out according to methods described in Wang Q, Rager JD, Weinstein K, Kardos PS, Dobson GL, Li J, Hidalgo IJ. Int. J. Pharm. 2008;288:349. doi: 10.1016/j.ijpharm.2004.10.007.
- 39.CYP3A4 inhibition assay was carried out according to methods described in Zientek M, Miller H, Smith D, Dunklee MB, Heinle L, Thurston A, Lee C, Hyland R, Fahmi O, Burdette D. J. Pharmacol. Toxicol. Methods. 2008;58:206. doi: 10.1016/j.vascn.2008.05.131. Kuresh AY, Lyons R, Payne L, Jones BC, Saunders K. J. Pharm. Biomed. Anal. 2008;48:92. doi: 10.1016/j.jpba.2008.05.011.
- 40.Morrison RD, Blobaum AL, Byers FW, Santomango TS, Bridges TM, Stec D, Brewer KA, Sanchez-Ponce R, Corlew MM, Rush R, Felts AS, Manka J, Bates BS, Venable DF, Rodriguez AL, Jones CK, Niswender CM, Conn PJ, Lindsley CW, Emmitte KA, Daniels JS. Drug Metab. Dispos. 2012;40:1834. doi: 10.1124/dmd.112.046136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.For the IV study, the blood samples were collected at 2, 7, 15, 30, 60, 120, 240, 420, and 1440 min after dose administration. For the PO study, blood samples were collected at 15, 30, 60, 120, 240, 420, and 1440 min after dose administration. For the tissue distribution study, rats were euthanized and decapitated at 60 min after dose administration and blood, hepatic portal vein, and brain samples were collected. Following protein precipitation, the supernatants of all plasma and brain homogenate samples were analyzed by means of LC-MS/MS. PK studies were approved by the Vanderbilt University Medical Center Institutional Animal Care and Use Committee.
- 42.(a) Njung’e K, Handley SL. Brit. J. Pharmacol. 1991;104:105. doi: 10.1111/j.1476-5381.1991.tb12392.x. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Broekkamp CL, Rijk HW, Joly-Gelouin D, Lloyd KL. Eur. J. Pharmacol. 1986;126:223. doi: 10.1016/0014-2999(86)90051-8. [DOI] [PubMed] [Google Scholar]
- 43.Thomas A, Burant A, Bui N, Graham D, Yuva-Paylor LA, Paylor R. Psychopharmacology. 2009;204:361. doi: 10.1007/s00213-009-1466-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Marble burying experiments were conducted in accordance with the National Institute of Health regulations of animal care covered in Principles of Laboratory Animal Care and were approved by the Vanderbilt University Medical Center Animal Care and Use Committee. For a detailed experimental procedure for the marble burying assay see reference 23b.
- 45.IP dosing has proven over time to be a convenient and consistent route of administration for our behavioral studies in mice.
- 46.Mice were euthanized and decapitated at predetermined time-points after dose administration and blood and brain samples were collected. Following protein precipitation, the supernatants of all plasma and brain homogenate samples were analyzed by means of LC-MS/MS. PK studies were approved by the Vanderbilt University Medical Center Institutional Animal Care and Use Committee. Average plasma concentrations: 3.99 μM (15 min), 3.00 μM (30 min), and 1.69 μM (60 min). Average brain concentrations: 6.21 μM (15 min), 3.69 μM (30 min), and 2.80 μM (60 min).