Abstract
Amyotrophic lateral sclerosis (ALS) is a severe neurodegenerative condition characterized by loss of motor neurons in the brain and spinal cord. Expansions of a hexanucleotide repeat (GGGGCC) in the noncoding region of the C9ORF72 gene are the most common cause of the familial form of ALS (C9-ALS), as well as frontotemporal lobar degeneration and other neurological diseases. How the repeat expansion causes disease remains unclear, with both loss of function (haploinsufficiency) and gain of function (either toxic RNA or protein products) proposed. Here, we report a cellular model of C9-ALS with motor neurons differentiated from induced pluripotent stem cells (iPSCs) derived from ALS patients carrying the C9ORF72 repeat expansion. No significant loss of C9ORF72 expression was observed, and knockdown of the transcript was not toxic to cultured human motor neurons. Transcription of the repeat was increased leading to accumulation of GGGGCC repeat-containing RNA foci selectively in C9-ALS motor neurons. Repeat-containing RNA foci co-localized with hnRNPA1 and Pur-α, suggesting that they may be able to alter RNA metabolism. C9-ALS motor neurons showed altered expression of genes involved in membrane excitability including DPP6, and demonstrated a diminished capacity to fire continuous spikes upon depolarization compared to control motor neurons. Antisense oligonucleotides (ASOs) targeting the C9ORF72 transcript suppressed RNA foci formation and reversed gene expression alterations in C9-ALS motor neurons. These data show that patient-derived motor neurons can be used to delineate pathogenic events in ALS.
Introduction
Amyotrophic lateral sclerosis (ALS) and frontotemporal lobar degeneration (FTLD) are neurodegenerative disorders that overlap in their clinical presentation, pathologic findings, and genetic origins (1–3). No treatments are currently available. ALS presents clinically as muscle wasting with stiffness and spasticity from loss of motor neurons in the spinal cord, whereas FTLD manifests most commonly with behavioral and language disturbances due to degeneration of regions of the frontal and temporal cortices (4). Both diseases commonly show accumulations of abnormal proteins including the DNA/RNA binding protein TDP-43 (5). Mutations in several genes cause both disorders, including TARDBP (which encodes TDP-43) and VCP, although these are responsible for very few cases (6, 7). Recently, expansions of a GGGGCC hexanucleotide repeat in the first intron/promoter of the C9ORF72 gene were reported to be the most commonly identified genetic cause of ALS and FTLD in both familial and sporadic cases in Caucasians (8–10). More recently, C9ORF72 repeat expansions were reported in other neurodegenerative diseases, including Alzheimer’s disease (11, 12) and Parkinson’s disease (13). The broad neurodegenerative phenotype and the high frequency of the mutations emphasize the need to develop treatments for C9ORF72 repeat expansion diseases.
A key remaining question is whether the repeat expansion in C9ORF72 leads to loss of function, gain of function, or both. Several lines of evidence suggest that the repeat expansion may suppress or alter the expression of the mutant allele. Decreased expression of C9ORF72 transcripts has been reported (8, 10), as has hypermethylation of the repeat containing allele (14). Knockdown of the C9ORF72 orthologue in zebrafish resulted in motor deficits (15). However, early reports also indicated that the repeat is transcribed and leads to accumulation of repeat-containing RNA foci in patient tissues (8). Subsequently, it was found that simple peptides could be generated by repeat-associated non-ATG dependent translation (16, 17). Both RNA foci and protein aggregates may produce a gain of function toxicity in neurons to promote neurodegeneration. Further supporting this gain of function is the fact that other mutations which would cause haploinsufficiency, such as early stop codons, have not been observed (18). A patient homozygous for the C9ORF72 repeat expansion had a phenotype similar to heterozygotes, rather than the more severe phenotype that would be expected for complete loss of function (19).
Here, we generated induced pluripotent stem cells (iPSCs) from patients with ALS caused by the C9ORF72 repeat expansion (C9-ALS) and differentiated them into motor neurons. Using a variety of methods, we observed that expression of the C9ORF72 was not significantly decreased in human motor neuron cultures from C9-ALS patients. Knockdown of all C9ORF72 transcripts was not toxic to iPSC-derived motor neurons from normal control subjects. Antisense oligonucleotides (ASOs) targeting the C9ORF72 transcript suppressed gain of function manifestations including formation of RNA foci, and corrected altered gene expression profiles.
Results
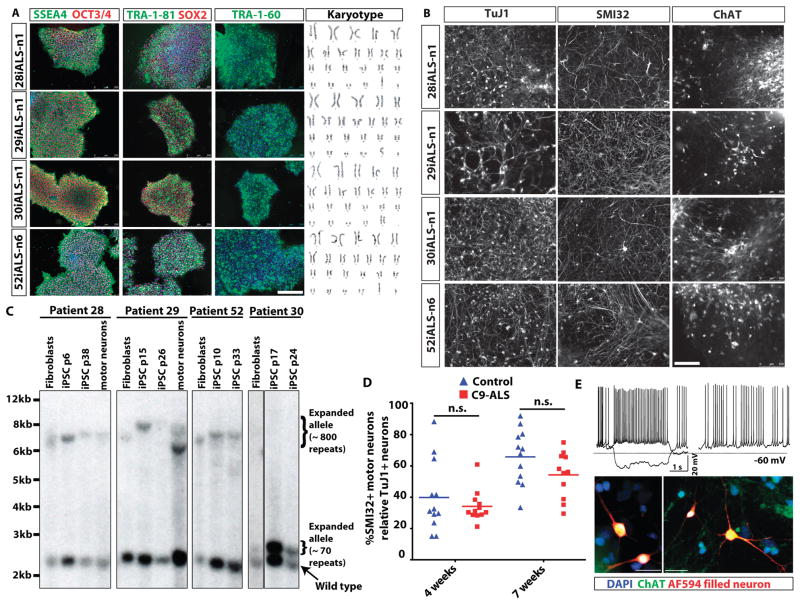
Skin fibroblasts were reprogrammed from four different C9ORF72 hexanucleotide expansion carriers who had either ALS, or ALS with FTLD (Table S1). A non-integrating system based on the oriP/EBNA1 (Epstein-Barr nuclear antigen-1) based episomal plasmid vector system was used to avoid potential deleterious effects of random insertion of proviral sequences into the genome (20–22). All iPSC lines expressed the pluripotency markers (SSEA4, TRA-1-81, OCT3/4, SOX2) along with a normal karyotype (Fig 1A). Pluripotency was further confirmed using PluriTest, a validated open-access bioinformatics pathway for assessing pluripotency using transcriptome profiling (23), alkaline phosphatase (marker of pluripotency), flow cytometry analysis of positive SSEA4 and OCT4+ marker expression, and spontaneous embryoid body differentiation assay to detect formation of the three germ layers (Fig. S1). All iPSC lines lacked expression of exogenous transgenes using qRT-PCR, and genomic PCR analysis demonstrating that the oriP/EBNA1 method generated “footprint-free” iPSC lines (Fig. S2). C9-ALS and control patient iPSC lines were then differentiated into motor neurons and associated support cells according to established protocols (21) as also illustrated in the schematic in Fig. S2C. Our differentiation protocol yielded OLIG2 and HB9 expressing motor neuron precursors (Fig. S2D) and cultures composed of normal appearing spinal motor neurons that were labeled with SMI32 and ChAT (Fig. 1B and S3A). At 7 weeks of differentiation all iPSC-derived motor neuron cultures comprised SMI32+ motor neurons (33–45%) and TuJ1+ pan-neurons (58–75%), while the remaining cells (20–30%) were labeled with GFAP (astrocyte marker) and Nestin (neural progenitor marker) (Fig. S3B). Consistent with their acquisition of motor neuron markers reflecting in vitro maturation, patch clamp recordings showed that motor neurons fired normal appearing spontaneous and depolarization-induced action potentials (Fig. 1E).
Fig. 1. Generation of C9ORF72 ALS patient-derived iPSCs and motor neurons.
(A) Pluripotency marker expression in iPSC lines derived from four ALS patients with a C9ORF72 repeat expansion. Immunostaining shows expression of embryonic and pluripotency stem cell surface antigens, SSEA4, TRA-1-60, TRA-1-81; and nuclear pluripotency markers OCT3/4 and SOX2. Karyotypes of the four C9ORF72 patient iPSC lines are shown on the right. (B) Efficient production of motor neurons from ALS C9ORF72 patient-derived iPSCs demonstrated by SMI32+ and ChAT+ immunostaining. TuJ1 is a pan-neuronal marker used to assess for total production of neurons. (C) Examination of hexanucleotide repeat lengths in fibroblasts, iPSCs and motor neuron cultures by Southern blot analysis, showing somatic instability of the repeat with both expansion (iPSCs in lines 28i,29i,52i) and contraction (motor neurons in iPSC line derived from patient 29). Passage numbers for iPSCs are shown (p). (D) Motor neuron survival over time was not altered in the iPSC-derived motor neuron cultures derived from four individual C9ORF72 ALS (C9-ALS) patients versus four control subjects (n=3 independent experiments) (1-way ANOVA, Tukey’s Multiple Comparison Test; Control vs. ALS, 95% confidence interval of difference, 4 weeks: −12.8 to 24.15 and 7 weeks: −7.36 to 30.42). Motor neuron counts are represented as a ratio of motor neuron (SMI32+) to pan-neuronal (TuJ1+) populations. (E) Properties of functional motor neurons were observed when iPSCs were differentiated into motor neurons (representative motor neuron from control iPSC line 83i control shown at day 69 of differentiation). Current injections of −10 pA and +5 pA (top left). Resting potential of the motor neuron was −60mV with 2.6 GigaOhm input resistance. Spontaneous activity of the same motor neuron (top right). Representative images confirming motor neuron identity of recorded neurons (bottom panels). Neurons filled with Alexa594 dye (red) during live electrophysiological recordings are shown post-fixation and counter-stained with motor neuron marker ChAT (green) confirming their identity.
To examine the stability of the GGGGCC repeat in C9-ALS patient motor neurons differentiated from iPSCs derived from patient fibroblasts, we performed Southern blot analysis (Fig. 1C). In three of the patient lines (28i, 29i, 52i), Southern blotting showed the expanded allele was 6–8kb, or ~800 repeats. In two lines there was a shift indicating somatic expansion of the repeat upon conversion of fibroblasts to iPSCs (29i and 52i), and line 29i showed a polymorphic contraction of the repeat upon differentiation into motor neurons (Fig. 1C). This may reflect the somatic variability observed across different patient brain regions (24), or may alternatively be due to clonal expansion of subclones with different-sized repeats. Interestingly, line 30i showed only ~70 repeats, which was stable during reprogramming of fibroblasts to iPSCs. Despite the shorter repeat, this individual exhibited early onset bulbar ALS with the shortest disease duration of the four patients, suggesting a sharp threshold for repeat length and disease. Additional genetic or environmental modifiers may have a strong influence on disease onset and severity (Table S1).
We previously observed that motor neurons from patients with spinal muscular atrophy, a childhood onset motor neuron disease, showed decreased survival in vitro (21, 25). Therefore, we examined motor neuron counts at two time points after differentiation, but saw no difference in the development or survival of motor neurons between C9-ALS patients versus controls (Fig. 1D). As neuronal counts become technically challenging due to confluent growth with longer time in culture, we cannot rule out that a survival phenotype may have emerged with further in vitro maturation.
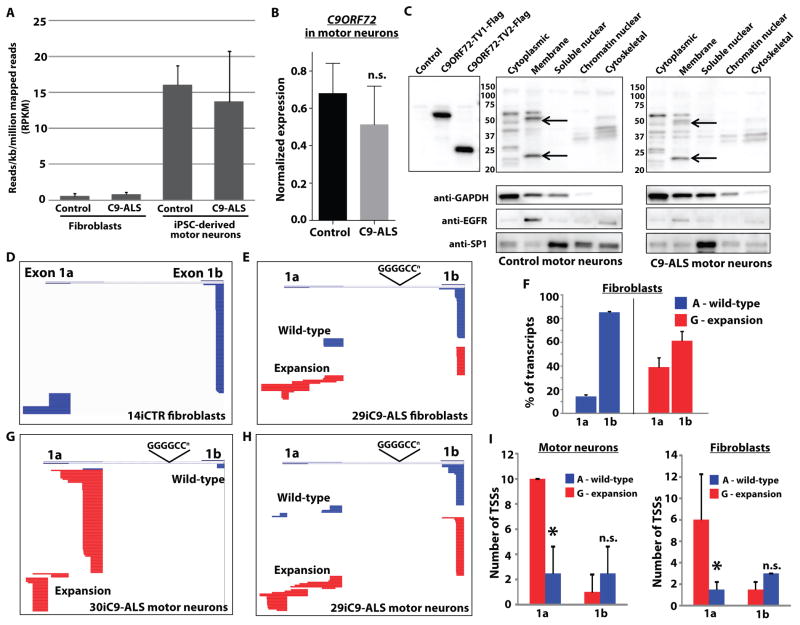
Early reports indicated that C9ORF72 gene expression is altered in patient tissue with decreased expression of either one (8) or all (10) isoforms. We first investigated expression from the C9ORF72 locus to examine the possibility of loss of function by performing RNA-seq analysis of both C9-ALS and control fibroblasts, and iPSC-derived motor neurons in culture. The normalized number of mapped sequence reads (RPKM) corresponding to all annotated exons of C9ORF72 was not different between C9-ALS patients and controls, in either fibroblasts or motor neuron cultures, with ~15-fold higher expression seen in neurons than in fibroblasts (Fig. 2A). Similar results were obtained using qR-PCR with primers to exon 2, common to all isoforms (Fig. 2B). Allele specific analysis from RNA-seq data indicated that both the wild-type and mutant allele were expressed similarly, and transcript specific analysis across two different annotation sets (Ensembl and Refseq) did not reveal differences between C9-ALS patients and controls (Fig. S4). Of note, sampling of exon 1a and 1b was relatively low in the RNA-seq reads possibly due to the high GC content and repetitive sequence in this region, so differential transcript analysis is driven primarily by exons 2–11. We further performed Western blot analysis of different cell fractions from iPSC-derived motor neuron cultures to investigate C9ORF72 protein levels. Commercially available C9ORF72 antibodies recognized isoforms 1 and 2 in whole cell lysates from transfected cells, with bands corresponding to these isoforms present in the membrane fraction of iPSC-derived motor neurons (Fig. 2C). This is consistent with the idea that C9ORF72 is a member of the DENN family of Rab-GEFs involved in membrane trafficking (26, 27). We confirmed the specificity of these bands by knockdown of C9ORF72 using antisense oligonucleotides (see below) (Fig. S5B). No difference in the level of C9ORF72 protein was observed in C9-ALS patient cells compared to controls (Fig. S5A).
Fig. 2. The hexanucleotide expansion does not alter expression of C9ORF72 but alters upstream exon utilization to promote transcription of the repeats.
(A) Reads per kilobase per million mapped reads (RPKM) from RNA sequencing of motor neurons differentiated from iPSCs derived from control individuals (n=4 independent subjects) and ALS patients with the C9ORF72 repeat expansions (C9-ALS, n=4 independent subjects) and fibroblasts, for all annotated transcripts from the C9ORF72 gene. Overall expression of C9ORF72 transcripts was not different between controls and C9ORF72 expansion carriers, with motor neuron cultures showing higher RPKM values than fibroblasts. (B) Quantitative RT-PCR in iPSC-derived motor neuron cultures from four control subjects and four C9ORF72 expansion carriers using primers in exon 2 (common to all C9ORF72 transcript variants), confirming equivalent expression of the C9ORF72 gene. Data are represented as mean ± SEM from n=3 independent experiments. n.s., not significant using student’s t-test. (C) Western blot of C9ORF72 protein in cellular fractions from iPSC-derived motor neuron cultures showing two bands in the membrane fraction running slightly smaller than the sizes of overexpressed long and short isoforms due to the presence of a Flag tag on these constructs. Representative blots showing similar levels of protein were observed in control (83iCTR) and C9ORF72 expansion (28iC9-ALS) motor neuron cultures indicating that the presence of the repeat did not alter overall protein levels. Data are representative of n=3 independent experiments. (D) Representative sequence alignments of upstream exon sequences (exons 1a or 1b) from 5′ RACE analysis of C9ORF72 transcripts in fibroblasts from a control subject (14iCTR). Stacked horizontal bars represent individual transcripts, with 100 transcripts sequenced for each sample. In control cells, transcripts containing exon 1b were most frequent, with little variability in transcriptional start sites for either exon 1a or exon 1b. (E) Alignment of 5′ RACE transcripts from fibroblasts from a C9ORF72 expansion patient (29iC9-ALS). Sequences derived from the wild-type or expansion allele were determined using SNP rs10757668 upstream of the RACE primer in exon 2. Expression of the wild-type allele (blue) was similar to control fibroblasts, whereas the expansion allele (red) showed more frequent usage of exon 1a and more variability in the transcriptional start site. (F) Percentage of transcripts containing exon 1a or 1b in C9ORF72 patient fibroblasts (29i and 30i) from the wild-type (A allele, blue) or expansion (G allele, red), showing an increase in the percentage of transcripts containing exon 1a derived from the expansion allele. *p<0.05, t-test, % of transcripts, wild-type vs. mutant allele. (G, H) Alignment of 5′ RACE from iPSC-derived motor neurons from two different C9ORF72 expansion ALS patients (30iC9-ALS and 29iC9-ALS), showing enhanced expression of the repeat (containing exon 1a), and variability in transcriptional start site usage by the mutant allele. (I) Comparison of the number of transcriptional start sites (TSSs) observed in fibroblasts from C9ORF72 patients 29i and 30i (right panel) and iPSC-derived motor neurons (left panel). The expansion allele (G, red) showed more TSSs for exon 1a transcripts than the wild-type allele (A, blue), whereas the number of TSSs for exon 1b was similar between the two alleles. *p<0.05, t-test, # of TSSs, wild-type (A) versus mutant (G).
The GGGGCC expansion in C9ORF72 occurs between the alternatively used exons 1a and 1b (8, 9). Therefore, the repeat is transcribed if exon 1a is utilized, but is located in the promoter region if exon 1b is utilized. As the RNA-seq showed low sampling of exons 1a and 1b and is dependent on existing transcript annotation to determine how the presence of the repeat influences upstream exon utilization, we performed 5′ RACE (rapid amplification of cDNA ends) of C9ORF72 transcripts in fibroblasts and iPSC-derived motor neuron cultures. Fibroblasts from a control subject showed transcripts predominantly containing exon 1b, with little variability in transcriptional start sites among exon 1a and 1b (Fig. 2D). Because of the presence of a single nucleotide polymorphism in exon 2 in ~17% of the population (rs10757668) (8), we were able to assign sequences as emanating from either the wild-type or expansion containing allele in two of the C9-ALS patient fibroblast (Fig 2E) and iPSC-derived motor neuron lines (Fig. 2G, H). The wild-type allele behaved similarly to control subjects, with predominant utilization of exon 1b. However, the mutant allele showed increased utilization of exon 1a compared to 1b in C9-ALS patients versus controls (Fig. 2F), and more variability in the location of the transcriptional start site (Fig 2I). Similar findings were present in iPSC-derived motor neurons from C9-ALS patients, with nearly selective utilization of exon 1a by the mutant allele in some cases (Fig. 2G). Expression of both isoforms was also observed in spinal cords from C9-ALS patients (Fig. S6). Together these data suggest that C9ORF72 expression is not significantly decreased in C9-ALS patient cells, but rather that there is a paradoxical shift toward transcription of the allele with the hexanucleotide repeat, supporting the notion that the C9ORF72 repeat expansion leads to gain of function, rather than loss of function.
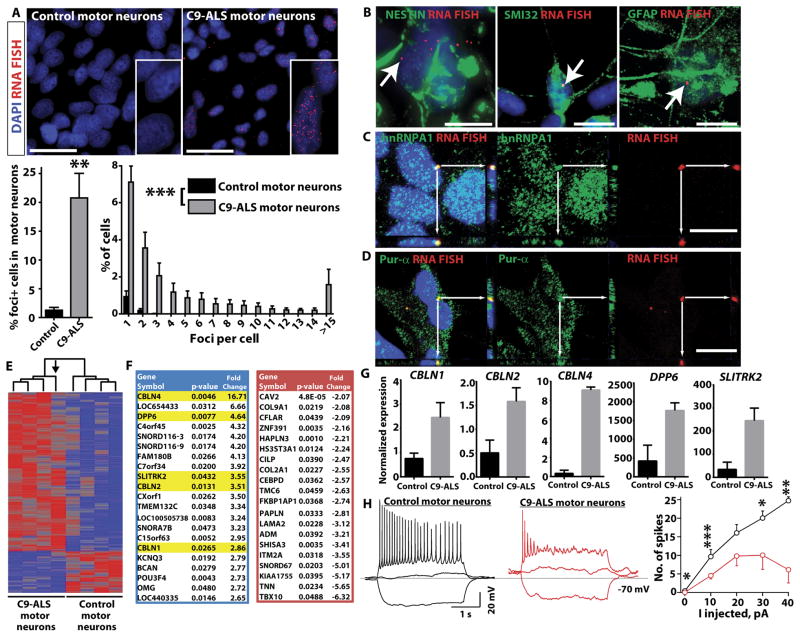
Given that the hexanucleotide repeat in C9ORF72 is transcribed, we next performed fluorescence in situ hybridization (FISH) to look for evidence of GGGGCC-containing RNA foci in C9-ALS patient iPSC-derived motor neurons. RNA foci were initially reported in C9-ALS and FTD patient tissues (8). Consistent with the observed transcription of the repeat, RNA foci were detected by FISH in ~20% cells in C9-ALS patient motor neuron cultures (Fig. 3A). Cells typically had 1–3 foci in the nucleus, but those with >15 foci were not uncommon and occasional cytoplasmic foci were also seen. Interestingly, the fewest RNA foci were observed in line 30i, which harbored only ~70 repeats (Fig. S7). Co-immunostaining with FISH revealed that foci were present in neuronal progenitors (nestin+), as well as motor neurons (SMI32+) and astroglial cells in the cultures (GFAP+) (Fig. 3B). RNA foci are directly involved in the pathogenesis of a number of repeat expansion diseases, including myotonic dystrophy type 1 (DM1), where they bind to RNA binding proteins and disrupt their function, leading to changes in gene expression and splicing (28). Therefore, we examined co-localization of RNA foci with a panel of different RNA-binding proteins. Confocal imaging revealed that RNA foci frequently co-localized with the hnRNPA1 and Pur-α (Fig. 3C, D), but not with hnRNPA3, hnRNPA2/B1, or TDP-43 or FUS RNA-binding proteins, which are mutated in rare cases of familial ALS (Fig. 3C, D; Fig. S8) (29). Recently, GGGGCC repeat associated non-ATG dependent (RAN) translation products were reported to be present in C9-ALS patient tissues (16, 17). However, we were not able to detect C9-RAN translation products in C9-ALS patient-derived motor neuron cultures (Fig. S9), or an increase in p62 positive inclusions, indicating that the presence of GGGGCC containing RNA foci does not always correlate with the production of insoluble dipeptides by RAN translation.
Fig. 3. iPSC-derived motor neuron cultures from C9-ALS patients develop GGGGCC RNA foci that bind to Pur-α and hnRNPA1.
(A) Representative images of fluorescence in situ hybridization (FISH) with an antisense probe to the GGGGCC repeat in iPSC-derived motor neuron cultures (lines 83iCTR and 52iC9-ALS). RNA foci were present in cells from all C9-ALS patients, but not in motor neuron cultures from control subjects. RNA foci were predominantly nuclear, but occasionally found in the cytoplasm as well. Scale bar = 25 μm. Histogram below shows the number of foci per cell. **p<0.01 unpaired t-test (two-tailed). ***control (all four subjects) vs. C9-ALS (all four subjects) p<0.0001 Mann Whitney test. (B) Representative images showing co-staining of GGGGCC FISH and markers of different cell types in iPSC-derived motor neuron cultures. RNA foci were present in the nuclei of neuronal precursors (nestin positive; line 29iC9-ALS shown), motor neurons (SMI32 positive; line 28iC9-ALS shown), and astroglial cells (GFAP positive; line 52iC9-ALS shown). Scale bar = 10 μm. (C, D) Representative images showing co-staining of GGGGCC FISH with RNA binding proteins. Co-localization of GGGGCC foci with hnRNPA1 and Pur-α was observed by confocal imaging. White arrows point to foci that stained for both RNA foci and hnRNPA1 or Pur-α in the same focal plane. Adjacent panels show the y-z and x-z axes confirming colocalization in 3 dimensions. Scale bar = 10 μm. C, line 52iC9-ALS; D, line 28iC9-ALS. (E) Heat map showing hierarchical clustering of differentially expressed genes identified by RNA-seq of iPSC-derived motor neuron cultures between four different C9-ALS patients (n=4; lines 28i, 29i, 30i, 52i) and four control subjects (n=4; lines 00i, 03i, 14i, 83i) with p< 0.05. The arrow indicates motor neuron cultures from the subject with the fewest GGGGCC repeats (~70 in line 30iC9-ALS), which clustered farthest from the three lines with larger repeats (~800). (F) Gene list of 20 highest upregulated (blue) and downregulated (red) genes in C9-ALS patient iPSC-derived motor neuron cultures versus controls. Genes highlighted in yellow include DPP6, implicated in prior ALS GWAS studies, and three members of the cerebellin family (CBLN1, 2, and 4). (G) qRT-PCR validation of differentially expressed genes highlighted in (F), all p<0.05, CTR (n=4 subjects) vs C9-ALS (n=4 subjects), t-test. (H) C9-ALS iPSC-derived motor neuron cultures are less excitable (n=184) than control iPSC-derived motor neuron cultures (n=137). Recordings were performed on motor neuron cultures that were between days 66–79 of differentiation. Representative traces are shown in response to current injections of −10, 0 and 10 pA into motor neurons derived from a control subject (black, iPSC line 83i) or C9-ALS patient (red, iPSC line 28i). Mean number of action potentials elicited as a function of current injection for control (black) and C9-ALS (red) cultured motor neurons. The graph (right) shows the number of spikes fired at different levels of current injection, with reduced numbers of spikes fired in C9-ALS iPSC-derived motor neurons compared to control iPSC-derived motor neurons (CTR, iPSC lines 14i and 83i, C9-ALS iPSC lines 28i and 52i; n=2 independent experiments). Resting potential and input resistance of the motor neurons is −68.5 mV with 3.4 GigaOhm for the control and −61.5 mV with 3.2 GigaOhm for the C9-ALS motor neurons. *p<0.05; **p<0.01; ***p<0.001; C9-ALS vs. control, unpaired t-test, two sample, unequal variance.
To examine alterations in gene expression in C9-ALS patient cells, we next analyzed the transcriptome by RNA-seq in C9-ALS iPSC-derived motor neuron cultures versus control motor neuron cultures (Fig. 3E, F and Figs. S10, S11). Hierarchical clustering of differentially expressed genes (p<0.05) showed that the three C9-ALS lines with ~800 repeats clustered closest to each other, whereas line 30i (~70 repeats) was most distinct within the C9-ALS cluster, suggesting that gene expression alterations correlated with repeat length (Fig. 3E). The list of the differentially expressed genes included DPP6, a gene identified in multiple prior GWA studies as being associated with sporadic ALS (30–32), the interacting potassium channel KCNQ3, and three members of the cerebellin family of proteins involved in synapse formation (33) (Fig. 3F). Functional pathway analysis of differentially expressed transcripts supported enrichment in genes involved in cell adhesion, synaptic transmission and neural differentiation (Table S2). Validation of a subset of these genes by quantitative RT-PCR confirmed their dysregulation, including DPP6 (Fig. 3G).
Given that expression of several genes involved in regulating membrane excitability (DPP6, KCNQ3) and synaptic transmission (CBLN1, CBLN2, CBLN4) were disrupted, we performed detailed electrophysiology on C9-ALS patient-derived motor neurons compared to controls (Fig. 3H; Fig. S12). We observed that C9-ALS patient-derived motor neurons showed decreased electrical excitability with the production of fewer spikes upon depolarization, consistent with the expected effect of increased KCNQ3 channel expression (34).
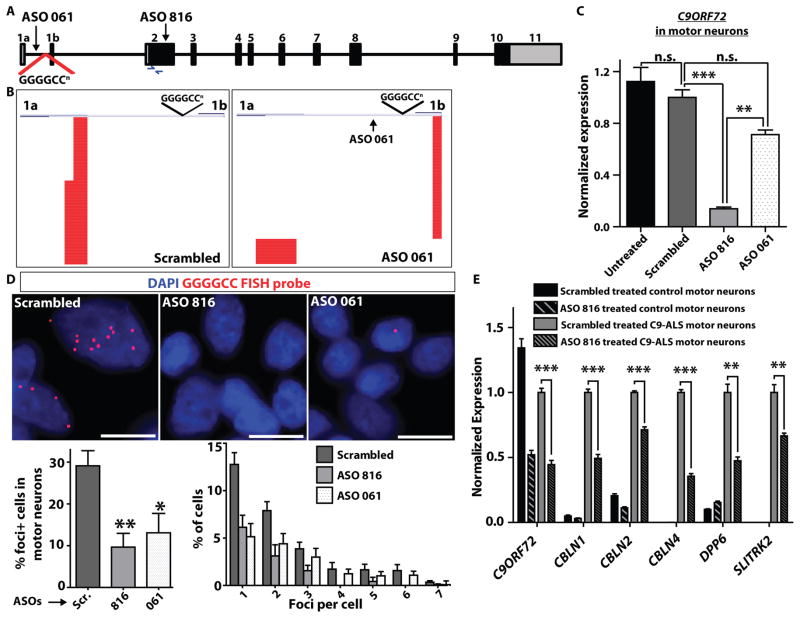
Antisense oligonucleotides (ASOs) have been used for gene-specific knockdown in diseases involving RNA or protein gain-of-function. Recent examples include ASO-mediated RNAse H-dependent degradation of CTG repeat containing “toxic” RNA transcripts in DM1 (35), and CAG repeat containing transcripts in Huntington’s disease (36). Given that we observed both transcription of the hexanucleotide repeat and formation of RNA foci with concomitant changes in gene expression in C9-ALS patient cells, we investigated whether ASOs targeting C9ORF72 could alter these disease-specific cellular phenotypes. We used two ASOs, one targeting exon 2 which is common to all transcripts (ASO816), and a second targeting the region in intron 1 adjacent to the repeat (ASO061) (Fig. 4A). Treatment of iPSC-derived motor neuron cultures with ASO816 led to knockdown of total C9ORF72 transcript levels by ~90% (Fig. 4B), with no observed toxicity to cultured motor neurons (Fig. S13). ASO061, which targets the first intron, led to a small decrease in C9ORF72 transcript levels (Fig. 4B) but altered upstream exon utilization leading to more exon 1b than exon 1a (repeat containing) transcripts, as determined by RACE analysis (Fig. 4C; Fig. S13). We next examined the effect of ASOs targeting C9ORF72 on the formation of GGGGCC-containing RNA foci. Both ASO816 (which knocks down overall C9ORF72 levels) and ASO061 (which specifically targets repeat-containing transcripts) led to suppression of RNA foci formation in C9-ALS patient iPSC-derived motor neurons (Fig. 4D). To determine if knockdown of C9ORF72 expression and RNA foci could reverse disease specific transcriptional changes, we performed qRT-PCR of DPP6, CBLN1, CBLN2, CBLN4, and SLITRK2 in iPSC-derived motor neurons from ALS patients and controls. Remarkably, motor neurons treated with ASO816 showed partial but significant correction (p<0.01) of the over-expression of these 5 genes observed in patient cells (Fig. 4E), with similar results obtained for ASO061 (Fig. S13). To assess reversal of transcriptional changes across all of the differentially expressed genes in C9-ALS patient iPSC-derived motor neuron cultures (Fig. 3F), we performed RNA-seq analysis of control and C9-ALS motor neurons, treated with either scrambled ASO or ASO816 (Fig. S13B). We observed concordance with the original RNA-seq analysis, with 93% of genes showing dysregulation in C9-ALS patient iPSC-derived motor neuron cultures compared to controls in both experiments. Furthermore, 50% of the upregulated genes, and 18% of downregulated genes, showed significant reversal toward a control cell phenotype after treatment with ASO816 (p<0.05) (Fig. S13B). These data provide further evidence that the hexanucleotide expansion causes gain of function toxicity as C9ORF72 knockdown corrected the transcriptional profile, rather than exaggerating altered gene expression.
Fig. 4. Knockdown of C9ORF72 with antisense oligonucleotides suppresses RNA foci in ALS motor neurons.
(A) Schematic diagram of C9ORF72 gene structure, showing the location of antisense oligonucleotides (ASO) 816 and 061, and primers for assaying C9ORF72 expression in exon 2. (B) Quantitative RT-PCR for total C9ORF72 in iPSC-derived motor neurons (C9-ALS lines 28iC9-ALS and 52iC9-ALS) treated with ASOs 816 and 061. ASO816, which targets the first coding exon common to all C9ORF72 isoforms, knocked down overall C9ORF72 levels by ~90%. ASO061 had a partial effect on total transcript levels. (1-way ANOVA Tukey’s Multiple Comparison Test, 95% CI of diff; untreated vs. scrambled (n.s.): −0.1713 to 0.4167; scrambled vs. ASO061 (n.s.): −0.006394 to 0.5816; scrambled vs. ASO816 (***): − 0.5671 to 1.155; ASO816 vs. ASO061 (**.):−0.8675 to −0.2796). n.s. = not significant. Data are represented as mean ± SEM from n=3 independent experiments. (C) 5′ RACE analysis (line 52iC9-ALS shown) to analyze 5′ exon utilization after treatment with either scrambled ASO (left) or ASO061 (right). While ASO061 did not alter total C9ORF72 transcript levels, it suppressed exon 1a containing transcripts, with a relative increase in exon 1b containing transcripts. (D) Representative images of RNA FISH on C9-ALS patient motor neuron cultures (lines 29iC9-ALS and 52iC9-ALS) treated with different ASOs (scrambled – “Scr.”, 816, 061) showing a marked suppression of RNA foci in ASO-treated cells. Scale bar = 10 μm. Graphs below show quantitation of the percentage of cells with foci (left), and a histogram showing the breakdown by foci per cell (right). ** = p<0.01; * = p<0.05; unpaired t-test (two-tailed). (E) Quantitative RT-PCR for genes that showed aberrant upregulation in C9-ALS iPSC-derived motor neuron cultures (DPP6, CBLN1, CBLN2, CBLN4 and SLITRK2) after treatment with scrambled ASO or ASO816 (lines 14iCTR and 52iC9-ALS shown). Error bars are mean ± sem. ***p<0.001; **p<0.01; unpaired t-test (two-tailed), scrambled vs. ASO816.
Discussion
Repeat expansions produce human disease through a variety of mechanisms. In fragile X syndrome and Friedreich’s ataxia, large repeats in non-coding regions lead to gene silencing and loss of function (37, 38), whereas in many spinocerebellar ataxias (SCA) and Huntington’s disease, expansions in gene coding regions predominantly lead to protein toxicity (39, 40). In repeat diseases including myotonic dystrophy (MD), expansions in non-coding transcribed regions can produce “toxic” RNA species, which sequester RNA binding proteins and disrupt gene splicing and transcription to mediate disease (41, 42). Interestingly, some non-coding repeats including those in DM1 and SCA8 were shown to produce non-ATG initiated peptides, which could also play a role in disease pathogenesis (43); similarly generated peptides were recently demonstrated in C9-ALS patient tissue (16, 17). Our data support gain of function as the mechanism of C9ORF72 repeat toxicity, as the mutant allele was preferentially transcribed rather than suppressed in ALS patient iPSC-derived motor neurons. Furthermore, knockdown of C9ORF72 to low levels (10% residual) in iPSC-derived motor neurons from control individuals was not toxic. The fact that RNA foci were present and co-localized with hnRNPA1 and Pur-α supports the notion that RNA toxicity contributes to C9ORF72 repeat expansion diseases. The GGGGCC repeat did not directly bind to TDP-43 but hnRNPA1 is a well characterized binding partner for TDP-43 (44). Therefore, sequestration of this protein by RNA foci could alter the effect of TDP-43 on its target RNAs thus providing a potential connection between TDP-43 and C9ORF72-related ALS. Notably, mutations in both hnRNPA1 and hnRNPA2/B1 were recently reported to cause motor neuron disease in humans (45), further supporting the notion that altered hnRNPA1 function can promote motor neuron degeneration. Additionally, Pur-α was recently shown to interact with GGGGCC-containing RNA foci, and to modulate the toxicity of these foci in a fly model (46). Given that multiple RNA binding proteins co-localize with the RNA foci, it remains to be determined whether the toxicity to motor neurons is primarily due to altered levels of an individual protein, or a combination of them. While it will take further work to determine whether the transcriptional alterations observed in C9-ALS patient iPSC-derived motor neurons are a direct or indirect consequence of altered RNA binding protein function, the fact that DPP6, a gene implicated in sporadic ALS in multiple independent GWA studies (30–32), was dysregulated suggests that these alterations reflect pathways directly relevant to ALS pathogenesis.
Several papers reported a decrease in the expression of C9ORF72 in patient tissue (8, 10, 14, 19) using qRT-PCR. While we observed a trend toward lower overall C9ORF72 mRNA levels via RNA-seq and qRT-PCR, it was not statistically significant. One reason for the discrepancy is likely to be that qRT-PCR cannot accurately sample all versions of exons 1a and 1b, which we observed were different and varied in location compared to the annotation databases used to make primer sets for qRT-PCR. Another reason is that some studies investigated autopsy material, which could show a decrease in C9ORF72 levels due to neuronal cell loss; cell lines derived from blood cells may not reflect expression levels in human motor neurons. Furthermore, we found that knockdown of C9ORF72 to very low levels had no impact on motor neuron survival. Finally, the altered gene expression profiles observed in C9-ALS patient cells was improved rather than worsened by C9ORF72 knockdown with ASOs, supporting the notion that these changes are due to gain of function of the C9ORF72 repeat rather than loss of function. Therefore, while it remains possible that the expansion leads to a small decrease in overall C9ORF72 expression, this does not appear to result in a functional deficit.
Although we did find evidence for toxic RNA foci in C9-ALS patient iPSC-derived motor neurons, we did not observe repeat-associated non-ATG dependent (C9-RAN) protein products in these cells. This is in contrast to another study (47), which observed C9-RAN positive material in neurons generated from patient iPSCs. One reason for the difference may be that this study generated generic neurons, rather than the motor neurons characterized here, and the production of RAN products could be neuronal cell type dependent. Regardless of whether C9-RAN products or toxic RNA granules (or both) are key to the pathogenesis of C9ORF72 repeat expansion diseases, our data support the possibility that knockdown of C9ORF72 by ASOs could reverse gain of function toxicity.
There are currently no effective treatments for ALS or FTD, and debate continues as to which model systems will most accurately predict success in subsequent clinical trials. Here, we have used iPSC technology to demonstrate robust patient-specific phenotypes in the disease relevant cell type. Given that the ASOs targeting the region adjacent to the C9ORF72 repeat were able to alter upstream exon utilization to block transcription of repeat containing RNA, it may be possible to suppress the toxic effects of C9ORF72 repeat transcription (whether RNA or protein mediated) while avoiding potential unforeseen consequences of knocking down overall C9ORF72 levels. Given the urgent need for effective drug development for ALS and FTD patients, and the relatively high frequency of C9ORF72 repeat expansions with an estimated 90,000 carriers in the U.K. (24), these data provide a basis for moving toward attempting antisense strategies to treat C9ORF72 repeat expansion diseases.
Materials and Methods
Study Design
In this study we utilized iPS cell and antisense oligonucleotide (ASOs) technologies to; a) determine whether expansions of a hexanucleotide repeat (GGGGCC) in the noncoding region of the C9ORF72 gene of ALS patient motor neurons cause disease through gain or loss of function mechanisms and, b) to discover potential therapeutics by targeting those disease mechanisms. We included four healthy control individuals and four C9ORF72 repeat expansion associated-ALS (C9-ALS) patients, generated non-integrating iPSC lines using episomal plasmid-based approach, and differentiated them into neural cultures containing motor neurons. We then determined in iPSC-derived motor neuron cultures from all 8 individuals (4 control and 4 C9-ALS); 1) C9ORF72 repeat stability by Southern Analysis, 2) motor neuron cell death vulnerability in C9-ALS cultures over time by performing cell counts, 3) C9ORF72 expression and C9-ALS disease-specific transcriptome profiles using RNA-sequencing and qRT-PCR, 4) expression and localization C9ORF72 protein by sub-cellular fractionation and Western analysis, 5) utilization of upstream C9ORF72 exons and transcription start sites by 5′ RACE-PCR, 6) formation and quantification of RNA foci structures and co-localization with RNA-binding proteins by RNA fluorescence in situ hybridization (FISH)/immunocytochemistry staining and confocal microscopy, 7) electrical excitability patterns of motor neurons upon depolarization by performing patch-clamp electrophysiology, and 8) rescue of the observed C9-ALS disease-specific cellular phenotypes by treating motor neuron cultures with ASOs targeting different regions of C9ORF72. Excluding electrophysiological assessments, all experiments described here were assessed in four control and four C9-ALS patient-derived motor neuron cultures in three or greater independent experimental replicates in a blinded fashion. Electrophysiology recording experiments were performed twice independently in a blinded method in motor neuron cultures from two control individuals and two C9-ALS patients.
Ethics Statement
Human control fibroblast cell lines were obtained from the Coriell Institute for Medical Research. The Coriell Cell Repository maintains the consent and privacy of the donor fibroblast samples. All the cell lines and protocols in the present study were carried out in accordance with the guidelines approved by institutional review boards at the Cedars-Sinai Medical Center and Washington University at St. Louis.
Generation of C9ORF72 ALS and healthy control iPSCs using Episomal Plasmids
Fibroblasts from C9ORF72 ALS patients (28iALS-n2, 29iALS-n1, 30-iALS-n1, 52iALS-n6) were derived at Washington University of St. Louis. Healthy control fibroblasts (00iCTR-n2: GM05400; 14iCTR-n6: GM03814; 83iCTR-n13: GM02183) were obtained from the Coriell Institute for Medical Research) or derived from healthy donors at Cedars-Sinai (03iCTR-n1). Reprogramming of the lines was performed using pCXLE-hUL, pCXLE-hSK, and pCXLE-hOCT3/4-shp53-F vectors (Addgene, adapted from previously published protocols (20)). Amaxa Human Dermal Fibroblast Nucleofector Kit was utilized to make the virus-free iPSC ALS lines. Briefly, fibroblasts (0.8 × 106 cells per nucleofection) were harvested and centrifuged at 200g for 5 minutes. The cell pellet was re-suspended carefully in Nucleofector Solution (VPD-1001, Lonza) and combined with episomal plasmids expression of six factors: OCT4, SOX2, KLF4, L-MYC, LIN28, and p53 shRNA, achieved by plasmid nucleofection (20). This method has a significant advantage over viral transduction, because genes do not integrate and are instead expressed episomally in a transient fashion. The cell/DNA suspension was transferred into the Nucleofector® and the U-023 program applied. All cultures were be maintained under norm-oxygen conditions (5%O2) during reprogramming, which further enhances the efficiency of iPS cell generation. The media was kept on for 48h and gradually changed to hiPSC media containing small molecules to enhance reprogramming efficiency. The small molecules used were, 1) sodium butyrate (0.5 mM), 2) glycogen synthase kinase 3β inhibitor of the Wnt/β-catenin signaling pathway (CHIR99021, 3 μM), 3) MEK pathway inhibitor (PD 0325901, 0.5 μM), 4) Selective inhibitor of TGF-β type I receptor ALK5 kinase, type I activin/nodal receptor ALK4 and type I nodal receptor ALK7 (A 83-01, 0.5 μM). Colonies with ES/iPSC-like morphology appeared 25–31 days later. Subsequently, colonies with the best morphology were picked and transferred to layers with standard hiPSC medium and BD Matrigel™ Matrix for feeder-independent maintenance of hiPSCs in chemically-defined mTeSR®1 medium. Three independent iPS cell clones will be picked from each reprogrammed fibroblast sample, further expanded and cryopreserved according to previously published protocols (48).
iPSC characterization
Rigorous characterization of iPSC cells was performed at the Cedars-Sinai iPS cell core using standard battery of pluripotency assays including, pluripotency surface and nuclear marker immunostaining and quantification by flow cytometry (> 80% SSEA4 and Oct3/4 double positivity), G-Band Karyotyping to ensure normal karyotypes, spontaneous embryoid body differentiation to judge germ layer formation capacity, gene-chip and bioinformatics-based PluriTest assay, quantitative RT-PCR for expression of endogenous pluripotency genes, and confirmation of absence of episomal plasmid genes by genomic DNA PCR, as previously described (20, 21, 23).
Motor neuron differentiation
The iPS cells were grown to near confluence under normal maintenance conditions prior to the start of the differentiation. Briefly, neural differentiation of iPSC colonies was induced by removal of mTeSR1 media and addition of defined neural differentiation media (SaND) media composed of IMDM supplemented with B27-vitamin A (2%) and N2 (1%).The cells were treated with this media for 6 days. The culture medium was replenished every 2–3 days. On day 6, the cultures were gently lifted from Matrigel by accutase treatment for 5 min at 37C. Single cell suspension at density of 10,000 cells/well was centrifuged in the presence of Matrigel and SaND media supplemented with the caudalizing factor, all-trans retinoic acid (RA; 0.1 μM), in sterilized 384-well PCR plates for formation of uniform sized neural aggregates After 2 days (day 8), neural aggregates were cultured in suspension low-attachment flasks grown for a further 9 days. To induce motor neuron differentiation, at day 17 post-iPSC stage, caudalized neural aggregates were placed in stage 1 motor neuron induction medium (Neurobasal, 2% B27 and 1% N2) in the presence of all-trans retinoic acid (RA; 0.1 μM) and ventralizing factor, purmorphamine (PMN; 1 μM), for further 8 days. Partially or fully dissociated caudo-ventralized neural aggergates spheres were then plated on poly-ornithine/laminin-coated coverslips in motor neuron maturation medium consisting of DMEM/F12, supplemented with 2% B27, RA (0.1 μM), PMN (1 μM), db-cAMP (1 μM), ascorbic acid (AA; 200 ng/ml), brain-derived neurotrophic factor (BDNF; 10 ng/ml) and glial cell line-derived neurotrophic factor (GDNF; 10 ng/ml) for a further 2–7 weeks. These conditions allowed for motor neuron differentiation under serum-free conditions. All differentiating cultures were maintained in humidified incubators at 37°C (5% CO2 in air).
Electrophysiology
Induced pluripotent human motorneurons derived from fibroblast of healthy or diseased patient were studied with a blinded experimental design at 9 to 10 weeks after differentiation. Cells were cultured on laminin coated coverslips and placed in extracellular solution (with composition in mM: 119 NaCl, 26 NaHCO3, 11 glucose, 2.5 KCl, 2.5 CaCl2, 1.3 MgCl2 and 1 NaH2PO4, pH 7.4 when gassed with 5% CO2 and 95% O2). Motor neurons were identified based on their characteristic trapezoidal shape with more than two dendritic processes were visualized with an upright microscope (Leica) using ×40 water immersion objective and constantly perfused with carbogen-bubbled extracellular solution at a rate of 2.8–3 ml per min. The majority of cells (43/60) elicited action potentials upon current injection, which were blocked by tetrodotoxin. Recordings were made with borosilicate glass pipettes with resistances between 4 to 5 M when filled with internal pipette solution (composition in mM: 135 KMSO4, 10 NaCl, 10 HEPES, 3 MgATP 0.3 Na2GTP, 0.1mM EGTA add 100 μM Alexa Flour 594 hydrazide (Molecular Probes) adjusted to pH 7.4. Whole-cell recordings were made using a Multiclamp 700B amplifier (Molecular Devices) in current-clamp mode. Each cell was subject to a series of increasing current injections from −10pA to 70pA in steps of 5 or 10 pA. Currents were filtered at 4 kHz and sampled at 20 kHz using a Digidata 1440 (Axon Instruments). Spike detection and analysis was performed using pClamp 10 and further analysis was carried in Excel or Igor software. Figures were made in Igor. The results are reported as mean ± SEM.
Immunocytochemistry
At the appropriate time point of differentiation in culture, plated cells were fixed in paraformaldehyde (PFA, 4% vol/vol) or chilled acetone:methanol (1:1), and rinsed in phosphate-buffered saline (PBS). Fixed cultures were blocked in 5–10% (vol/vol) goat or donkey serum with 0.2% (vol/vol) Triton X-100 and incubated with primary antibodies (Table S2). After incubation with the primary antibodies, cultures were rinsed in PBS and incubated in species specific AF488 or AF594-conjugated secondary antibodies. Nuclei were counterstained with Hoechst 33258 (0.5 μg/ml; Sigma) and mounted on glass slides using GelTol Aqueous Mounting Medium (Immunotech). Visualization was performed at 10X, 20X, 63X magnification (Nikon/Lecia) and quantification of positive cells was completed using Metamorph Offline software (Molecular Devices).
Plasmids
C9ORF72 cDNA transcript variant 1 (NM_145005.5) and transcript variant 2 (NM_018325.1) were purchased from Origene (pCMV6-C9ORF72-TV1-myc-DDK, cat. # RC222418 and pCMV6-C9ORF72-TV2-myc- DDK, cat. # RC209700).
ASOs
The antisense oligonucleotides were added at a concentration of 3μM to the motor neuron maturation media only on the designated first day of treatment. Only a single treatment was performed. Knockdown was observed after 7 days and this knockdown persisted through at least 14 days without additional treatment. The timepoint for assessment of mRNA knockdown, protein knockdown, foci reduction, and ASO reversal were all performed at 14 days after ASO treatment. The media was changed normally for the duration of treatment until the cells were fixed. Target sequences were as follows: control ASO- 141923 – CCTTCCCTGAAGGTTCCTCC; Exon 2 ASO-576816 – GCCTTACTCTAGGACCAAGA; Isoform specific ASO-577061 – TACAGGCTGCGGTTGTTTCC.
Primary antibodies
The following antibodies were used for Western Blot analysis: anti-TARDBP (Proteintech Group, 10782-2-AP), anti-C9ORF72 (GeneTex, GTX119776), anti-GAPDH (Sigma-Aldrich, G8795), anti-EGFR (Thermo Scientific, PA1-1110), and anti-SP1 (Millipore, 07-645). The anti-C9RANT antibody was previously described (16).
RNA isolation and Real-time quantitative PCR (RT-PCR)
RNA was collected from motor neuron cultures using PureLinkTM RNA mini kit (Ambion). RNA (1 μg) was first DNase treated and then reverse transcribed to cDNA with oligo(dT) using the Promega Reverse Transcriptase System. qPCR reactions were performed in triplicate using SYBR Green master mix (Applied Biosystems), genes of interest were normalized to RPL13 ribosomal Protein L13A, and calculated by 2^-ΔΔCT method (Schmittgen and Livak, 2008). PCR products were amplified using the following primers: C9ORF72 using the forward primer 5′-TGTGACAGTTGGAATGCAGTGA-3′ and the reverse primer 5′-GCCACTTAAAGCAATCTCTGTCTTG-3′, RPL13 Ribosomal Protein L13A using the forward 5′-CCTGGAGGAGAAGAGGAAAGAGA-3′ and the reverse 5′-TGAGGACCTCTGTGTATTTGTCAA-3′, CBLN1 using the forward 5′-TCAGAACGCAGCACTTTCATC-3′ and the reverse 5′-TTTAGCATGAGGCTCACCTGT-3′, CBLN2 using the forward 5′-CGACCGGTGCTTTTAAGGGT-3′ and the reverse 5′-CGAAGTTGCTCCAAACGCC-3′, CBLN4 using the forward 5′-CAGATCCTGGTGAATGTGGGT-3′ and the reverse 5′-AGTTAACCTGGATAGTTTGGCTCT-3′, and SLITRK2 using forward 5′-CCAAGTCTCCTGTGCCTCTC-3′ and the reverse 5′-CAGGTCAGAGATATTAGTGA-3′. Each PCR cycle consisted of 95°C for 10 min, 95°C 30 s -> 58°C for 60 s, for 50 cycles, and 72°C 5 min. The melting curve was measured and recorded from 65°C to 95°C in increments of 0.05°C to 0.5°C.
Western and dot blot analysis
293T cells were maintained in DMEM plus 10% FBS, 1% Glutamax, and 1% Non-essential amino acids. Transient transfection was performed in a 6 well plate using 1 μg of DNA and 3 μl of Mirus transfection reagent. 293T cell were transfected with C9ORF72 transcript variant 1 and 2 as positive controls for the identification of endogenous C9ORF72 in the motor neuron cultures. Whole cell lysates from 293T cells were washed twice with 1X PBS and lysed in buffer containing 50 mM Tris-HCl (pH 8.5), 100 mM NaCl, 5 mM MgCl2,1 mM EDTA, 0.5% NP40, 1 mM PMSF, and a complete protease inhibitor pellet (Roche).
Subcellular Protein Fractionation
Motor neuron cultures were washed twice with ice cold 1X PBS and collected by centrifugation 3000 rpm for 5′. Approximately 20 μl of cells from 3 control and 3 ALS motor neuron cultures were fractionated in triplicate following the protocol in the Subcellular Protein Fractionation Kit for Cultured Cells from Thermo Scientific. Soluble/insoluble Fractionation: Motor neuron cultures were washed twice with ice cold 1X PBS and collected in lysis buffer (10mM Tris (pH 7.4), 1% Triton X-100, 150 mM NaCl, 10% Glycerol, and 0.2 mM PMSF, Roche complete protease mini and phosphoSTOP pellets). Cells collected were first sheared in a glass dounce homogenizer then lysed on ice for 60 min before centrifugation at 15,000×g for 20′ at 4°C. Supernatant was collected as the detergent-soluble fraction. The pellet was washed 3x with lysis buffer and centrifuged at 15,000×g for 5′ each at 4°C. The pellet was resuspended in lysis buffer supplemented with 4% SDS, sonicated 3x, and boiled for 30 min. and collected as the detergent-insoluble fraction. Soluble/insoluble fractionation protocol was previously described (YM Kim, The J. Neuro. Sci, 2011) with minor changes in protease/phosphostase inhibitors and the addition of sonication to resuspend the detergent-insoluble pellets. For dot blot analysis for C9RANT products from human brain, tissue was processed as previously reported (16).
All protein concentrations were determined in using the BCA protein assay kit (Thermo), separated with SDS-PAGE, transferred onto PVDF membrane and western blot analysis performed using standard protocols (BIO RAD). 10–30 μg protein extracts were denatured in Laemmli sample buffer followed by 5 minutes of boiling and then resolved on either a 10% or Any KD gel (BIO RAD). After electrophoresis (150 V for 1:30 hours), the proteins were transferred using BIO RAD’s Trans-Blot® Turbo™ Transfer System and Transfer Pack to a 0.2μm PVDF membrane, with constant current of 2.5A up to 25V for 5 or 7 min. The membranes were then blocked in 5% nonfat dry milk TBST solution for 1 hour at room temperature, and incubated overnight at 4°C with the primary antibodies. The membranes were washed three times with TBST, incubated with horseradish peroxidase–linked goat anti-mouse or rabbit antibodies (Jackson ImmunoResearch) for 1 hours at room temperature and then washed three times in TBST. Detection of the immunoreactive bands was performed with the SuperSignal West Dura Extended Duration Substrate (Thermo) and chemiluminescent detected with the BIO RAD imaging system. PVDF membranes were stripped with One Minute® Plus Western Stripping Buffer (GM Biosciences) and reprobed with the appropriate subcellular antibody markers.
Quantitation and Statistical Analysis
Western blot quantitation preformed with ImageJ 1.46r software (National Institutes of Health). Experiments were performed in triplicate and values are presented as mean +/− SEM. All statistical evaluations were performed with GraphPad Prism software using unpaired t test to compare two groups. Prizm software (GraphPad software, La Jolla, CA) was used for all statistical analyses. All counting data from immunocyto/histochemical analyses and cell survival were expressed as mean values ± SEM and analyzed by two-tailed t-test or two-way ANOVA with Bonferonni post hoc test. Differences were considered significant when p<0.05
Southern blot quantification of C9ORF72 expansion size
10 μg of genomic DNA was isolated from either patient-derived cell lines (fibroblasts, iPSCs, or motor neuron cultures). Fragment electrophoresis at 20V for 24 hours was performed on a 0.8% SeaKem GTG agarose gel (Takara) with 1x TBE, with subsequent transfer to GeneScreen Plus nylon membranes (PerkinElmer). A 590-bp probe containing the smaller published probe gave improved sensitivity and was generated by PCR using the following primers: forward 5′-AAATTGCGATGACTTTGCAGGGGACCGTGG and reverse 5′-GCTCTCACAGTACTCGCTGAGGGTGAACAA). After gel purification, the probe was labeled with 32P-dCTP (PerkinElmer) using the Random Primed DNA labeling kit (Roche) and purified using Ambion NucAway spin columns (Life Technologies). Hybridization was carried out overnight at 68°C in Perfect Hyb Plus buffer (Sigma) containing 100μg/ml salmon sperm DNA (Life Technologies/Invitrogen). Filters were washed twice for 5 minutes at room temperature with 2x SSC+0.1% SDS, and twice for 20 minutes at 68°C with 0.2x SSC+0.1% SDS. BioMax MS film (Kodak) was exposed with an intensifying screen at −80°C for 5 days.
Rapid amplification of cDNA ends (5′ RLM-RACE) analysis
Total RNA was isolated from fibroblasts and precursor motor neuron cultures using the PureLink™ RNA Mini Kit as per the manufacturer’s protocol (Ambion BY Life Technologies, Cat. 12183018A Carlsbad, CA). 5′-RLM-RACE was performed using the GeneRacerTM kit (Invitrogen, Cat. L1502-02 Carlsbad, CA) with the manufacturer’s instructions modified as follows: 2 μg of total RNA was used for each sample. RNA was treated with calf intestinal phosphatase (CIP) to remove the 5′ phosphates from any truncated mRNA. Dephosphorylated RNA was treated with tobacco acid pyrophosphates (TAP) to remove the 5′ cap from full-length mRNA, leaving a 5′ phosphate. The GeneRacer RNA™ oligomer was then ligated to the 5′end of the mRNA using T4 RNA ligase. First-stand cDNA was synthesized using SuperScript™ III-reverse-transcriptase using random hexamer primers. From this reaction, 1 μg of cDNA was used to amplify the 5′ ends for sequencing using Platinum Taq DNA Polymerase High Fidelity (Invitrogen, Cat.12532-016, Carlsbad, CA), the GeneRacer 5′ primer (5′-CGACTGGAGCACGAGGACACTGA-3′) and a C9ORF72 gene-specific primer (5′-AACTGGAATGGGGATCGCAGCACAT-3′) with cycling parameters as described in the GeneRacerTM kit manual: 1 cycle of 94°C for 2 min, then 35 cycles of 94°C for 24 s, 55°C for 30 s and 72°C for 1 min, then 1 cycle of 72°C for 2 min and held at 4. The unused primers and nucleotides were removed using two different techniques for the fibroblast and precursor motor neurons. The fibroblast primary PCR products were treated with EXOSAP-IT (Affymetrix, Cat. 78200, Santa Clara, CA). One microliter of the EXOSAP-IT fibroblast amplification products were diluted into 10 μl of water and electrophoretically separated in a 2% agarose gel incorporating ethidium bromide and visualized three band products (469, 474 and 574 bp) by ultraviolet light transillumination. Once confirmed, the secondary PCR was performed using GoTaq (Promega, Cat. PRM8297, Madison, Wisconsin), internal primers [GeneRacer (5′-GGACACTGACATGGACTGAAGGAGTA-3′) C9ORF72 gene-specific (5′-GTGATGTCGACTCTTTGCCCACCGC-3′)] and 1 μg of the primary PCR. The secondary PCR cycling parameter for fibroblasts is as indicated: 1 cycle of 94°C for 2 min, then 15 cycles of 94°C for 24 s, 55°C for 30 s and 72°C for 1 min, then 1 cycle of 72°C for 2 min.
More extensive methods were taken when purifying the precursor motor neurons. It was indicative that ExoSAP-IT was unable to remove the excess primers and nucleotides by the multiple bands that were present on the 3% agarose gel after solely treated with EXOSAP-IT. The primary PCR products were gel purified before proceeding with the nested PCR. Fifteen microliters of the primary PCR product was ran on a 1.5% gel and bands located between 400–600bp were removed. One microliter of the 12ul elution was used as a template in the nested PCR. In addition, a hot start touchdown PCR method was used to minimize mispriming and extension, increase specificity and reduce background amplification on the secondary PCR of the motor neurons. The secondary PCR cycling parameter for precursor motor neurons was modified as follows: 1 cycle of 94°C for 2 min, then 5 cycles of 94°C for 20 s and 72°C for 1 min, 94°C for 30 sec and 70°C for 1 min, then 6 cycle of 94°C for 30 s, 68°C for 30 s decreasing by two degrees every 30 seconds and 68°C for 30 s, then 14 cycles of 94°C for 30 s, 65°C for 30 s and 68°C for 30 s, then 1 cycle of 68°C for 10 min and finally 4°C for holding.
Fifteen micrograms of the secondary PCR products, overall, were ran on 1.5% agarose gel and bands located between 150–300bp (163, 168 and 268bp) were collected and eluted in 12 ul water. One microliter of the gel purified secondary PCR product was cloned using pCR®4-TOPO vector (Life Technologies, Cat.K4575-01, Carlsbad, CA) and transformed in TOP 10 cells to grow overnight on Kanamycin agar plates. Colonies were hand-picked and sent to GeneWiz sequencing for analysis.
Cell Counting
Several pictures were taken in a non-biased fashion of each of the slides. The total cell count was obtained by counting hoescht staining using the Metamorph nuclei counting program. The other marker cell counts were done by the investigators. Methods for counting were standardized.
RNA-seq and microarray analysis
For RNA-seq analysis, iPSC cultures were differentiated to motor neurons as above and harvested using an Ambion extraction kit. mRNA was isolated, and library preparation and sequencing was performed on the Illumina Hi-Seq platform for 100bp paired end reads. The resultant reads were aligned to the hg19 build of the human genome using BOWTIE, and imported to Partek software for gene annotation and differential expression analysis.
Fluorescence in situ hybridization (FISH) and FISH/immunocytochemistry staining
The cells were grown on 4-well chamber slide (Lab-Tek II chamber slide system, Cat#154917, Thermo Fischer Scientific, Rochester, NY, USA) and fixed with 4% paraformaldehyde (Cat#15714; Electron Microscopy Sciences, Hatfield, PA, USA) in phosphate buffered saline (PBS). The cells were then permeabilized with diethylpyrocarbonate (DEPC)-PBS/0.2% Triton X-100 (Fisher Scientific, Pittsburgh, PA, USA; Cat #BP151) and washed with DEPC-PBS. Four hundred μl of hybridization buffer comprising of 50% formamide (IBI Scientific, Peosta, IA, USA; Cat #IB72020), DEPC-2xSSC (300 mM sodium chloride, 30 mM sodium citrate, pH 7.0), 10% w/v dextran sulfate (Sigma-Aldrich, St. Louis, MO, USA; Cat #D8960), and DEPC-50 mM sodium phosphate, pH 7.0) was added to each slide. The probe sequence was LNA oligonucleotide #500150 (/5TYE563/CCCCGGCCCCGGCCCC; Exiqon, Inc; Woburn, MA, USA). Glass cover slip was applied and slides placed in a slide holder (Microscope slide folder, Thermo Fischer Scientific Inc., Pittsburg, PA, USA, Cat #22-244-026) and prehybridised for 30 min at 66°C. The hybridization buffer was then allowed to drain off and 400 μl of 40nM probe mix in hybridization buffer was added to each of the slides. Cover slip was re-applied and hybridization performed for 3 hours at 66°C in the dark. The samples were then rinsed once in DEPC-2xSSC/0.1% Tween-20 (Fisher Scientific, Pittsburgh, PA, USA; Cat #BP337) at room temperature and in DEPC-0.1xSSC for three times at 65°C. Finally, incubation with 1 μg/μl of DAPI (4′, 6-diamidino-2-phenylindole, dihydrochloride; Molecular Probes, Inc., Eugene, OR, USA, Cat #D21490) was performed and the slides were mounted using ProLong Gold antifade reagent (Molecular Probes, Inc., Eugene, OR, USA, Cat #P36931). The processed slides were stored overnight at 4°C in the dark before performing microscopic analysis. To investigate into the cell types harboring RNA foci, we added a modified immunofluorescence protocol to the hybridized specimens. RNA FISH was done as mentioned above up to the steps of post hybridization DEPC-0.1xSSC washes. The slides were blocked for 15 min with 10% Horse Serum in Tris buffered saline/0.5% Triton X-100 and then incubated overnight at 4°C in a humid chamber with one of the following antibodies: anti-nestin (rabbit polyclonal, 1:1000 dilution, EMD Millipore, Billerica, MA, USA, cat #AB5922), anti-neurofilament H non-phosphorylated (SMI32; mouse monoclonal, 1:1000, Covance, Princeton, NJ, USA, Cat #SMI-32R), anti-glial fibrillary acidic protein (GFAP; rabbit polyclonal, 1:1000, Dako, Carpinteria, CA, USA, Cat #Z0334), anti-N-terminal TARDP (rabbit polyclonal, 1:1000, Proteintech Group, Chicago, IL, USA, Cat #10782-2-AP), anti-hnRNP A1(mouse monoclonal, 1:1000, GeneTex, Irvine, CA, USA, Cat #GTX25832), or anti-fused in sarcoma (FUS; rabbit polyclonal, 1:100, MBL Int., Co., Woburn, MA, USA, Cat #JM-3771-100). AlexaFluor 488 conjugated secondary antibodies (goat anti-rabbit IgG, cat #A11034 or goat anti-mouse IgG, cat #A11029, Molecular Probes, Inc., Eugene, OR, USA) were used at a 1:1000 dilution for 30 min at room temperature. Incubation with DAPI was done as described above and the slides were mounted with Pro-Long Gold antifade reagent. The processed slides were stored overnight at 4°C in the dark before performing microscopic analysis.
Supplementary Material
Fig. S1. Characterization of C9ORF72 patient and control iPSCs.
Fig. S2. Confirmation of absence of exogenous pluripotency genes in C9ORF72 patient iPSCs and generation of motor neuron precursors.
Fig. S3. Generation of control subject motor neuron cultures from iPSCs.
Fig. S4. RNA-seq analysis of SNP rs1075766 for identification of wild-type and expansion alleles, and differential C9ORF72 transcript analysis from Refseq and Ensembl annotations.
Fig. S5. C9ORF72 protein and antibody characterization.
Fig. S6. 5′ RACE and qRT-PCR analysis of C9ORF72 in human ALS patient spinal cords.
Fig. S7. RNA foci quantitation in iPSC-derived motor neuron cultures from individual patients.
Fig. S8. Characterization of binding of GGGGCC RNA foci to known heterogeneous nuclear ribonucleoproteins (hnRNPs) and ALS-related factors in C9-ALS motor neuron cultures by confocal imaging.
Fig. S9. C9RANT and p62 inclusions were not observed in C9-ALS motor neuron cultures.
Fig. S10. Summary of RNA-seq analysis in iPSC-derived motor neurons from C9-ALS patients versus controls.
Fig. S11. Hierarchical clustering analysis of RNA-sequencing data in iPSC-derived motor neurons from C9-ALS patients versus controls.
Fig. S12. Electrophysiological properties of control and C9-ALS patient derived motor neurons separated by individual subject.
Fig. S13. Absence of any ASO toxicity in motor neuron cultures and reversal of transcription profiles by RNA-sequencing in iPSC-derived motor neurons.
Table S1. Clinical information on subjects with C9ORF72 hexanucleotide expansions and controls used for iPS cell lines in this study.
Table S2. Functional pathway analysis of differentially expressed genes in iPSC-derived motor neuron cultures from C9-ALS patients versus controls.
Acknowledgments
We thank Renate Lewis and Bryan Traynor for help with Southern blotting, Paul Cooper for assistance with PCR genotyping of C9ORF72 repeat patients, Charles A. Thornton for providing technical advice on performing RNA FISH.
Funding: This work was supported by National Institutes of Health (NIH) Grants NS055980 and NS069669 (R.H.B), NIH-U24NS07837 (C.N.S.), and the California Institute for Regenerative Medicine grant RT2-02040 (to C.N.S., D.S.). R.H.B. holds a Career Award for Medical Scientists from the Burroughs Wellcome Fund. Analytical work was partially supported by the UCLA Muscular Dystrophy Core Center funded by the National Institute of Arthritis, Musculoskeletal, and Skin Disorders (P30 AR057230) within the Center for Duchenne Muscular Dystrophy at UCLA LP work was supported by Mayo Clinic Foundation; National Institutes of Health/National Institute on Aging [R01 AG026251 (LP), National Institutes of Health/National Institute of Neurological Disorders and Stroke [R21 NS074121 (TG), R01 NS063964 (LP); R01 NS077402 (LP), and R21 NS084528 (LP)]; National Institute of Environmental Health Services [R01 ES20395 (LP)]; Amyotrophic Lateral Sclerosis Association (LP).
Footnotes
Author Contributions: D.S., J.G.O., S.G., M.S., A.K.M.G.M., S.B., S.C., M.B., T.G., L.P., P.M., T.S.O., J.R., M.H., F.R., F.B., C.N.S., and R.H.B. participated in the planning, design, and interpretation of experiments. D.S. and M.S. performed iPSC culture, motor neuron differentiation and survival analysis, J.G.O. performed RAN product and C9ORF72 expression experiments, P.M. performed electrophysiology experiments, S.C., S.G. and S.B. performed 5′RACE experiments and plasmid cloning, A.K.M.G.M. performed FISH and immunocytochemistry experiments, and M.H. performed Southern blots; D.S., C.N.S. and R.H.B. wrote the manuscript.
Competing interests: FB and FR are employees of Isis Pharmaceuticals and hold stock options in the company. FB serves on the scientific advisory board of the Experimental Therapeutic Centre, Singapore. FB has submitted patents related to this work regarding the design and use of antisense oligonucleotides targeting the C9ORF72 transcript.
References
- 1.Lomen-Hoerth C, Anderson T, Miller B. The overlap of amyotrophic lateral sclerosis and frontotemporal dementia. Neurology. 2002 Oct 8;59:1077. doi: 10.1212/wnl.59.7.1077. [DOI] [PubMed] [Google Scholar]
- 2.Chen-Plotkin AS, Lee VM, Trojanowski JQ. TAR DNA-binding protein 43 in neurodegenerative disease. Nat Rev Neurol. 2010 Apr;6:211. doi: 10.1038/nrneurol.2010.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ravits J, et al. Deciphering amyotrophic lateral sclerosis: what phenotype, neuropathology and genetics are telling us about pathogenesis. Amyotrophic lateral sclerosis & frontotemporal degeneration. 2013 May;14(Suppl 1):5. doi: 10.3109/21678421.2013.778548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Seeley WW. Selective functional, regional, and neuronal vulnerability in frontotemporal dementia. Curr Opin Neurol. 2008 Dec;21:701. doi: 10.1097/WCO.0b013e3283168e2d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Neumann M, et al. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science. 2006 Oct 6;314:130. doi: 10.1126/science.1134108. [DOI] [PubMed] [Google Scholar]
- 6.Johnson JO, et al. Exome sequencing reveals VCP mutations as a cause of familial ALS. Neuron. 2010 Dec 9;68:857. doi: 10.1016/j.neuron.2010.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Al-Chalabi A, et al. The genetics and neuropathology of amyotrophic lateral sclerosis. Acta Neuropathol. 2012 Sep;124:339. doi: 10.1007/s00401-012-1022-4. [DOI] [PubMed] [Google Scholar]
- 8.DeJesus-Hernandez M, et al. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron. 2011 Oct 20;72:245. doi: 10.1016/j.neuron.2011.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Renton AE, et al. A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron. 2011 Oct 20;72:257. doi: 10.1016/j.neuron.2011.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gijselinck I, et al. A C9orf72 promoter repeat expansion in a Flanders-Belgian cohort with disorders of the frontotemporal lobar degeneration-amyotrophic lateral sclerosis spectrum: a gene identification study. Lancet Neurol. 2012 Jan;11:54. doi: 10.1016/S1474-4422(11)70261-7. [DOI] [PubMed] [Google Scholar]
- 11.Harms M, et al. C9orf72 hexanucleotide repeat expansions in clinical Alzheimer disease. JAMA neurology. 2013 Jun;70:736. doi: 10.1001/2013.jamaneurol.537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cacace R, et al. C9orf72 G4C2 repeat expansions in Alzheimer’s disease and mild cognitive impairment. Neurobiol Aging. 2013 Jun;34:1712 e1. doi: 10.1016/j.neurobiolaging.2012.12.019. [DOI] [PubMed] [Google Scholar]
- 13.Nuytemans K, et al. C9ORF72 Intermediate Repeat Copies Are a Significant Risk Factor for Parkinson Disease. Annals of human genetics. 2013 Jul 12; doi: 10.1111/ahg.12033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xi Z, et al. Hypermethylation of the CpG Island Near the GC Repeat in ALS with a C9orf72 Expansion. Am J Hum Genet. 2013 May 22; doi: 10.1016/j.ajhg.2013.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ciura S, et al. Loss of function of C9orf72 causes motor deficits in a zebrafish model of Amyotrophic Lateral Sclerosis. Ann Neurol. 2013 May 30; doi: 10.1002/ana.23946. [DOI] [PubMed] [Google Scholar]
- 16.Ash PE, et al. Unconventional Translation of C9ORF72 GGGGCC Expansion Generates Insoluble Polypeptides Specific to c9FTD/ALS. Neuron. 2013 Feb 20;77:639. doi: 10.1016/j.neuron.2013.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mori K, et al. The C9orf72 GGGGCC Repeat Is Translated into Aggregating Dipeptide-Repeat Proteins in FTLD/ALS. Science. 2013 Feb 7; doi: 10.1126/science.1232927. [DOI] [PubMed] [Google Scholar]
- 18.Harms MB, et al. Lack of C9ORF72 coding mutations supports a gain of function for repeat expansions in amyotrophic lateral sclerosis. Neurobiol Aging. 2013 Sep;34:2234 e13. doi: 10.1016/j.neurobiolaging.2013.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fratta P, et al. Homozygosity for the C9orf72 GGGGCC repeat expansion in frontotemporal dementia. Acta Neuropathol. 2013 Sep;126:401. doi: 10.1007/s00401-013-1147-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Okita K, et al. A more efficient method to generate integration-free human iPS cells. Nat Methods. 2011 May;8:409. doi: 10.1038/nmeth.1591. [DOI] [PubMed] [Google Scholar]
- 21.Sareen D, et al. Inhibition of apoptosis blocks human motor neuron cell death in a stem cell model of spinal muscular atrophy. PLoS ONE. 2012;7:e39113. doi: 10.1371/journal.pone.0039113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.HDi Consortium. Induced pluripotent stem cells from patients with Huntington’s disease show CAG-repeat-expansion-associated phenotypes. Cell stem cell. 2012 Aug 3;11:264. doi: 10.1016/j.stem.2012.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Muller FJ, et al. A bioinformatic assay for pluripotency in human cells. Nat Methods. 2011 Apr;8:315. doi: 10.1038/nmeth.1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Beck J, et al. Large C9orf72 Hexanucleotide Repeat Expansions Are Seen in Multiple Neurodegenerative Syndromes and Are More Frequent Than Expected in the UK Population. Am J Hum Genet. 2013 Feb 19; doi: 10.1016/j.ajhg.2013.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ebert AD, et al. Induced pluripotent stem cells from a spinal muscular atrophy patient. Nature. 2009 Jan 15;457:277. doi: 10.1038/nature07677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang D, Iyer LM, He F, Aravind L. Discovery of Novel DENN Proteins: Implications for the Evolution of Eukaryotic Intracellular Membrane Structures and Human Disease. Frontiers in genetics. 2012;3:283. doi: 10.3389/fgene.2012.00283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Levine TP, Daniels RD, Gatta AT, Wong LH, Hayes MJ. The product of C9orf72, a gene strongly implicated in neurodegeneration, is structurally related to DENN Rab-GEFs. Bioinformatics. 2013 Feb 15;29:499. doi: 10.1093/bioinformatics/bts725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Osborne RJ, et al. Transcriptional and post-transcriptional impact of toxic RNA in myotonic dystrophy. Hum Mol Genet. 2009 Apr 15;18:1471. doi: 10.1093/hmg/ddp058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lagier-Tourenne C, Polymenidou M, Cleveland DW. TDP-43 and FUS/TLS: emerging roles in RNA processing and neurodegeneration. Hum Mol Genet. 2010 Apr 15;19:R46. doi: 10.1093/hmg/ddq137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van Es MA, et al. Genetic variation in DPP6 is associated with susceptibility to amyotrophic lateral sclerosis. Nat Genet. 2008 Jan;40:29. doi: 10.1038/ng.2007.52. [DOI] [PubMed] [Google Scholar]
- 31.Blauw HM, et al. A large genome scan for rare CNVs in amyotrophic lateral sclerosis. Hum Mol Genet. 2010 Oct 15;19:4091. doi: 10.1093/hmg/ddq323. [DOI] [PubMed] [Google Scholar]
- 32.Kwee LC, et al. A high-density genome-wide association screen of sporadic ALS in US veterans. PLoS ONE. 2012;7:e32768. doi: 10.1371/journal.pone.0032768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wei P, et al. The Cbln family of proteins interact with multiple signaling pathways. J Neurochem. 2012 Jun;121:717. doi: 10.1111/j.1471-4159.2012.07648.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cavaliere S, Gillespie JM, Hodge JJ. KCNQ channels show conserved ethanol block and function in ethanol behaviour. PLoS ONE. 2012;7:e50279. doi: 10.1371/journal.pone.0050279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wheeler TM, et al. Targeting nuclear RNA for in vivo correction of myotonic dystrophy. Nature. 2012 Aug 2;488:111. doi: 10.1038/nature11362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kordasiewicz HB, et al. Sustained therapeutic reversal of Huntington’s disease by transient repression of huntingtin synthesis. Neuron. 2012 Jun 21;74:1031. doi: 10.1016/j.neuron.2012.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Todd PK, Paulson HL. RNA-mediated neurodegeneration in repeat expansion disorders. Ann Neurol. 2010 Mar;67:291. doi: 10.1002/ana.21948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Koeppen AH. Friedreich’s ataxia: pathology, pathogenesis, and molecular genetics. J Neurol Sci. 2011 Apr 15;303:1. doi: 10.1016/j.jns.2011.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Williams AJ, Paulson HL. Polyglutamine neurodegeneration: protein misfolding revisited. Trends Neurosci. 2008 Oct;31:521. doi: 10.1016/j.tins.2008.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Orr HT. Cell biology of spinocerebellar ataxia. J Cell Biol. 2012 Apr 16;197:167. doi: 10.1083/jcb.201105092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wheeler TM, Thornton CA. Myotonic dystrophy: RNA-mediated muscle disease. Curr Opin Neurol. 2007 Oct;20:572. doi: 10.1097/WCO.0b013e3282ef6064. [DOI] [PubMed] [Google Scholar]
- 42.Echeverria GV, Cooper TA. RNA-binding proteins in microsatellite expansion disorders: mediators of RNA toxicity. Brain Res. 2012 Jun 26;1462:100. doi: 10.1016/j.brainres.2012.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zu T, et al. Non-ATG-initiated translation directed by microsatellite expansions. Proc Natl Acad Sci U S A. 2011 Jan 4;108:260. doi: 10.1073/pnas.1013343108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Buratti E, et al. TDP-43 binds heterogeneous nuclear ribonucleoprotein A/B through its C-terminal tail: an important region for the inhibition of cystic fibrosis transmembrane conductance regulator exon 9 splicing. J Biol Chem. 2005 Nov 11;280:37572. doi: 10.1074/jbc.M505557200. [DOI] [PubMed] [Google Scholar]
- 45.Kim HJ, et al. Mutations in prion-like domains in hnRNPA2B1 and hnRNPA1 cause multisystem proteinopathy and ALS. Nature. 2013 Mar 28;495:467. doi: 10.1038/nature11922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xu Z, et al. Expanded GGGGCC repeat RNA associated with amyotrophic lateral sclerosis and frontotemporal dementia causes neurodegeneration. Proc Natl Acad Sci U S A. 2013 May 7;110:7778. doi: 10.1073/pnas.1219643110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Almeida S, et al. Modeling key pathological features of frontotemporal dementia with C9ORF72 repeat expansion in iPSC-derived human neurons. Acta Neuropathol. 2013 Sep;126:385. doi: 10.1007/s00401-013-1149-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yu J, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007 Dec 21;318:1917. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Characterization of C9ORF72 patient and control iPSCs.
Fig. S2. Confirmation of absence of exogenous pluripotency genes in C9ORF72 patient iPSCs and generation of motor neuron precursors.
Fig. S3. Generation of control subject motor neuron cultures from iPSCs.
Fig. S4. RNA-seq analysis of SNP rs1075766 for identification of wild-type and expansion alleles, and differential C9ORF72 transcript analysis from Refseq and Ensembl annotations.
Fig. S5. C9ORF72 protein and antibody characterization.
Fig. S6. 5′ RACE and qRT-PCR analysis of C9ORF72 in human ALS patient spinal cords.
Fig. S7. RNA foci quantitation in iPSC-derived motor neuron cultures from individual patients.
Fig. S8. Characterization of binding of GGGGCC RNA foci to known heterogeneous nuclear ribonucleoproteins (hnRNPs) and ALS-related factors in C9-ALS motor neuron cultures by confocal imaging.
Fig. S9. C9RANT and p62 inclusions were not observed in C9-ALS motor neuron cultures.
Fig. S10. Summary of RNA-seq analysis in iPSC-derived motor neurons from C9-ALS patients versus controls.
Fig. S11. Hierarchical clustering analysis of RNA-sequencing data in iPSC-derived motor neurons from C9-ALS patients versus controls.
Fig. S12. Electrophysiological properties of control and C9-ALS patient derived motor neurons separated by individual subject.
Fig. S13. Absence of any ASO toxicity in motor neuron cultures and reversal of transcription profiles by RNA-sequencing in iPSC-derived motor neurons.
Table S1. Clinical information on subjects with C9ORF72 hexanucleotide expansions and controls used for iPS cell lines in this study.
Table S2. Functional pathway analysis of differentially expressed genes in iPSC-derived motor neuron cultures from C9-ALS patients versus controls.