SUMMARY
Heart failure (HF) is driven by the interplay between master regulatory transcription factors and dynamic alterations in chromatin structure. While pathologic gene transactivation in this context is known to be associated with recruitment of histone acetyl-transferases and local chromatin hyperacetylation, the role of epigenetic reader proteins in cardiac biology is unknown. We therefore undertook a first study of acetyl-lysine reader proteins, or bromodomains, in HF. Using a chemical genetic approach, we establish a central role for BET-family bromodomain proteins in gene control during HF pathogenesis. BET inhibition potently suppresses cardiomyocyte hypertrophy in vitro and pathologic cardiac remodeling in vivo. Integrative transcriptional and epigenomic analyses reveal that BET proteins function mechanistically as pause-release factors critical to activation of canonical master regulators and effectors that are central to HF pathogenesis and relevant to the pathobiology of failing human hearts. This study implicates epigenetic readers in cardiac biology and identifies BET co-activator proteins as therapeutic targets in HF.
INTRODUCTION
Heart failure (HF) is a leading cause of healthcare expenditures, hospitalization and mortality, in modern society (Hill and Olson, 2008; Roger et al., 2012). HF occurs when the heart is unable to maintain organ perfusion at a level sufficient to meet tissue demand, and results in fatigue, breathlessness, multi-organ dysfunction, and early death. Existing pharmacotherapies for individuals afflicted with HF, such as beta adrenergic receptor antagonists and inhibitors of the renin-angiotensin system, generally target neurohormonal signaling pathways. While such therapies have improved survival in HF patients, residual morbidity and mortality remain unacceptably high (Roger et al., 2012). In light of this unmet clinical need, the elucidation of novel mechanisms involved in HF pathogenesis holds the promise of identifying new therapies for this prevalent and deadly disease.
In response to diverse hemodynamic and neurohormonal insults, the heart undergoes pathologic remodeling, a process characterized by increased cardiomyocyte (CM) volume (hypertrophy), interstitial fibrosis, inflammatory pathway activation, and cellular dysfunction culminating in contractile failure (Sano et al., 2002; van Berlo et al., 2013). The pathologic nature of this process has been validated in large epidemiologic studies, which demonstrate the presence of chronic cardiac hypertrophy to be a robust predictor of subsequent HF and death (Hill and Olson, 2008; Levy et al., 1990). While hypertrophic remodeling may provide short-term adaptation to pathologic stress, sustained activation of this process is maladaptive and drives disease progression (Hill and Olson, 2008). Studies over the past decade have clearly demonstrated that inhibition of specific pro-hypertrophic signaling effectors exert cardioprotective effects even in the face of persistent stress. Together, these data provide a cogent rationale that targeting the hypertrophic process itself can be beneficial without compromising contractile performance (Hill and Olson, 2008; van Berlo et al., 2013).
Hemodynamic and neurohormonal stressors activate a network of cardiac signal transduction cascades that ultimately converge on a defined set of transcription factors (TFs), which control the cellular state of the CM (Hill and Olson, 2008; Lee and Young, 2013; van Berlo et al., 2013). Studies in animal models have implicated several master TFs that drive HF progression (e.g. NFAT, GATA4, NFκB, MEF2, c-Myc) via induction of pathologic gene expression programs that weaken cardiac performance (Maier et al., 2012; van Berlo et al., 2011; Zhong et al., 2006). In addition to stimulus-coupled activation of DNA-binding proteins, changes in cell state occur through an interplay between these master regulatory TFs and changes in chromatin structure (Lee and Young, 2013). Notably, stress pathways activated in HF are associated with dynamic remodeling of chromatin (McKinsey and Olson, 2005; Sayed et al., 2013), including global changes in histone acetylation and DNA methylation. As alterations in higher-order chromatin structure modulate the net output of multiple, simultaneously activated transcriptional networks (Lee and Young, 2013; Schreiber and Bernstein, 2002), manipulation of cardiac gene expression via targeting chromatin-dependent signal transduction represents a potentially powerful therapeutic approach to abrogate pathologic gene expression and HF progression.
Transcriptional activation is associated with local N-ε-acetylation of lysine sidechains on the unstructured amino-terminal tail of histone proteins (Schreiber and Bernstein, 2002). Dynamic positioning of acetyl-lysine (Kac) arises from the interplay of so-called epigenetic “writers” (histone acetyltransferases or HATs) and epigenetic “erasers” (histone deacetylases or HDACs). Context-specific recognition of Kac at regions of actively transcribed euchromatin is mediated by epigenetic “reader” proteins possessing a Kac-recognition module or bromodomain (Filippakopoulos et al., 2012). Molecular recognition of Kac by bromodomain-containing proteins serves to increase the effective molarity of transcriptional complexes promoting chromatin remodeling, transcriptional initiation and elongation (Dawson et al., 2012). Elegant studies over the past decade have implicated both epigenetic writers (e.g. EP300) (Wei et al., 2008) and erasers (e.g. HDACs) (Montgomery et al., 2007; Trivedi et al., 2007; Zhang et al., 2002) in cardiac development and disease. In contrast, little is known about epigenetic readers in cardiac biology.
Members of the bromodomain and extraterminal (BET) family of bromodomain-containing reader proteins (BRD2, BRD3, BRD4, and testis-specific BRDT) associate with acetylated chromatin and facilitate transcriptional activation by recruitment of co-regulatory complexes such as mediator (Jiang et al., 1998) and the positive transcription elongation factor b (P-TEFb) (Jang et al., 2005; Yang et al., 2005). Recently, we developed a first-in-class potent, selective bromodomain inhibitor, JQ1, which displaces BET bromodomains from chromatin resulting in suppression of downstream signaling events to RNA polymerase II (Pol II) (Delmore et al., 2011; Filippakopoulos et al., 2010). We and others have utilized this chemical genetic tool to probe BET function in a number of developmental and disease contexts, such as cancer (Delmore et al., 2011; Filippakopoulos et al., 2010), HIV infection (Banerjee et al., 2012), and spermatogenesis (Matzuk et al., 2012). The role of BET bromodomains proteins in the heart remains unknown.
In this study, we report that BETs are critical effectors of pathologic cardiac remodeling via their ability to co-activate defined stress-induced transcriptional programs in the heart. An important mechanism by which BETs drive pathologic gene induction is via their ability to promote transcriptional pause release and elongation, thereby co-activating multiple master TFs known to initiate and promote HF. The elucidation of BET function in the heart implicates epigenetic reader proteins in cardiac biology and HF pathogenesis. Moreover, use of chemical biology to specifically probe the role of BET bromodomain-containing proteins in the myocardium suggests that targeted manipulation of chromatin-based signal transduction might be harnessed for therapeutic gain in heart disease.
RESULTS
BET bromodomains are cell-autonomous regulators of pathologic cardiomyocyte hypertrophy in vitro
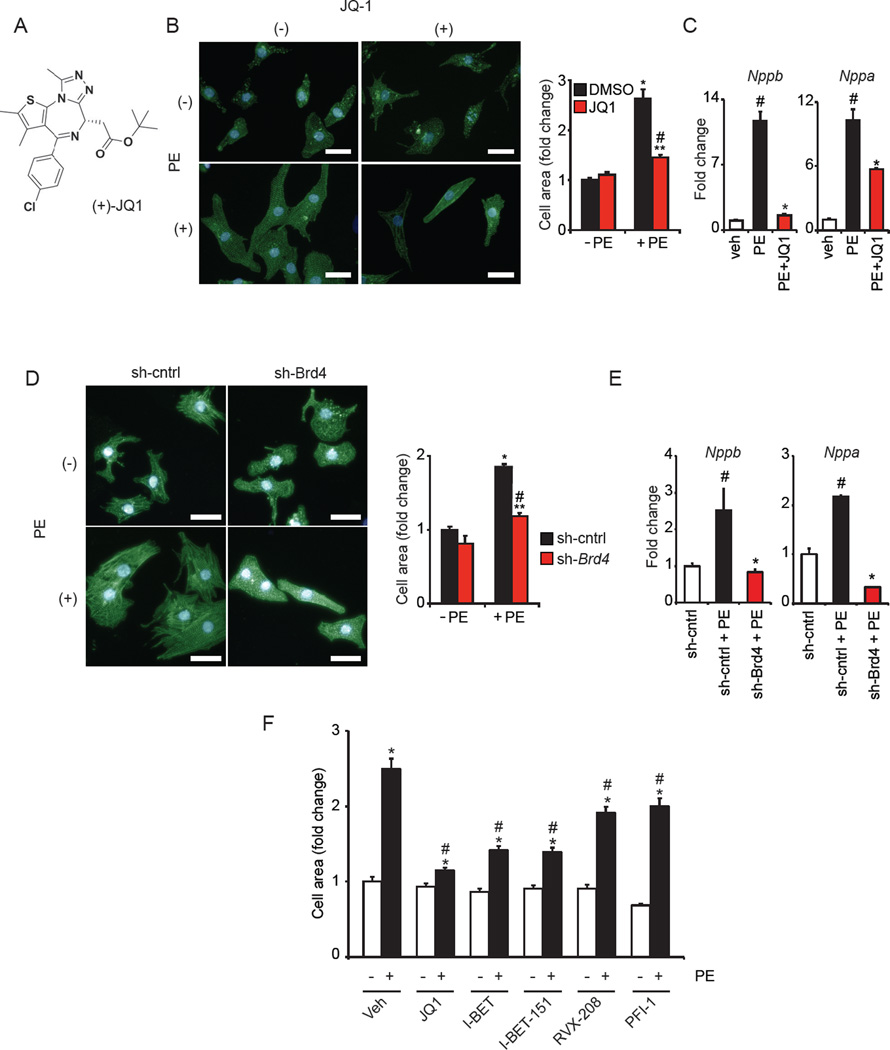
As pathologic cardiac hypertrophy features coordinate transcriptional activation of numerous master regulatory TFs, we hypothesized that BET bromodomains would function as co-activator proteins in this disease process. We first assessed the expression patterns of BETs in the heart. Analysis of neonatal rat ventricular cardiomyocytes (NRVM) and adult mouse ventricular tissue revealed that Brd2, Brd3, and Brd4 are transcribed, with Brd4 emerging as most highly expressed (Figure S1A-B). Western blots in NRVM, mouse heart tissue and human heart tissue confirmed abundant BRD4 expression (Figure S1C) and immunofluorescence staining of NRVM demonstrated BRD4 to be nuclear localized (Figure S1D). First using a chemical biology approach, we leveraged the validated small-molecule probe of BET bromodomain function, JQ1 (Figure 1A), in the established NRVM model in vitro (Simpson et al., 1982). Nanomolar doses of JQ1 significantly blocked phenylephrine (PE) mediated cellular hypertrophy (Figure 1B) and pathologic gene induction (Figure 1C). In a similar manner, knockdown of Brd4 in NRVM (Figure S1E) also attenuated PE mediated hypertrophic growth (Figure 1D) and pathologic gene induction (Figure 1E). We next assessed a number of structurally dissimilar BET inhibitors (I-BET, I-BET-151, RVX-208, PFI-1; chemical structures shown in Figure S1F) for their ability to inhibit CM hypertrophy. At equimolar doses, we found that inhibition of agonist-induced CM hypertrophy was indeed a class effect of BET inhibitors, with the relative potency of these compounds correlating with their known IC50 against BRD4 (Filippakopoulos et al., 2010). Together, these data implicate BET bromodomain proteins as putative cell autonomous regulators of pathologic CM hypertrophy, and identify potent anti-hypertrophic effects of the small molecule BET inhibitor JQ1 in vitro.
Figure 1. BET bromodomain inhibition blocks CM hypertrophy in vitro.
(A) (+)-JQ1 structure. (B) Representative image of NRVM treated ± JQ1 (250 nM) and PE (100 µM) for 48h with cell area quantification. *P<0.05 vs. DMSO −PE. **P<0.05 vs. JQ1 −PE. #P<0.05 vs. DMSO +PE. (C) qRT-PCR of NRVM treated with JQ1 (500nM) and PE (100µM, 48h, N=4). #P<0.05 vs. veh, *P<0.05 vs. PE. (D) Representative image of NRVM infected with Ad-sh-Brd4 or sh-cntrl treated ± PE (100µM, 48h) with cell area quantification. *P<0.05 vs. sh-cntrl −PE. **P<0.05 vs. sh-Brd4 −PE. #P<0.05 vs. sh-cntrl +PE. (E) qRT-PCR of NRVM during BRD4 knockdown ± PE (100 µM, 48h, N=4). #P<0.05 vs. sh-cntrl, *P<0.05 vs. sh-cntrl+PE. (F) Cell area of NRVM treated with indicated BET inhibitors (500 nM) ± PE (100 µM, 48h). *P<0.05 vs. –PE control for indicated compound. #P<0.05 vs. veh +PE. Bar=30µM. Data shown as mean ± SEM. See also Figure S1.
BETs are required for induction of a pathologic gene expression program in cardiomyocytes
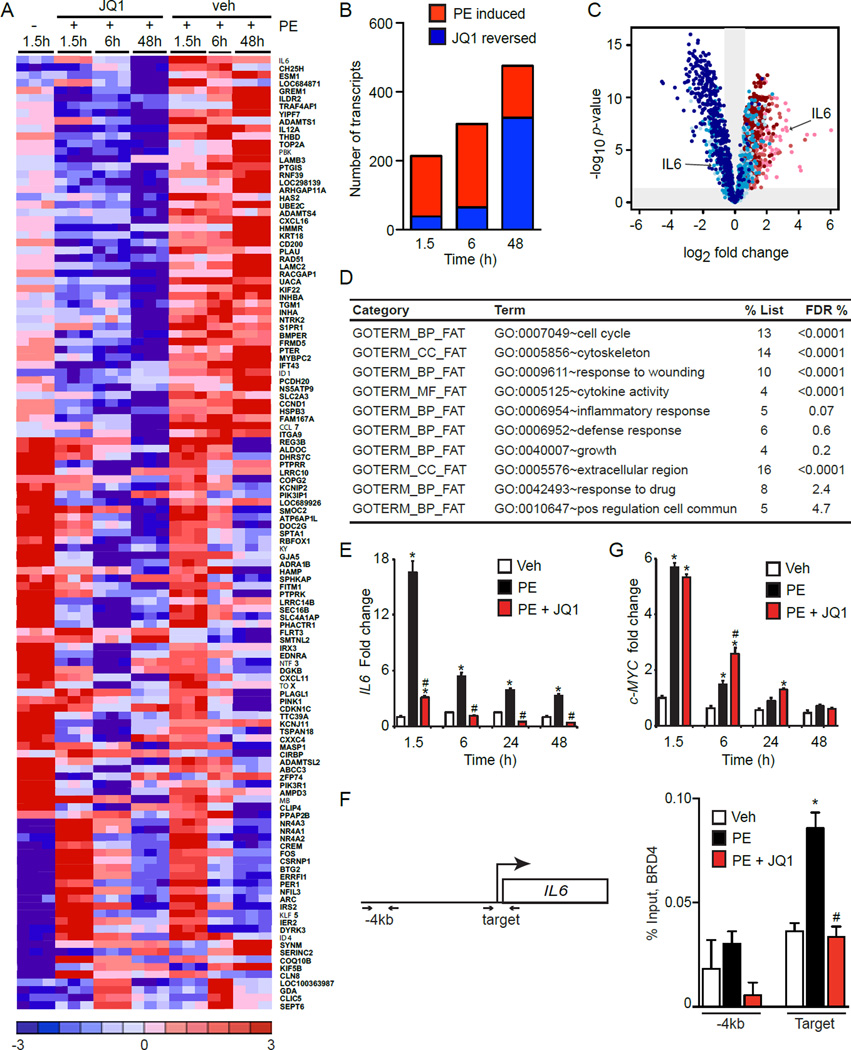
To determine the transcriptional effects of BET bromodomain inhibition during CM hypertrophy, we performed gene expression profiling (GEP) studies in cultured NRVM at baseline and after PE stimulation (1.5, 6, 48 hours) in the presence or absence of JQ1. These three time-points capture induction of early response genes such as c-Myc (Zhong et al., 2006), as well as the final hypertrophic expression program. Assessment of differentially expressed transcripts revealed three major clusters: genes that were PE inducible and suppressed by JQ1, genes that were PE inducible and unaffected by JQ1, and genes that were PE suppressed and unaffected by JQ1. A heat map of genes selected based on the highest magnitude of PE-mediated change illustrates each of these clusters (Figure 2A; full list of differential transcripts provided in Table S1). Global analysis of GEPs revealed that PE stimulation resulted in the cumulative induction of over 450 genes and that JQ1 abrogated induction of a substantial subset of PE-inducible genes. Statistically significant transcriptional effects were evident as early as 1.5 hours and increased over time (Figure 2B-C), consistent with the emerging role of BETs as co-activators of inducible gene expression programs (Nicodeme et al., 2010). Functional pathway analysis of PE-inducible transcripts that were suppressed by JQ1 revealed that BETs facilitate expression of a host of biological processes known to be involved in pathologic CM activation, including cytoskeletal reorganization, extracellular matrix production, cell-cycle reentry, paracrine/autocrine stimulation of cellular growth, and pro-inflammatory signaling (Figure 2D) (Song et al., 2012). Using the pro-hypertrophic cytokine IL6 as a representative target (Figure 2C), we confirmed by qRT-PCR that JQ1 significantly attenuated PE-mediated induction (Figure 2E). Activity of BETs during pathologic stress was not due to PE-mediated increases in BET expression (Figure S2A). Chromatin immunoprecipitation (ChIP) studies demonstrated BRD4 occupancy at the IL6 locus and an increase in BRD4 enrichment after 90 min of PE stimulation that was blocked by BET inhibition (Figure 2F). Interestingly, BET bromodomain inhibition did not affect PE-mediated induction of Myc (Figure 2G), an established transcriptional driver of pathologic cardiac hypertrophy (Zhong et al., 2006). Prior studies from our laboratory and others have implicated BET bromodomains in neoplastic MYC transcription in hematopoietic tumors (Delmore et al., 2011; Zuber et al., 2011). Collectively, these in vitro data (Figures 1–2) demonstrate that BET bromodomain containing proteins regulate CM hypertrophy in a cell-autonomous manner via co-activation of a broad, but specific transcriptional program.
Figure 2. BET regulated transcriptional programs during CM hypertrophy in vitro.
(A) Selected heat map of differentially expressed (DE) transcripts. NRVM treated with 500nM JQ1, 100µM PE. (B) Global analysis of DE transcripts showing induction of genes by PE with time (red) and progressive reversal of PE-mediated gene induction by JQ1 (blue). (C) Volcano plot showing fold change (x-axis) effect of PE+JQ1 versus PE+vehicle (shades of blue) on all genes upregulated at any time-point by PE vs. veh (shades of red). Progression from lighter to darker shading represents increasing time (1.5h, 6h, 48h). (D) Functional pathway analysis (DAVID) of PE-induced genes that were JQ1-reversed. FDR <5% considered statistically significant. (E) IL6 qRT-PCR in NRVM treated with JQ1 (500nM) and PE (100µM, N=4). *P<0.05 vs. veh, #P<0.05 vs. PE. (F) BRD4 ChIP-qPCR in NRVM treated with JQ1 (500nM) and PE (100µM) for 90min. Target and nontarget (-4kb region) primers depicted. N=3, *P<0.05 vs. veh, #P<0.05 vs. PE. (G) c-MYC qRT-PCR in NRVM treated with JQ1 (500 nM) and PE (100µM, N=4). *P<0.05 vs. veh, #P<0.05 vs. PE. Data shown as mean ± SEM. See also Figure S2 and Table S1.
BET Bromodomain inhibition arrests pathologic hypertrophy and heart failure in vivo
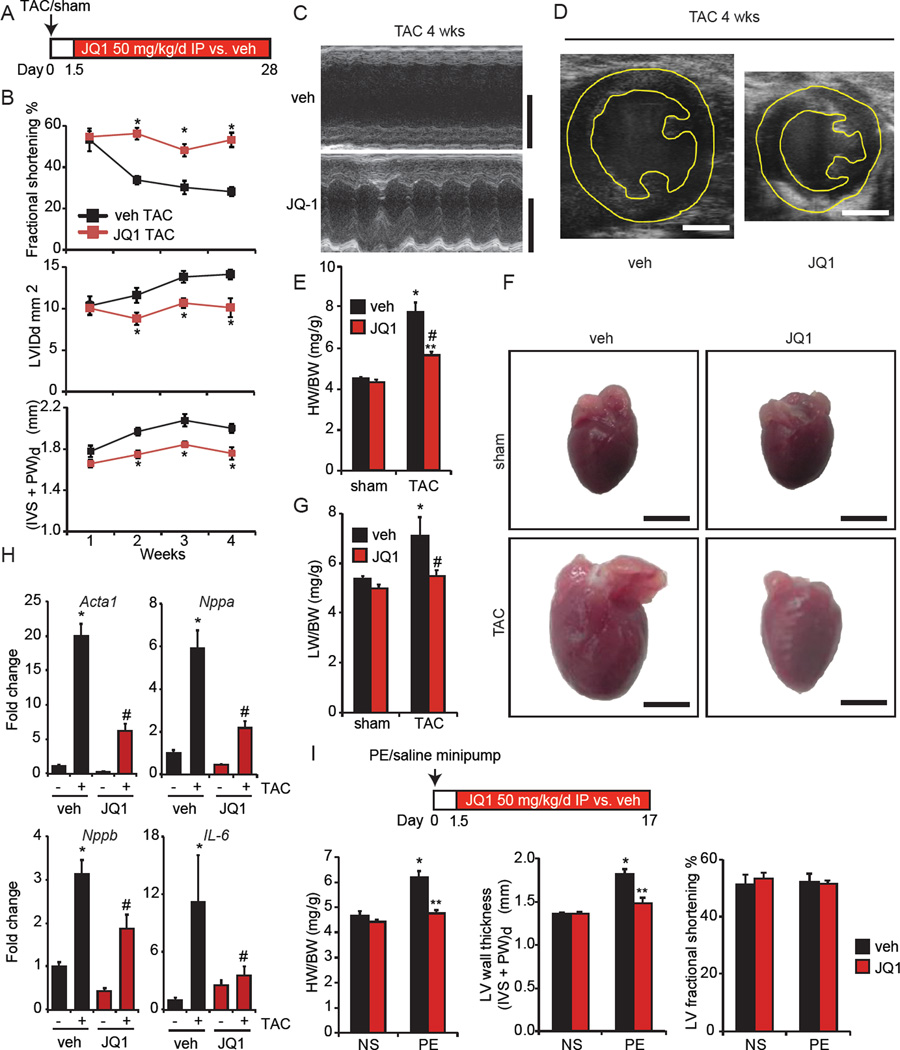
Given these observations in cultured CMs, we hypothesized that BETs might regulate pathologic cardiac remodeling in the intact organism. We leveraged the favorable therapeutic index of JQ1, which has previously been shown to potently inhibit BET bromodomain function in adult mice without significant toxicity when administered chronically at 50 mg/kg/day (Filippakopoulos et al., 2010). Independently, we confirmed the lack of overt toxicity by treating mice with JQ1 for 2–3 weeks, observing no effect on endurance exercise capacity (Figure S3A), a metric of global cardiometabolic health. For in vivo studies, we first used transverse aortic constriction (TAC), a thoroughly characterized model of cardiac pressure-overload, which provides focal hemodynamic stress to the heart and recapitulates several cardinal aspects of pathologic hypertrophy and HF in humans (Rockman et al., 1991). In our hands, adult mice subject to TAC develop concentric left ventricular hypertrophy (LVH) by 7–10 days and progress to advanced HF after 3–4 weeks.
We performed TAC or sham surgery followed by administration of JQ1 (50 mg/kg/day vs. vehicle control) beginning 1.5 days after initiation of TAC (Figure 3A). Serial echocardiography showed that JQ1 protected against TAC-mediated LV systolic dysfunction, cavity dilation, and wall thickening with effects that were sustained out to 4 weeks (Figure 3B-D, Figure S3B, Video S1–S2). JQ1 treatment also inhibited pathologic cardiomegaly (Figure 3E; representative photos in Figure 3F), pulmonary congestion (Figure 3G), and myocardial expression of canonical hypertrophic marker genes (Figure 3H) after TAC. JQ1 was well tolerated, as evidenced by normal activity (Video S3–S4), and lack of significant mortality or weight loss compared to vehicle treated mice (data not shown). In addition, JQ1 had no adverse effect on LV structure or function in sham treated mice (Figure 3E-G and Figure S3B). Importantly, JQ1 does not affect systemic blood pressure (Figure S3C). Furthermore, the protective effects of JQ1 in the TAC model were not associated with differences in the pressure gradient across the aortic constriction (Figure S3D).
Figure 3. BET Bromodomain inhibition with JQ1 potently attenuates pathologic cardiac hypertrophy and HF in vivo.
(A) Experimental protocol. (B) Echocardiographic parameters during TAC (N=7). LVIDd is LV end diastolic area, (IVS + PW)d is sum thickness of the interventricular septum and posterior LV wall at end diastole. *P<0.05 vs. veh TAC. (C) Representative M-mode tracings and (D) end-diastolic 2D images at 4wks TAC. Bar=2 mm. (E) Heart/body weight (HW/BW) ratios, 4wks. *P<0.05 vs. sham veh. #P<0.05 vs. TAC veh. **P<0.05 vs. sham JQ1. (F) Representative photos of freshly excised whole hearts. Bar=3mm. (G) Lung/body weight (LW/BW) ratios, 4wks TAC (N=7 TAC, N=5 sham). *P<0.05 vs. sham veh. #P<0.05 vs. TAC veh. (H) qRT-PCR in mouse hearts (N=5–7). *P<0.05 vs. sham veh. #P<0.05 vs. TAC veh. (I) PE infusion (75 mg/kg/day) and JQ1 administration (N=7 PE, N=5 normal saline). *P<0.05 vs. NS veh. **P<0.05 vs. PE veh. Data shown as mean ± SEM. See also Figure S3 and Supplemental Videos 1–4.
In addition to hemodynamic stress, excessive neurohormonal activation is also a central driver of pathologic cardiac hypertrophy (Hill and Olson, 2008; van Berlo et al., 2013). Therefore, we next assessed whether JQ1 could ameliorate pathology in a second mouse model of neurohormonally-mediated cardiac hypertrophy. Mice were implanted with osmotic minipumps delivering phenylephrine (PE, 75mg/kg/day vs. saline) followed by JQ1 or vehicle administration beginning 1.5 days after minipump installation. This infusion protocol produces robust concentric LVH in 2–3 weeks, but does not cause significant LV cavity dilation or depression of LV systolic function in wild type mice. Concordant with our TAC results above, JQ1 potently suppressed the development of pathologic cardiac hypertrophy during chronic PE infusion, without any compromise in LV systolic function (Figure 3I).
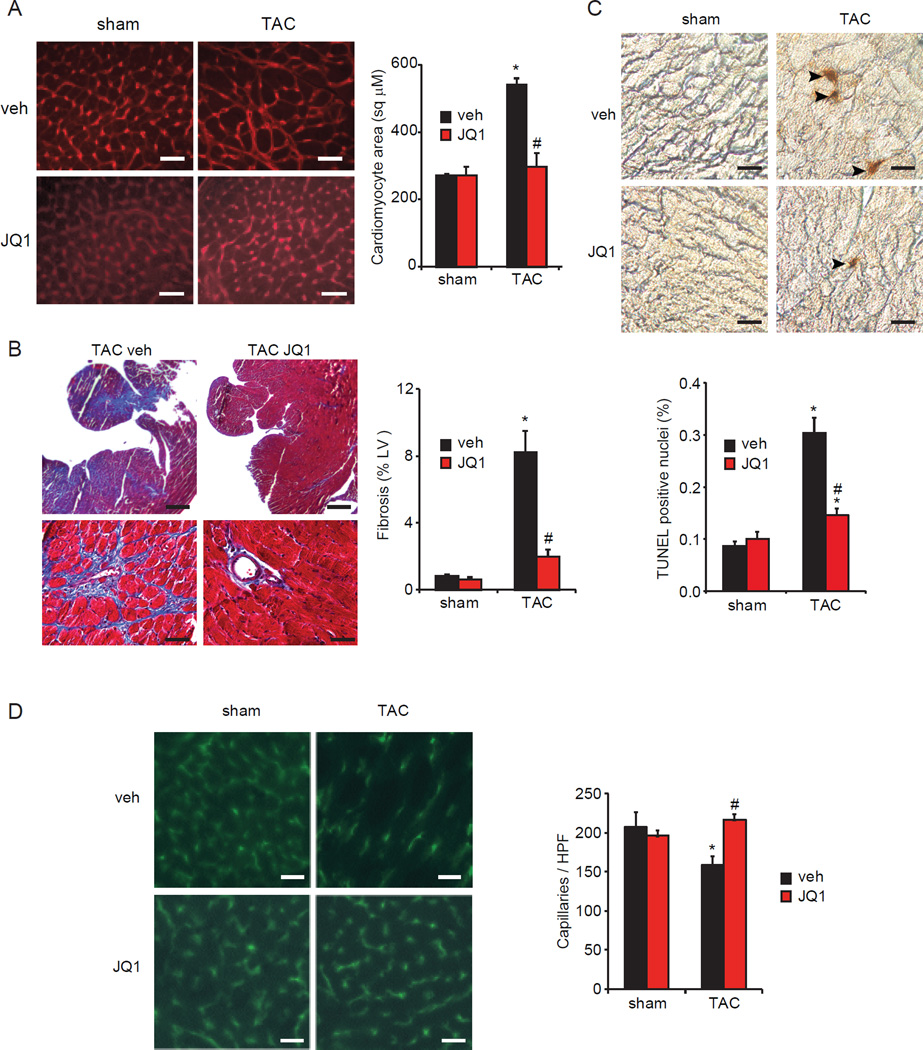
In addition to favorable effects on cardiac function, we assessed whether JQ1 also ameliorated cardinal histopathologic features of HF in vivo. Analysis of mouse heart tissue demonstrated that JQ1 significantly attenuated the development of CM hypertrophy (Figure 4A), myocardial fibrosis (Figure 4B), apoptotic cell death (Figure 4C), and capillary rarefaction (Figure 4D) typically seen after 4 weeks of TAC (Sano et al., 2007; Song et al., 2010).
Figure 4. BET Bromodomain inhibition attenuates cardinal histopathologic features of HF.
(A) CM area quantification in LV sections. Bar=30µm. (B) Trichrome staining and quantification of fibrotic area. Bar=400µm (top), 40µm (bottom). (C) TUNEL staining of heart sections with quantification of TUNEL-positive nuclei. Bar=20 µm. (D) PECAM-1 immunofluorescence staining of LV sections with quantification of myocardial capillary density. HPF, 400X high powered field. Bar=30 µm. For panels A-D: N=3–4, 4wk time-point, *P<0.05 vs. sham veh, #P<0.05 vs. TAC veh. Data shown as mean ± SEM.
BET inhibition suppresses a pathologic cardiac gene expression program in vivo
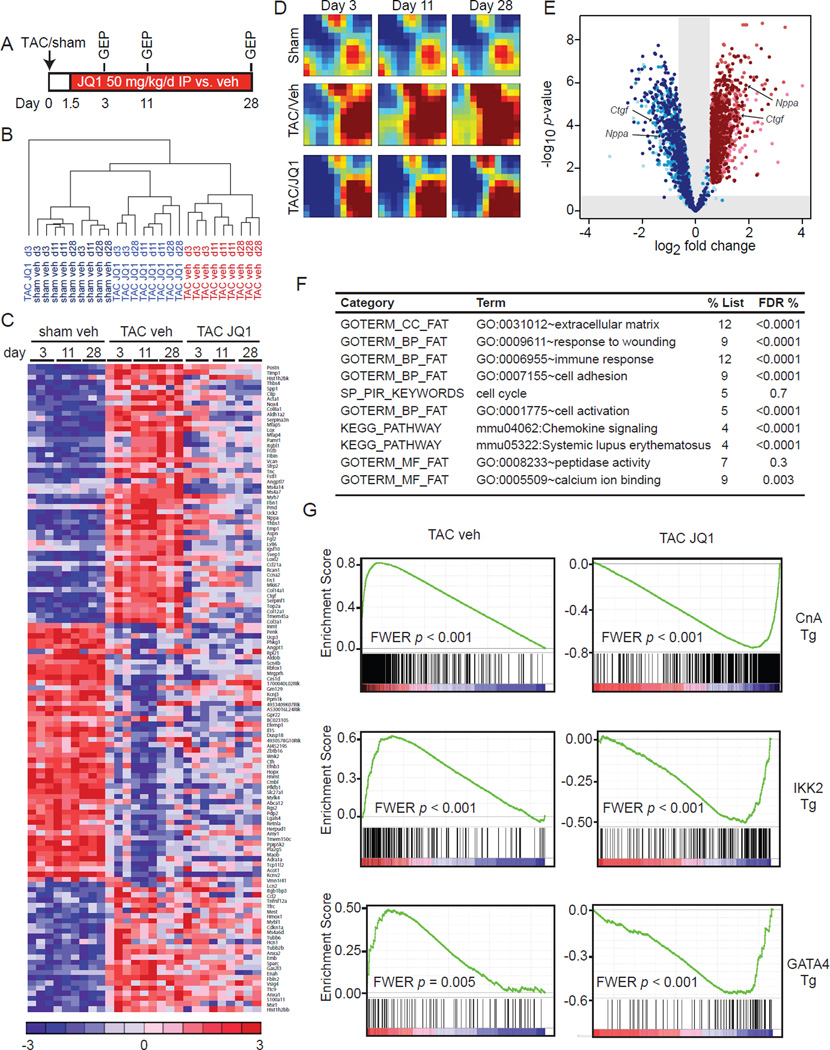
Using the TAC model, we performed detailed transcriptional analysis in 3 groups (sham-vehicle, TAC-vehicle, and TAC-JQ1) at 3 time-points (Figure 5A): 3 days (to reflect early events that occur prior to the onset of hypertrophy), 11 days (established hypertrophy), and 28 days (advanced pathologic remodeling with signs of HF). Unsupervised hierarchical clustering revealed that the TAC-veh group had a distinct GEP signature that evolved with time when compared to the sham-veh group (Figure 5B). In contrast, TAC-JQ1 clustered with the sham group and displayed no significant temporal change despite continuous exposure to TAC (Figure 5B). Hence, JQ1 treatment suppressed the evolution of a broad pathologic gene expression program in the heart, with effects evident as early as 3 days post-TAC. Similar to our studies in isolated CMs (Figure 2), global GEP analysis revealed three major clusters: genes that were TAC-inducible and suppressed by JQ1, those that were TAC inducible and unaffected by JQ1, and those that were TAC suppressed and unaffected by JQ1. A representative heat map of genes (selected for the highest magnitude of TAC-mediated change) highlights each of these three clusters (Figure 5C; full list of differential transcripts provided in Table S2). TAC did not significantly alter myocardial expression of Brd2, Brd3, or Brd4 (Figure S4A).
Figure 5. BETs co-activate a broad, but specific transcriptional program in the heart during TAC.
(A) Protocol for GEP experiment. (B) Unsupervised hierarchical clustering of GEPs. (C) Heatmap of selected genes. Full list of DE genes in Table S2. (D) GEDI plots showing temporal evolution of gene clusters. (E) Volcano plot showing fold change effect of TAC+JQ1 vs. TAC-veh (shades of blue) on all genes upregulated at any time-point by TAC-veh vs. sham-veh (shades of red). Progression from lighter to darker shading represents increasing time (3d, 11d, 28d) (F) DAVID analysis of genes that were TAC-induced and JQ1-reversed. FDR <5% considered statistically significant. (G) GSEA for TAC-veh and TAC-JQ1 against three independent GEPs derived from CM-specific activation of canonical pro-hypertrophic transcriptional effectors in vivo: Calcineurin-NFAT (driven by a constitutively active Calcineurin A transgene (Bousette et al., 2010), NFκB driven by an IKK2 trasngene (Maier et al., 2012) and transgenic GATA4 overexpression (Heineke et al., 2007)). FWER p<0.250 represents statistically significant enrichment. Data representative for all three time-points. Plots shown for 28d time-point. Data shown as mean ± SEM. See also Figure S4 and Table S2.
To visualize the global transcriptional effects of TAC and BET bromodomain inhibition in the model over time, we performed Gene Expression Dynamics Inspector (GEDI) analysis (Eichler et al., 2003). While the sham mosaic remained temporally invariant, TAC resulted in progressive induction of gene clusters over time, indicated by increased signal in numerous tiles within the mosaic (Figure 5D). BET bromodomain inhibition disrupted the temporal evolution of this TAC-induced, pathologic transcriptional program with a mosaic signature that more closely resembled the sham group (Figure 5D). A volcano plot showing fold change effect of TAC-JQ1 versus TAC-vehicle (shades of blue) on the set of all genes upregulated at any time-point by TAC (shades of red) is shown in Figure 5E. These data illustrate the potent and statistically significant effects of JQ1 in suppressing TAC-mediated gene induction. Additionally, comparison of fold change effect of TAC-JQ1 vs. sham-vehicle (shades of blue) on the set of all genes upregulated at any time-point by TAC (shades of red) shows that JQ1 administration reversed TAC-mediated gene induction to levels similar to those in the sham treated hearts (Figure S4B). Functional pathway analysis of TAC-inducible transcripts that were suppressed by JQ1 showed enrichment for key biological processes involved in pathologic myocardial remodeling and HF progression in vivo (Figure 5F) (Song et al., 2012; Zhao et al., 2004). Importantly, these functional terms aligned with the data from isolated NRVM (Figure 2D) and represent pathologic processes universally observed in advanced human HF (Hannenhalli et al., 2006).
Given the broad effects on myocardial gene expression seen with JQ1, we hypothesized that BETs enable pathologic gene induction via their ability to coordinately co-activate multiple TF pathways in vivo. Using gene set enrichment analysis (GSEA) (Subramanian et al., 2005), we compared our set of TAC-inducible genes that were suppressed by BET inhibition, against compendia of TF signatures. Specifically, we studied the Broad Institute Molecular Signatures Database C3 motif gene sets (Xie et al., 2005), as well as three functional, in vivo signatures of CM-specific transcriptional effectors : Calcineurin-NFAT (Bousette et al., 2010), NFκB (Maier et al., 2012) and GATA4 (Heineke et al., 2007). These analyses revealed that the TAC induced gene expression profile was positively enriched for IRF and ETS consensus binding motifs (FWER p < 0.0001), as well as for myocardial signatures that result from Calcineurin, NFκB, and GATA4 activation (Figure 5G). Conversely, the effect of JQ1 demonstrated strong negative enrichment for these same TF signatures (Figure 5G; FWER p < 0.0001 for IRF and ETS motifs). In contrast, while TAC was strongly correlated with both c-Myc and E2F signatures, there was no significant correlation between c-Myc/E2F and JQ1 effect at any time-point (Figure S4C). Consistent with our NRVM studies, we also found that JQ1 had no effect on TAC-mediated induction of Myc expression in vivo (Figure S4D). Hence, the GSEA data support a model in which BET bromodomains facilitate gene induction via co-activation of specific myocardial TF networks.
We next compared the set of TAC-inducible genes that were suppressed by JQ1 against validated gene expression profiles of advanced non-ischemic and ischemic HF in humans (Hannenhalli et al., 2006). This analysis demonstrated that targets of BETs in the mouse TAC model overlapped in a statistically significant manner with the set of genes induced in human HF (Figure S4E; χ2 < 5 × 10−14). Interestingly, the vast majority (89%) of these targets were common to both ischemic and non-ischemic human HF (Figure S4F). Thus, inasmuch as the gene expression profiles of mice subjected to TAC overlap with that of advanced HF in a human cohort, we found the principal transcriptional targets of BET bromodomains in mice were also relevant in human disease.
BET bromodomain inhibition abrogates transcriptional pause release genome-wide during pathologic hypertrophy in vivo
To establish the mechanism by which BET bromodomain inhibition impairs global transactivation of pathologic transcriptional programs concomitantly activated during pressure overload, we performed ChIP coupled with high throughput genome sequencing (ChIP-seq) on heart tissue from mice subjected to sham versus TAC surgery, with and without JQ1 treatment. For heart tissue ChIP-seq, a 4 day time-point was chosen to capture changes in chromatin state that occur during the initial phase of hypertrophic growth in the TAC model (~3–7 days). Experimental data obtained for BRD4 and Pol II were integrated with publicly available data from Bing Ren and colleagues, which provided epigenomic landscapes for murine cardiac euchromatin (Shen et al., 2012). Genome-wide data for Pol II enrichment in our sham operated mice demonstrated excellent statistical agreement with Pol II enrichment curated from the literature (R2 = 0.65; Figure S5A-B).
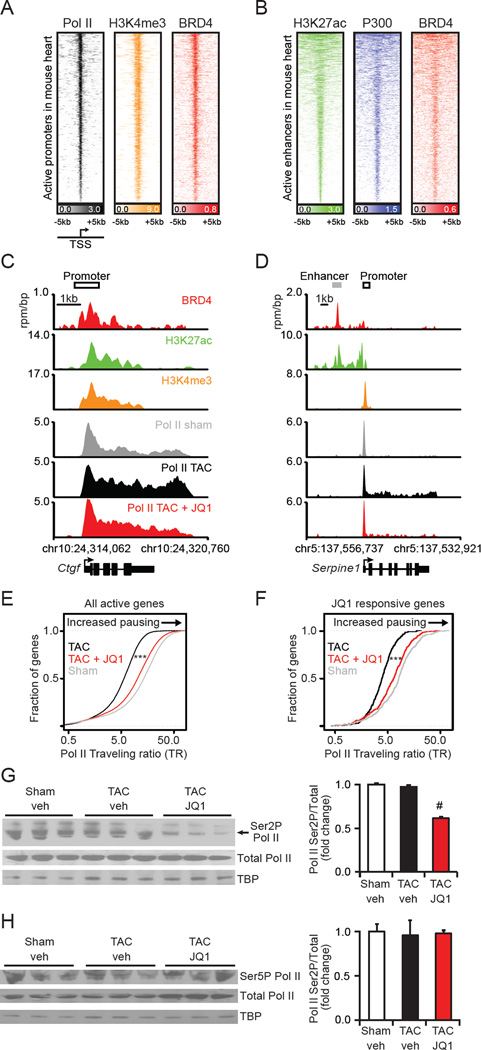
First, we determined the sites of genome-wide localization of BRD4 in the murine heart. Strong enrichment was observed at promoter regions of actively transcribed genes, as identified by enrichment for histone 3 lysine 4 trimethylation (H3K4me3) and Pol II (Figure 6A). Rank-ordering of all transcriptionally active promoters by Pol II occupancy identified global binding of BRD4 at sites of transcriptional initiation. Recently, BET bromodomains have also been shown to bind to enhancer elements in the eukaryotic genome (Loven et al., 2013; Zhang et al., 2012). Binding of BRD4 to enhancer elements was assessed by rank-ordering regions of enrichment for histone 3 lysine 27 acetylation (H3K27ac), and comparing BRD4 enrichment to that of P300, a known enhancer factor of functional significance in cardiac hypertrophy (Wei et al., 2008). Importantly, BRD4 binds to the vast majority of active enhancers in the murine cardiac genome (Figure 6B).
Figure 6. BET bromodomain inhibition abrogates transcriptional pause release genomewide in pathologic hypertrophy.
(A) Heatmap of Pol II (black), H3K4me3 (orange), and BRD4 (red) levels at active promoters ranked by Pol II levels in sham treated heart samples. Each row shows ±5kb centered on H3K4me3 peak. Rows ordered by average Pol II at the promoter. (B) Heatmap of H3K27ac (green), P300 (blue), and BRD4 (red) levels at active enhancers ranked by H3K27ac levels in adult heart. Each row shows ±5kb centered on H3K27ac peak. Rows ordered by amount of H3K27ac at enhancer. Color scaled intensities of A-B are in units of reads per million per base pair (rpm/bp). (C-D) Gene tracks at (C) Ctgf and (D) Serpine1 gene in heart. BRD4 (red) is from sham treated hearts. H3K27ac (green) and H3K4me3 (orange) derived from published landscapes of wild type mouse hearts (Shen et al., 2012) that are age/sex/strain-matched to our sham treated hearts. Pol II are from either sham (grey), TAC (black), or TAC+JQ1 treated (red) heart. x-axis shows genomic position. y-axis shows ChIP-seq occupancy (rpm/bp). (E-F) Empirical cumulative distribution plots of Pol II traveling ratios (TR) (Rahl et al., 2010) for genes that are transcriptionally active in either sham or TAC treated hearts (E) and genes that are TAC-induced and reversed by JQ1 (F). Differences in TR distribution between TAC and TAC+JQ1 treated hearts are statistically significant (*** Welch’s two-tailed t test, p < 2×10−16). (G-H) Western blots with densitometry of heart tissue nuclear extracts from sham, TAC, and TAC+JQ1 treated hearts for total Pol II or indicated phosphoforms (N=3; #P<0.05 vs. TAC veh). Data shown as mean ± SEM. See also Figure S5 and Table S3.
BRD4, through its C-terminal domain (CTD), physically associates with and activates CDK9 (Bisgrove et al., 2007; Jang et al., 2005; Yang et al., 2005), a core component of the P-TEFb complex which functions as an elongation-promoting Pol II CTD kinase (Peterlin and Price, 2006). Hyper-activation of P-TEFb and consequent increases in global transcription are considered a hallmark of pathologic cardiac hypertrophy in multiple systems (Espinoza-Derout et al., 2009; Sano et al., 2002; Yoshikawa et al., 2012). Inhibition of CDK9 function in NRVM has been shown to attenuate endothelin-1 mediated hypertrophy in vitro (Sano et al., 2002). These observations provided us with a rationale to explore the effect of BET bromodomain inhibition on transcriptional elongation in the TAC model in vivo.
BET bromodomain inhibition by JQ1 has been shown to displace BRD4 and P-TEFb from chromatin leading to a decrease in Pol II elongation at active genes (Loven et al., 2013). Gene-specific effects on Pol II occupancy were explored at canonical mediators and effectors of pathologic cardiac remodeling. As shown for representative genes in Figure 6C-D, sham-treated hearts feature a pronounced Pol II enrichment peak at the TSS of Ctgf and Serpine1 (SerpinE1/PAI-1), with only modest evidence of downstream elongation (gray wiggle plots). Following TAC, transcriptional activation (as shown in Figure 5C and Table S2) was accompanied by a shift in the distribution of gene bound Pol II towards a higher relative occupancy in the elongating gene body region (black wiggle plots; Figure 6C-D). BET bromodomain inhibition markedly attenuated Pol II occupancy in the elongating region, leading to higher relative Pol II occupancy at the TSS initiation site (red wiggle plots), consistent with its ability to attenuate transcription elongation. Additional representative Pol II enrichment plots for the Ace, Bgn, Thbs1, and Xirp2 loci are shown in Figure S5C.
Global effects of BET bromodomain inhibition on initiation and elongation were further examined by calculating the Pol II traveling ratio at all actively transcribed genes, and on the subset of genes induced by TAC. Traveling ratio is a validated measure of transcriptional pause release that compares promoter and elongating gene body occupancy levels of Pol II to quantify the ratio of paused to elongating Pol II (Lin et al., 2012; Wade and Struhl, 2008). BET bromodomain inhibition of TAC treated hearts led to a higher ratio of promoter to elongating gene body Pol II compared to TAC at all active genes (Figure 6E) and at TAC-induced genes that were reversed by JQ1 (Figure 6F). A metagene analysis of Pol II enrichment among actively transcribed genes verified the change in the ratio of promoter to elongating gene body Pol II by JQ1 (Figure S5D). The effects of JQ1 on transcriptional elongation in the TAC model were reproduced in an independent in vivo biological replicate, as shown in Figure S5E.
Given the high variability in dynamic range between heart tissue samples ChIP-seq (Figure S5F) and the relative nature of metrics such as the traveling ratio, we examined global levels of initiating and elongating Pol II to further quantify the effects of BET bromodomain inhibition. We observed a specific decrease in the elongation-specific serine 2 Pol II phosphoform (Ser2P) upon JQ1 treatment (Figure 6G) with little change to the initiation-specific serine 5 Pol II phosphoform (Ser5P) (Figure 6H). JQ1 mediated suppression of Pol II Ser2P abundance was recapitulated in the in vitro NRVM model (Figure S5G). Interestingly, we also observed significant upregulation of the P-TEFb inhibitory protein HEXIM1 (Espinoza-Derout et al., 2009; Peterlin and Price, 2006; Yoshikawa et al., 2012) during JQ1 treatment, suggesting an additional mechanism by which BET bromodomain inhibition may inhibit transcriptional elongation (Figure S5H-K). We conclude that transcriptional elongation is an important mechanism by which BET bromodomain inhibition attenuates gene expression programs activated during pathologic cardiac hypertrophy.
DISCUSSION
Our current work implicates BET bromodomain reader proteins as essential transcriptional co-activators of a pathologic gene expression program that drives CM hypertrophy and HF progression. Gene-expression profiling and ChIP-seq studies reveal that BET proteins function, in part, by promoting transcriptional pause release during pathologic stress. In broadest terms, the data presented here directly implicate epigenetic readers in cardiac biology and identify BET bromodomain proteins as potential therapeutic targets in heart disease.
Pathologic cardiac hypertrophy ensues by a collaborative interplay between master regulatory TFs and dynamic changes in chromatin structure (Lee and Young, 2013). TFs including NFAT, GATA4, and NFκB (Maier et al., 2012; van Berlo et al., 2011), as well as histone modifying enzymes such as EP300 and HDAC2 (Trivedi et al., 2007; Wei et al., 2008; Zhang et al., 2002) activate a CM gene expression program that results in cellular dysfunction. Dynamic and global changes in histone 3 lysine 9 acetylation, a chromatin mark for gene promoters and enhancers, have been observed genome-wide in the adult mouse heart after pressure overload (Sayed et al., 2013). Despite these fundamental observations, the mediators in the heart that link the activity of master regulatory TFs and histone acetylation to Pol II dynamics and global transcriptional anabolism are poorly understood.
Here, we establish that BET bromodomain reader proteins function critically in chromatin-mediated signal transduction to Pol II, co-activating pathologic gene expression in the heart. GSEA reveals that BET inhibition antagonizes multiple TF outputs known to be causal in HF pathogenesis including NFAT, NFκB and GATA4, suggesting that BET bromodomain proteins co-activate a broad transcriptional network involving multiple TFs. Importantly, we find that BET bromodomain proteins do not directly affect Myc mRNA levels or function in the heart – a striking contrast to observations in hematopoietic tumors, where BETs are required for c-Myc expression and activity (Delmore et al., 2011; Zuber et al., 2011). ChIP-seq analysis reveals that BRD4 co-occupies active promoters with Pol II (as defined by H3K4me3), and active gene enhancers (as defined by H3K27ac) in the adult mouse heart and that cardiac pressure overload induces Pol II pause release and transcriptional elongation within four days. BET bromodomain inhibition suppresses transcriptional pause release during pressure overload in vivo, and attenuates expression of the pathologic HF gene program. Together, these data demonstrate that BET bromodomain reader proteins are indispensable co-activators in pathologic gene expression in the heart that function, in part, through their ability to promote Pol II pause release and transcriptional elongation.
Activation of the P-TEFb complex, a central effector of pause release and transcriptional elongation (Lee and Young, 2013), has been observed in pathologic cardiac hypertrophy (Sano et al., 2002). Previous studies in NIH 3T3 cells have demonstrated that BRD4 interacts with P-TEFb and facilitates stimulus-coupled recruitment of P-TEFb to target promoters (Patel et al., 2013). Activity of CDK9, a core constituent of the P-TEFb complex, is also increased during hypertrophic stress and is required for agonist-induced hypertrophy in NRVM (Sano et al., 2002). CM-specific overexpression of the Cdk9-activating protein CyclinT1 results in cardiac hypertrophy in vivo (Sano et al., 2007). In addition, deficiency of HEXIM-1, a nuclear protein which sequesters the P-TEFb in an inactive complex, has been shown to potentiate pathologic hypertrophy in vivo (Espinoza-Derout et al., 2009). In our studies, localization of BRD4 with promoter-enhancer elements and the suppression of pause release with BET bromodomain inhibition in the TAC model (Figure 6) suggest BETs facilitate pause release at these gene loci.
The robust induction of HEXIM1 expression we observed following BET bromodomain inhibition (Figure S5H-K) may also serve to suppress P-TEFb activity and reduce Pol II pause release. Concordant reductions in Ser2 phosphorylation of the Pol II CTD (Figure 6G and S5G), a specific target of the P-TEFb complex, both in vitro and in vivo support these mechanisms of action. Defining the extent to which BETs alter transcriptional elongation through direct interactions with components of the PTEF-b complex (Bisgrove et al., 2007) or regulation of HEXIM-1 expression represent important avenues of future investigation.
While evidence presented here does implicate BETs in the regulation of P-TEFb function and Pol II pause release during TAC, we cannot exclude additional effects of BET bromodomains on Pol II initiation at specific gene promoters. Consistent with our findings, other work has previously identified that Pol II pause release is a dominant mechanism for gene induction in both the developing heart and in the adult mouse heart during pressure overload, with de novo Pol II recruitment occurring at only 5% of induced genes in the TAC model (Sayed et al., 2013). Potential roles for BET bromodomain proteins in locus-specific Pol II initiation in the heart, or in other aspects of mRNA processing, are the subject of ongoing research by our group. Furthermore, the very recent discovery of asymmetrically loaded BRD4 on a critical subset of state-specific enhancers termed super-enhancers (Loven et al., 2013) raises the possibility that such super-enhancers might also be active in the stressed myocardium and contribute to BET target specificity.
HF is known to progress via pathologic crosstalk between CMs and cardiac fibroblasts (van Berlo et al., 2013). While the TAC model of HF provides a relatively focal stress to the heart, and JQ1 attenuates pathologic remodeling without effects on blood pressure or hemodynamic load (Figure S3C-D), we recognize that BET bromodomain inhibition in vivo may act on cardiac fibroblasts and other cell types that populate the stressed myocardium, in addition to the observed effects on CMs. We show here that BET bromodomain inhibition or Brd4-knockdown in isolated NRVM both attenuate pathologic CM hypertrophy in vitro (Figure 1). In addition, we find that Brd4 is the highest expressed BET gene in isolated CMs and in adult heart tissue. These data identify a cell autonomous role for BRD4 in CMs in vitro and suggest that this protein might be an important target of BET bromodomain inhibitors in the heart in vivo. Future studies using cell-type and temporally restricted gene-deletion of Brd4 and other BET family members in adult mice will help annotate their gene- and tissue-specific functions in experimental models of HF.
In conclusion, this study implicates a family of conserved epigenetic reader proteins as essential components of the transcriptional machinery that drives pathologic cardiac remodeling and HF. BET bromodomain proteins function as master regulatory TF co-activators that regulate pathologic pause release in the failing heart. The chemical biological approach leveraged here in rodent models of pathologic hypertrophy and HF provides a rationale for developing drug-like BET bromodomain inhibitors as investigational therapeutic agents in heart disease.
EXPERIMENTAL PROCEDURES
Animal Models
All protocols concerning animal use were approved by the IACUC at Case Western Reserve University and conducted in accordance with the NIH Guide for the Care and Use of Laboratory Animals. All models were conducted in C57Bl/6J mice (Jackson Laboratories), which were maintained in a pathogen-free facility with standard light/dark cycling and access to food and water ad libitum.
Human samples
LV samples from healthy human hearts were obtained were obtained as described (Hannenhalli et al., 2006; Margulies et al., 2005) in accordance with the Investigation Review Committee at the Hospital of the University of Pennsylvania, Philadelphia, PA. Nuclear protein was extracted using the NE-Per kit (Thermo Scientific #78833) according to manufacturer’s instructions. Gene expression profiles from left ventricles obtained from non-failing versus failing human hearts were curated from a published dataset (Hannenhalli et al., 2006).
Statistical analysis
Data are reported as mean ± standard error. The statistical methods used in analysis of microarray and ChIP-Seq data are detailed separately above. Comparison of means between two groups was analyzed using a two-tailed Student's t-test with Bonferroni correction for multiple comparisons. For all analyses, P values <0.05 were considered significant.
Supplementary Material
HIGHLIGHTS.
BET bromodomains co-activate cardiac gene expression programs
BET inhibition blocks pathologic cardiac remodeling in vitro and in vivo
BRD4 occupies active cardiac enhancers and promoters
BET inhibition blocks pathologic transcriptional elongation in the adult heart
ACKNOWLEDGEMENTS
We are grateful to R. Young and P. Rahl for stimulating discussions, M. Berkeley and the late E. Fox (DFCI Microarray Core) for assistance with microarray experiments, Avery Whitlock for illustrations, and Tom Volkert, Jennifer Love, and Sumeet Gupta at the Whitehead Genome Core for assistance with genome sequencing. This research was supported by an NIH-R01 DK093821 and NIH-K08 HL086614, Individual Biomedical Research Award from The Hartwell Foundation, and the Visconsi Research Scholars Fund (S.M.H.); NIH-K08 CA128972, the Burroughs-Wellcome Fund, the Damon-Runyon Cancer Research Foundation, the Richard and Susan Smith Family Foundation, and the Next Generation Award (J.E.B.); U.S. Department of Defense (C.Y.L.); NIH-T32 HL105338 (M.A.A.); NIH-R01 HL105993 (K.B.M. and T.P.C); NIH-K08 HL105678 (J.D.B.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Banerjee C, Archin N, Michaels D, Belkina AC, Denis GV, Bradner J, Sebastiani P, Margolis DM, Montano M. BET bromodomain inhibition as a novel strategy for reactivation of HIV-1. Journal of leukocyte biology. 2012;92:1147–1154. doi: 10.1189/jlb.0312165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bisgrove DA, Mahmoudi T, Henklein P, Verdin E. Conserved P-TEFb-interacting domain of BRD4 inhibits HIV transcription. Proc Natl Acad Sci U S A. 2007;104:13690–13695. doi: 10.1073/pnas.0705053104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bousette N, Chugh S, Fong V, Isserlin R, Kim KH, Volchuk A, Backx PH, Liu P, Kislinger T, MacLennan DH, et al. Constitutively active calcineurin induces cardiac endoplasmic reticulum stress and protects against apoptosis that is mediated by alpha-crystallin-B. Proc Natl Acad Sci U S A. 2010;107:18481–18486. doi: 10.1073/pnas.1013555107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson MA, Kouzarides T, Huntly BJ. Targeting epigenetic readers in cancer. N Engl J Med. 2012;367:647–657. doi: 10.1056/NEJMra1112635. [DOI] [PubMed] [Google Scholar]
- Delmore JE, Issa GC, Lemieux ME, Rahl PB, Shi J, Jacobs HM, Kastritis E, Gilpatrick T, Paranal RM, Qi J, et al. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell. 2011;146:904–917. doi: 10.1016/j.cell.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichler GS, Huang S, Ingber DE. Gene Expression Dynamics Inspector (GEDI): for integrative analysis of expression profiles. Bioinformatics. 2003;19:2321–2322. doi: 10.1093/bioinformatics/btg307. [DOI] [PubMed] [Google Scholar]
- Espinoza-Derout J, Wagner M, Salciccioli L, Lazar JM, Bhaduri S, Mascareno E, Chaqour B, Siddiqui MA. Positive transcription elongation factor b activity in compensatory myocardial hypertrophy is regulated by cardiac lineage protein-1. Circ Res. 2009;104:1347–1354. doi: 10.1161/CIRCRESAHA.108.191726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filippakopoulos P, Picaud S, Mangos M, Keates T, Lambert JP, Barsyte-Lovejoy D, Felletar I, Volkmer R, Muller S, Pawson T, et al. Histone recognition and large-scale structural analysis of the human bromodomain family. Cell. 2012;149:214–231. doi: 10.1016/j.cell.2012.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filippakopoulos P, Qi J, Picaud S, Shen Y, Smith WB, Fedorov O, Morse EM, Keates T, Hickman TT, Felletar I, et al. Selective inhibition of BET bromodomains. Nature. 2010;468:1067–1073. doi: 10.1038/nature09504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannenhalli S, Putt ME, Gilmore JM, Wang J, Parmacek MS, Epstein JA, Morrisey EE, Margulies KB, Cappola TP. Transcriptional genomics associates FOX transcription factors with human heart failure. Circulation. 2006;114:1269–1276. doi: 10.1161/CIRCULATIONAHA.106.632430. [DOI] [PubMed] [Google Scholar]
- Heineke J, Auger-Messier M, Xu J, Oka T, Sargent MA, York A, Klevitsky R, Vaikunth S, Duncan SA, Aronow BJ, et al. Cardiomyocyte GATA4 functions as a stress-responsive regulator of angiogenesis in the murine heart. J Clin Invest. 2007;117:3198–3210. doi: 10.1172/JCI32573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill JA, Olson EN. Cardiac plasticity. N Engl J Med. 2008;358:1370–1380. doi: 10.1056/NEJMra072139. [DOI] [PubMed] [Google Scholar]
- Jang MK, Mochizuki K, Zhou M, Jeong HS, Brady JN, Ozato K. The bromodomain protein Brd4 is a positive regulatory component of P-TEFb and stimulates RNA polymerase II-dependent transcription. Mol Cell. 2005;19:523–534. doi: 10.1016/j.molcel.2005.06.027. [DOI] [PubMed] [Google Scholar]
- Jiang YW, Veschambre P, Erdjument-Bromage H, Tempst P, Conaway JW, Conaway RC, Kornberg RD. Mammalian mediator of transcriptional regulation and its possible role as an end-point of signal transduction pathways. Proc Natl Acad Sci U S A. 1998;95:8538–8543. doi: 10.1073/pnas.95.15.8538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee TI, Young RA. Transcriptional regulation and its misregulation in disease. Cell. 2013;152:1237–1251. doi: 10.1016/j.cell.2013.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy D, Garrison RJ, Savage DD, Kannel WB, Castelli WP. Prognostic implications of echocardiographically determined left ventricular mass in the Framingham Heart Study. N Engl J Med. 1990;322:1561–1566. doi: 10.1056/NEJM199005313222203. [DOI] [PubMed] [Google Scholar]
- Lin CY, Loven J, Rahl PB, Paranal RM, Burge CB, Bradner JE, Lee TI, Young RA. Transcriptional amplification in tumor cells with elevated c-Myc. Cell. 2012;151:56–67. doi: 10.1016/j.cell.2012.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loven J, Hoke HA, Lin CY, Lau A, Orlando DA, Vakoc CR, Bradner JE, Lee TI, Young RA. Selective inhibition of tumor oncogenes by disruption of superenhancers. Cell. 2013;153:320–334. doi: 10.1016/j.cell.2013.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier HJ, Schips TG, Wietelmann A, Kruger M, Brunner C, Sauter M, Klingel K, Bottger T, Braun T, Wirth T. Cardiomyocyte-specific IkappaB kinase (IKK)/NF-kappaB activation induces reversible inflammatory cardiomyopathy and heart failure. Proc Natl Acad Sci U S A. 2012;109:11794–11799. doi: 10.1073/pnas.1116584109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margulies KB, Matiwala S, Cornejo C, Olsen H, Craven WA, Bednarik D. Mixed messages: transcription patterns in failing and recovering human myocardium. Circ Res. 2005;96:592–599. doi: 10.1161/01.RES.0000159390.03503.c3. [DOI] [PubMed] [Google Scholar]
- Matzuk MM, McKeown MR, Filippakopoulos P, Li Q, Ma L, Agno JE, Lemieux ME, Picaud S, Yu RN, Qi J, et al. Small-Molecule Inhibition of BRDT for Male Contraception. Cell. 2012;150:673–684. doi: 10.1016/j.cell.2012.06.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinsey TA, Olson EN. Toward transcriptional therapies for the failing heart: chemical screens to modulate genes. J Clin Invest. 2005;115:538–546. doi: 10.1172/JCI24144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery RL, Davis CA, Potthoff MJ, Haberland M, Fielitz J, Qi X, Hill JA, Richardson JA, Olson EN. Histone deacetylases 1 and 2 redundantly regulate cardiac morphogenesis, growth, and contractility. Genes Dev. 2007;21:1790–1802. doi: 10.1101/gad.1563807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicodeme E, Jeffrey KL, Schaefer U, Beinke S, Dewell S, Chung CW, Chandwani R, Marazzi I, Wilson P, Coste H, et al. Suppression of inflammation by a synthetic histone mimic. Nature. 2010;468:1119–1123. doi: 10.1038/nature09589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel MC, Debrosse M, Smith M, Dey A, Huynh W, Sarai N, Heightman TD, Tamura T, Ozato K. BRD4 coordinates recruitment of pause-release factor P-TEFb and the pausing complex NELF/DSIF to regulate transcription elongation of interferon stimulated genes. Mol Cell Biol. 2013 doi: 10.1128/MCB.01180-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterlin BM, Price DH. Controlling the elongation phase of transcription with P-TEFb. Mol Cell. 2006;23:297–305. doi: 10.1016/j.molcel.2006.06.014. [DOI] [PubMed] [Google Scholar]
- Rahl PB, Lin CY, Seila AC, Flynn RA, McCuine S, Burge CB, Sharp PA, Young RA. c-Myc regulates transcriptional pause release. Cell. 2010;141:432–445. doi: 10.1016/j.cell.2010.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockman HA, Ross RS, Harris AN, Knowlton KU, Steinhelper ME, Field LJ, Ross J, Jr, Chien KR. Segregation of atrial-specific and inducible expression of an atrial natriuretic factor transgene in an in vivo murine model of cardiac hypertrophy. Proc Natl Acad Sci U S A. 1991;88:8277–8281. doi: 10.1073/pnas.88.18.8277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roger VL, Go AS, Lloyd-Jones DM, Benjamin EJ, Berry JD, Borden WB, Bravata DM, Dai S, Ford ES, Fox CS, et al. Executive summary: heart disease and stroke statistics--2012 update: a report from the American Heart Association. Circulation. 2012;125:188–197. doi: 10.1161/CIR.0b013e3182456d46. [DOI] [PubMed] [Google Scholar]
- Sano M, Abdellatif M, Oh H, Xie M, Bagella L, Giordano A, Michael LH, DeMayo FJ, Schneider MD. Activation and function of cyclin T-Cdk9 (positive transcription elongation factor-b) in cardiac muscle-cell hypertrophy. Nat Med. 2002;8:1310–1317. doi: 10.1038/nm778. [DOI] [PubMed] [Google Scholar]
- Sano M, Minamino T, Toko H, Miyauchi H, Orimo M, Qin Y, Akazawa H, Tateno K, Kayama Y, Harada M, et al. p53-induced inhibition of Hif-1 causes cardiac dysfunction during pressure overload. Nature. 2007;446:444–448. doi: 10.1038/nature05602. [DOI] [PubMed] [Google Scholar]
- Sayed D, He M, Yang Z, Lin L, Abdellatif M. Transcriptional regulation patterns revealed by high resolution chromatin immunoprecipitation during cardiac hypertrophy. J Biol Chem. 2013;288:2546–2558. doi: 10.1074/jbc.M112.429449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber SL, Bernstein BE. Signaling network model of chromatin. Cell. 2002;111:771–778. doi: 10.1016/s0092-8674(02)01196-0. [DOI] [PubMed] [Google Scholar]
- Shen Y, Yue F, McCleary DF, Ye Z, Edsall L, Kuan S, Wagner U, Dixon J, Lee L, Lobanenkov VV, et al. A map of the cis-regulatory sequences in the mouse genome. Nature. 2012;488:116–120. doi: 10.1038/nature11243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson P, McGrath A, Savion S. Myocyte hypertrophy in neonatal rat heart cultures and its regulation by serum and by catecholamines. Circ Res. 1982;51:787–801. doi: 10.1161/01.res.51.6.787. [DOI] [PubMed] [Google Scholar]
- Song HK, Hong SE, Kim T, Kim do H. Deep RNA sequencing reveals novel cardiac transcriptomic signatures for physiological and pathological hypertrophy. PloS one. 2012;7:e35552. doi: 10.1371/journal.pone.0035552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song X, Kusakari Y, Xiao CY, Kinsella SD, Rosenberg MA, Scherrer-Crosbie M, Hara K, Rosenzweig A, Matsui T. mTOR attenuates the inflammatory response in cardiomyocytes and prevents cardiac dysfunction in pathological hypertrophy. American journal of physiology Cell physiology. 2010;299:C1256–C1266. doi: 10.1152/ajpcell.00338.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trivedi CM, Luo Y, Yin Z, Zhang M, Zhu W, Wang T, Floss T, Goettlicher M, Noppinger PR, Wurst W, et al. Hdac2 regulates the cardiac hypertrophic response by modulating Gsk3 beta activity. Nat Med. 2007;13:324–331. doi: 10.1038/nm1552. [DOI] [PubMed] [Google Scholar]
- van Berlo JH, Elrod JW, Aronow BJ, Pu WT, Molkentin JD. Serine 105 phosphorylation of transcription factor GATA4 is necessary for stress-induced cardiac hypertrophy in vivo. Proc Natl Acad Sci U S A. 2011;108:12331–12336. doi: 10.1073/pnas.1104499108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Berlo JH, Maillet M, Molkentin JD. Signaling effectors underlying pathologic growth and remodeling of the heart. J Clin Invest. 2013;123:37–45. doi: 10.1172/JCI62839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wade JT, Struhl K. The transition from transcriptional initiation to elongation. Curr Opin Genet Dev. 2008;18:130–136. doi: 10.1016/j.gde.2007.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei JQ, Shehadeh LA, Mitrani JM, Pessanha M, Slepak TI, Webster KA, Bishopric NH. Quantitative control of adaptive cardiac hypertrophy by acetyltransferase p300. Circulation. 2008;118:934–946. doi: 10.1161/CIRCULATIONAHA.107.760488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X, Lu J, Kulbokas EJ, Golub TR, Mootha V, Lindblad-Toh K, Lander ES, Kellis M. Systematic discovery of regulatory motifs in human promoters and 3' UTRs by comparison of several mammals. Nature. 2005;434:338–345. doi: 10.1038/nature03441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Yik JH, Chen R, He N, Jang MK, Ozato K, Zhou Q. Recruitment of P-TEFb for stimulation of transcriptional elongation by the bromodomain protein Brd4. Mol Cell. 2005;19:535–545. doi: 10.1016/j.molcel.2005.06.029. [DOI] [PubMed] [Google Scholar]
- Yoshikawa N, Shimizu N, Maruyama T, Sano M, Matsuhashi T, Fukuda K, Kataoka M, Satoh T, Ojima H, Sawai T, et al. Cardiomyocyte-specific overexpression of HEXIM1 prevents right ventricular hypertrophy in hypoxia-induced pulmonary hypertension in mice. PloS one. 2012;7:e52522. doi: 10.1371/journal.pone.0052522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang CL, McKinsey TA, Chang S, Antos CL, Hill JA, Olson EN. Class II histone deacetylases act as signal-responsive repressors of cardiac hypertrophy. Cell. 2002;110:479–488. doi: 10.1016/s0092-8674(02)00861-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Prakash C, Sum C, Gong Y, Li Y, Kwok JJ, Thiessen N, Pettersson S, Jones SJ, Knapp S, et al. Bromodomain-containing protein 4 (BRD4) regulates RNA polymerase II serine 2 phosphorylation in human CD4+ T cells. J Biol Chem. 2012;287:43137–43155. doi: 10.1074/jbc.M112.413047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao M, Chow A, Powers J, Fajardo G, Bernstein D. Microarray analysis of gene expression after transverse aortic constriction in mice. Physiological genomics. 2004;19:93–105. doi: 10.1152/physiolgenomics.00040.2004. [DOI] [PubMed] [Google Scholar]
- Zhong W, Mao S, Tobis S, Angelis E, Jordan MC, Roos KP, Fishbein MC, de Alboran IM, MacLellan WR. Hypertrophic growth in cardiac myocytes is mediated by Myc through a Cyclin D2-dependent pathway. EMBO J. 2006;25:3869–3879. doi: 10.1038/sj.emboj.7601252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuber J, Shi J, Wang E, Rappaport AR, Herrmann H, Sison EA, Magoon D, Qi J, Blatt K, Wunderlich M, et al. RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature. 2011;478:524–528. doi: 10.1038/nature10334. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.