Abstract
Oral bacterial biofilms trigger chronic inflammatory responses in the host that can result in the tissue destructive events of periodontitis. However, the characteristics of the capacity of specific host cell types to respond to these biofilms remain ill-defined. This report describes the use of a novel model of bacterial biofilms to stimulate oral epithelial cells and profile select cytokines and chemokines that contribute to the local inflammatory environment in the periodontium. Monoinfection biofilms were developed with Streptococcus sanguinis, Streptococcus oralis, Streptococcus gordonii, Actinomyces naeslundii, Fusobacterium nucleatum, and Porphyromonas gingivalis on rigid gas-permeable contact lenses. Biofilms, as well as planktonic cultures of these same bacterial species, were incubated under anaerobic conditions with a human oral epithelial cell line, OKF4, for up to 24 h. Gro-1α, IL1α, IL-6, IL-8, TGFα, Fractalkine, MIP-1α, and IP-10 were shown to be produced in response to a range of the planktonic or biofilm forms of these species. P. gingivalis biofilms significantly inhibited the production of all of these cytokines and chemokines, except MIP-1α. Generally, the biofilms of all species inhibited Gro-1α, TGFα, and Fractalkine production, while F. nucleatum biofilms stimulated significant increases in IL-1α, IL-6, IL-8, and IP-10. A. naeslundii biofilms induced elevated levels of IL-6, IL-8 and IP-10. The oral streptococcal species in biofilms or planktonic forms were poor stimulants for any of these mediators from the epithelial cells. The results of these studies demonstrate that oral bacteria in biofilms elicit a substantially different profile of responses compared to planktonic bacteria of the same species. Moreover, certain oral species are highly stimulatory when in biofilms and interact with host cell receptors to trigger pathways of responses that appear quite divergent from individual bacteria.
Keywords: Oral bacteria, Biofilms, Epithelial cells, Cytokines, Chemokines
1. Introduction
Host gingival tissues respond to an array of microbial challenges in the oral cavity that are crucial for maintaining homeostasis within this constantly infected environment. These responses comprise a wide array of pro- and anti-inflammatory molecules produced by resident cells of the periodontium [1,2], as well as response molecules derived from infiltrating inflammatory and immune cells in the tissues [3,4]. While these innate and adaptive immune responses are generated to protect the host from the microbial burden juxtaposed to the tissues, magnitude of the burden and chronicity of the inflammatory response can result in both soft and hard tissue damage defined as periodontitis [5].
Many in vitro studies have reduced the complex in situ microbial infection into individual proposed pathogenic or commensal species as planktonic challenges of host cells [6–8]. These types of studies have frequently focused on a particular host target molecule [9,10], emphasized molecular aspects of Toll-like receptor engagement [11,12] and/or developed data to identify intracellular signaling pathways that account for the response profile of the cells [13]. Moreover, it appears clear that the net result of these host–bacterial interactions as maintaining health or manifesting disease is reflected in the relative distribution and abundance of a wide range of biomolecules with competing and complementary activities in the tissue microenvironment [14,15]. Many of these conclusions have resulted from studies that attempt to sample the oral environment and compare the patterns of various responses in health and disease [16], during progression of the periodontal lesion [17], or following clinically successful therapy of disease [18]. While providing a robust snapshot of the characteristics of the compendium of responses that can occur in the oral cavity, these types of studies are not able to delineate details of the microorganisms that have a predilection to elicit the particular responses or the relative contribution of individual host cell types to these response profiles.
It is clear that the bacteria inhabiting the oral cavity in contact with oral tissues reside in complex multispecies biofilms [19]. These biofilm structures arise via interaction with host substrates [20,21] and accrue and mature based upon the oral environment and specific interactions among individual species of bacteria [22,23]. However, there are very few reports that have evaluated the response profiles of specific host cells to oral bacteria in biofilms. Recently, Guggenheim and colleagues used a hydroxyapatite disc model to prepare oral multispecies biofilms and used these to challenge epithelial cell cultures [24,25]. While numerous species were used to create the biofilms, of the nine species used, Porphyromonas gingivalis and Fusobacterium nucleatum made up a rather small proportion of the overall microbial composite at a level approximating 1% of the total. Thus, while the architecture of these very complex biofilms was described using confocal scanning laser microscopy, it remains undetermined the density of the bacteria that interacted with the individual cells, nor the species that may have been primary participants in this process. Nevertheless, this report did document a range of host responses molecules produced in response to the biofilm challenge that occurred under aerobic conditions over 24 h. The primary findings were an apparent increase in apoptosis and degradation of IL-1β, IL-6 and IL-8 cytokines that were elicited from the epithelial cells that was predicted to be related to the presence of proteases, such as those produced by P. gingivalis in these biofilms.
The present report describes our use of a novel biofilm model, created on rigid gas permeable contact lens (RGPL) material [26,27] that were used to challenge oral epithelial cell cultures in an anaerobic environment that may better reflect the subgingival sulcus. We evaluated patterns of cell responses to single species biofilms, compared with planktonic challenge with the same species, to elucidate unique features of the cellular responses to the biofilm challenge.
2. Materials and methods
2.1. Bacteria and culture conditions
P. gingivalis (FDC381), F. nucleatum ATCC 25586, Actinomyces naeslundii ATCC 49840, and Streptococcus gordonii ATCC 10558 were cultured in Brain Heart Infusion (Becton Dickinson and Company, Sparks, MD) medium supplemented with 5 μg hemin ml−1 and 1 μg menadione ml−1 under anaerobic conditions (85% N2, 10% H2, 5% CO2) at 37 °C. Streptococcus sanguinis ATCC 10556 and Streptococcus oralis ATCC 10557 were grown in Trypticase yeast extract salts (TYS) medium under anaerobic conditions. All bacterial strains used in this study have been described previously and were obtained from the ATCC, except P. gingivalis (FDC381) [26].
2.2. Biofilm growth conditions
Biofilms were grown on rigid gas permeable lenses (RGPLs) (Advanced Vision Technologies, Golden, CO) as previously described [26]. Briefly, prior to biofilm formation, RGPLs were coated with 1% fetal bovine serum (FBS; Invitrogen) to support the adherence of bacteria and incubated at room temperature until dry. Each RGPL was inoculated with a 5 ml of monospecies planktonic culture at 0.3 OD A600 in a single well of a 6-well polystyrene tissue culture plate (BD Falcon, Franklin Lakes, NJ) and incubated in an anaerobic chamber for 3 days for development of biofilms under static conditions. At each 24-h interval spent media was replenished by fresh medium. After incubation, RGPLs with adherent biofilms were washed in 1X PBS twice to remove loosely adherent cells and used in subsequent epithelial cell challenge. Biofilms grown on three additional RGPLs were used for bacterial enumeration by qPCR analysis as described previously [26].
2.3. OKF4 cell growth and bacterial challenge
An immortalized epithelial cell line OKF4 (Rheinwald 2002) was cultured in keratinocyte serum free medium (Invitrogen, Carlsbad, CA) and seeded into 48-well tissue culture plates (Costar, Cambridge, MA) at a density of 105 cells per well in a 1 ml volume and allowed to adhere for 24 h in a 5% CO2 chamber at 37 °C to form a confluent monolayer. Planktonic, biofilm and control treatments were each carried out in six wells in 1 ml/well fresh media and continuously incubated for 6 h under anaerobic conditions (85% N2, 5% CO2, and 10% H2). For planktonic challenge, overnight cultures were harvested by centrifugation and resuspended in keratinocyte medium. A 108, 107 and 106 cells/well challenge corresponding to a multiplicity of infection (MOI) at 1000:1, 100:1 and 10:1 was used to stimulate the OKF4 cells. The estimated MOI for the planktonic challenge represents the numbers of bacteria that could be predicted to be in association with the OKF4 cells interacting with the biofilms. Three day old biofilms grown on contact lens were overlaid with biofilm-side facing the epithelial cells. OKF4 cells with or without overlaid RGPL were used as controls for the biofilm or planktonic bacterial challenges, respectively. OKF4 cell supernatants from each of two wells were pooled and stored at −80 °C for cytokine determination. Previous studies with this model system demonstrated that these biofilm challenges did not result in any obvious toxicity or cell death, as determined by both metabolic activity and housekeeping gene expression [27].
2.4. Inhibition of biofilm growth during the course of challenge
In order to constrain the biofilms from replicating in the keratinocyte media during the 24 h challenge with OKF4 cells, biofilms were treated with green fluorescent nucleic acid stain SYTO 24. SYTO 24 was chosen as it yielded lowest optical density with highest fluorescence intensity values for S. gordonii, A. naeslundii and P. gingivalis indicating that this stain inhibited replication while not affecting viability [26]. S. oralis and S. sanguinis biofilms were also treated with SYTO 24 to inhibit replication. Prior to challenging OKF4 cells, biofilms were immersed in 10 μg/ml SYTO 24 stain in keratinocyte media for 5 min. after which they were immersed in 1X PBS twice to remove excess stain. F. nucleatum was not treated with any SYTO stains as it did not replicate in keratinocyte media.
2.5. Detection of cytokines/chemokines
The level of cells by 24 h was determined using a multiplexed beadlyte kit (R & D systems, Minneapolis, MN, USA) and a Luminex IS100 (Luminex, Inc., Austin, TX) instrument. The mean ± standard error of the mean of the planktonic and biofilm stimulation of OKF4 cells was compared with unchallenged and RGPL overlaid OKF4 cells, respectively. Statistical comparison of the data was accomplished using an ANOVA on ranks test with Dunn’s test for multiple comparisons for multiple comparisons (SigmaStat 3.5; Systat Software, Inc., Chicago, IL).
3. Results
3.1. Characteristics of cytokine responses
An array of cytokines and cell communication factors produced by epithelial cells and having some potential role in responses to oral bacterial challenge was evaluated and included, IL-1α, IL-6, and TGFα. The results in Fig. 1A–C demonstrate the responses of the epithelial cells to challenge with the range of oral bacteria. Biofilms of A. naeslundii and all three streptococci provided minimal stimulus of IL-1α, while the planktonic challenge with these microorganisms appeared to inhibit production of this cytokine. The F. nucleatum biofilms significantly upregulated production of IL-1α, while P. gingivalis biofilms significantly inhibited this response. Of the planktonic bacteria only P. gingivalis appeared to have the capacity to induce IL-1α in this system.
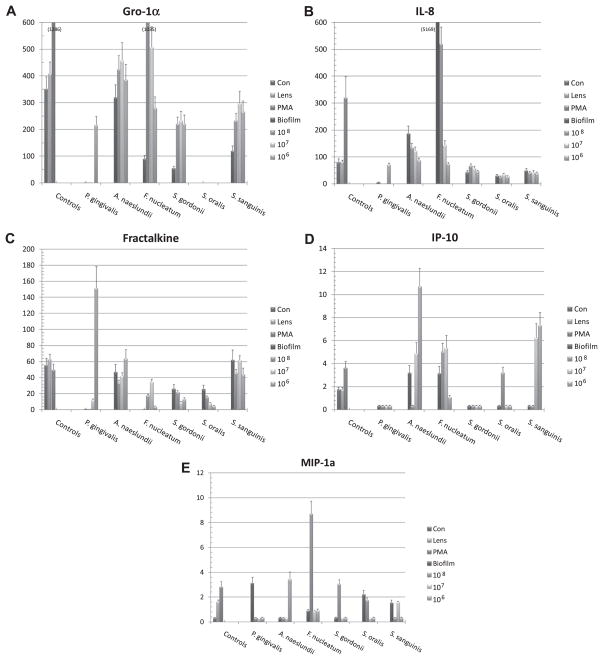
Fig. 1.
A–C: Levels of cytokines elicited by challenge of oral epithelial cells with biofilms or planktonic forms of various oral bacteria. Each bar denotes mean level of triplicate determinations for each condition and the vertical brackets signify 1 SD. The asterisk (*) denotes significantly different than controls (Con for planktonic; Lens for biofilms) at least at p < 0.05. The # signifies significantly greater that other stimuli for the particular microorganism at p < 0.05.
The response of the epithelial in producing IL-6 was quite limited across these bacterial species. Only biofilms and planktonic A. naeslundii and F. nucleatum elicited this cytokine response, with the biofilms of each species inducing significantly greater quantities of IL-6 compared to the planktonic challenge.
As was noted with IL-1α, all of the bacterial biofilms inhibited the production of TGFα, with P. gingivalis totally eliminated. Planktonic bacterial challenge with P. gingivalis and S. sanguinis elicited elevated levels of this cell communication factor.
3.2. Characteristics of chemokine responses
This investigation also profiled a range of chemokines, Gro-1α, IL-8, Fractalkine, IP-10, and MIP-1α, RANTES and MCP-1 that are produced by epithelial cells and could be expected to provide early warning signals to the local tissue environment and the host immune system. The results in Fig. 2A–E provide a profile of these chemokine responses by the epithelial cells, with the exception of RANTES and MCP-1 which provided minimal detectable levels. Gro-1α production was inhibited by biofilms challenge with P. gingivalis, F. nucleatum and all three streptococci. Only planktonic challenge with A. naeslundii and F. nucleatum showed an induction of this chemokine. Interestingly, among the oral streptococci, S. oralis was completely nonreactive as a stimulus for this chemokine.
Fig. 2.
A–E: Levels of chemokines elicited by challenge of oral epithelial cells with biofilms or planktonic forms of various oral bacteria. Each bar denotes mean level of triplicate determinations for each condition and the vertical brackets signify 1 SD. The asterisk (*) denotes significantly different than controls (Con for planktonic; Lens for biofilms) at least at p < 0.05. The # signifies significantly greater that other stimuli for the particular microorganism at p < 0.05.
Significantly elevated production of IL-8 was observed with biofilms of A. naeslundii and F. nucleatum, as well as following challenge with planktonic versions of these species. In contrast, no effect was observed with the oral streptococci, and P. gingivalis inhibited the production of this critical chemokine.
The levels of Fractalkine were generally unaffected by challenge with all of the oral bacteria, except the lowest dose of planktonic P. gingivalis. A similar selective effect was observed with IP-10 and MIP-1α. Biofilms of A. naeslundii and F. nucleatum stimulated production of IP-10, as well as planktonic challenge with these species. Biofilms of P. gingivalis and the oral streptococci inhibited IP-10 production. Interestingly, with MIP-1α the biofilms of P. gingivalis increased the levels of this chemokine, as did selective doses of planktonic challenge with each of the other species of bacteria.
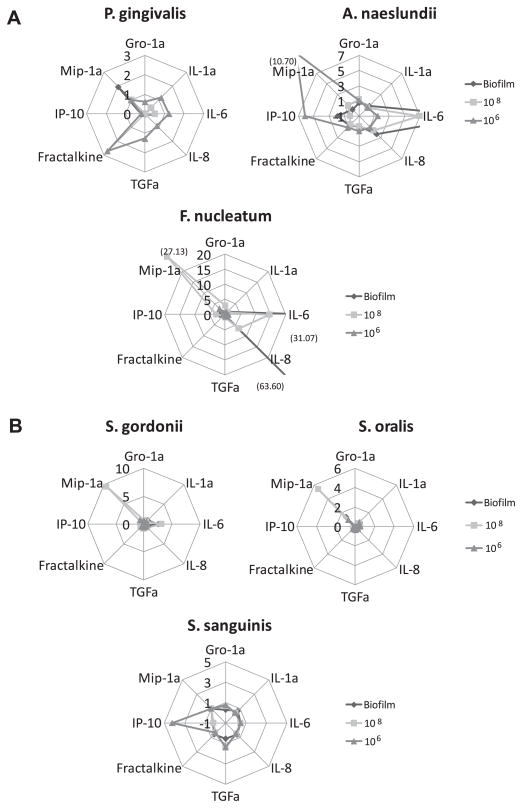
3.3. Profiles of cytokine/chemokine production by epithelial cells
Fig. 3A and B provides a summary of the patterns of responses of the epithelial cells to the individual biofilms and high and low dose challenge of the planktonic bacteria. The high dose planktonic challenge is similar to the overall numbers of the species in the biofilms, while the low dose planktonic challenge estimates the number of the bacteria in the layered biofilms that might be expected to be able to interact directly with the epithelial cells. Clearly, the profiles of stimulation are unique to each species, and the biofilms are unique compared to these doses of the planktonic stimulus across the species. The biofilms of A. naeslundii and F. nucleatum appear to be the most active with regards to breadth and quantity of mediators induced. In contrast, P. gingivalis biofilms were minimally reactive, and in most cases inhibited production of the cytokines and chemokines. Fig. 3B shows that generally the oral streptococci in any form were not particularly active in stimulating the epithelial cells. However, of interest was that while these are considered highly related species, there were distinct differences in the patterns of mediators induced by challenge with the planktonic forms of the bacteria. As noted, the streptococcal biofilms were not active in stimulating cytokines/chemokines and in most cases appeared to inhibit basal production by the epithelial cells.
Fig. 3.
A and B: Summary of cytokine and chemokine response profiles for monospecies biofilms compared with response patterns for 108 or 106 planktonic forms for each bacterial species. The line peaks denote fold difference in response compared to control cell basal levels (planktonic comparison) or cells overlaid with a bacterial free RGPL (biofilms comparison). Numbers in parentheses denote fold changes beyond the scale of the graph.
4. Discussion
This report provides some of the first data documenting the patterns of cytokines and chemokines that are induced in oral epithelial cells following challenge with monospecies oral bacterial biofilms. It is clear that the oral bacterial ecology is acquired early in life and evolves over time with the host to form a complex of numerous genera and species occupying the various ecological niches in the oral cavity as biofilms [28,29]. Moreover, it is clear that with the two major oral diseases being infections, the characteristics of the microbial ecology changes when transitioning from health to disease [30]. Many investigations have reported the responses of various host cells, e.g. epithelial cells, fibroblasts, lymphocytes, following challenge with oral microorganisms in planktonic form from individual species [7,31–33]. Reducing the system even further, various studies have targeted selected biomolecules or structures of the oral bacteria, that in an isolated in vitro system can also stimulate an array of host responses [34,35]. Specifically, with respect to the characteristics of the interactions examined in this study, P. gingivalis, F. nucleatum, Actinomyces species, and oral streptococci have been shown to elicit IL-1α, IL-1β, IL-6, IL-8, IL-10, IL-8, LL37, HBD 1,2,3 from oral epithelial cells [31]. As noted in a number of these reports, the characteristics of the host cell responses to individual oral bacteria varies significantly across genera and species, as well as showing some differences when considering individual microorganisms as oral pathogens or commensals [7,8,31]. While these studies have some value in understanding the biology of host–bacterial interactions that occur in vivo, the challenge does not reflect the primary organization of the oral bacteria into complex multispecies biofilms that stimulate both non-immune and immune cells in situ. Recent reports have emphasized the important differences in oral microbial interactions with host tissues and cells when these bacteria exist in complex biofilms. Thus, it could be anticipated that these responses would vary due to interbacterial metabolic and virulence synergisms within biofilms. Additionally, differences in host responses would be expected related to the variations in bacterial gene expression profiles that contribute to the biofilm structure, enhanced resistance to antimicrobials, and physiologic microenvironments within the biofilms [36–40]. This study provides some foundational knowledge using a novel biofilm model system to demonstrate unique features of host response patterns to challenge across a range of oral bacteria and when comparing biofilms to planktonic forms of the bacteria.
A recent study has reported that complex multispecies biofilms stimulate a range of mediators from epithelial cells including IL-1β, IL-6, IL-8 [24] and RANKL/OPG [25]. These nine species biofilms provide support to the underlying premise that host cellular responses to biofilms versus planktonic bacteria are substantively different. However, these studies do not shed light on the characteristics of individual bacteria in these biofilms, whether or not they are even specifically interacting with the epithelial cell monolayer, and in general, these multispecies biofilms were vastly dominated by a very limited number of species [24]. Consequently, there remain unanswered questions regarding the stimulatory capacity of individual bacteria in biofilms, how they compare with planktonic stimuli, and how these responses would be altered in the presence of a complex multispecies biofilm.
This report provides details on the use of a novel rigid gas-permeable lens material to build bacterial biofilms that were used to challenge oral epithelial cells. For this investigation we selected a group of cytokines and chemokines that have been reported to be induced by various stimuli interacting with a range of epithelial cell types [41,42]. IL-1α is a cytokine produced by a range of cell types, with important functions in enhancement of inflammation and host defence [43]. IL-6 is a pleiotropic pro-inflammatory cytokine that regulates several biological functions including inflammatory responses and bone biology [44]. TGFα is produced by macrophages and keratinocytes, is closely related to epidermal growth factor (EGF), and can also bind to the EGF receptor leading to epithelial development [45]. The results demonstrated that each of these cytokines/growth factors was differentially induced by biofilms of the oral bacteria. Clearly, IL-1α was the most responsive to the biofilms, while IL-6 was selectively elicited by only A. naeslundii and F. nucleatum biofilms, and TGFα levels were inhibited below basal cell production by biofilms of all the oral bacteria. As was predicted the biofilm stimulation patterns were distinct from the planktonic bacteria, and in a number of instances, the level of stimulation was greater than planktonic challenge with approximately a two logs higher number of the bacteria.
Anticipating that a major role of the epithelial cells is to provide an “early warning” signal to the host inflammatory and innate and adaptive immune responses, we examined a range of chemokines that would be predicted to have a principle role for inducing the emigration of protective cells and biomolecules into the infected oral sites. IL-8 is a primary chemokine for attracting neutrophils into sites of inflammation, and is a hallmark of the chronic inflammatory response in periodontitis [46]. Gro-1α is a chemokine with multiple functions associated with atherosclerosis, angiogenesis and many inflammatory conditions [47]. Soluble Fractalkine is a potent chemoattractant for T cells and monocytes, although cell-bound Fractalkine promotes adhesion of leukocytes to activated endothelial cells [48]. IP-10 is secreted by monocytes, endothelial cells, epithelial cells and fibroblasts in response to interferon (IFN)γ. It is a chemoattractant for monocytes/macrophages, T cells, NK cells, and dendritic cells, and as with Fractalkine promotes T cell adhesion to endothelial cells [49]. MIP-1α is one of a family of chemokines produced by stimulated macrophages and epithelial cells in response to infection. This chemokine activates granulocytes contributing to acute inflammatory responses [50] and induces the synthesis of a range of pro-inflammatory molecules [51]. As was noted with the cytokines, the patterns of responses of the chemokines to the oral microbial biofilms were quite varied. Only IL-8 and IP-10 appeared to show elevated response levels to the biofilms, and this was limited to A. naeslundii and F. nucleatum. Additionally, with IP-10, planktonic forms of the A. naeslundii were significantly more stimulatory, while the F. nucleatum biofilms were most active for the chemokines compared to planktonic bacteria. While we also examined MCP-1 and RANTES in this study, neither of these analytes provided sufficient signal to identify any role they might have in the epithelial cell responses to these oral biofilms.
Our findings using planktonic forms of a range of species are consistent with those previous reports regarding epithelial cell responses. Of some interest was the variation in the dose response of a number of the mediators towards challenge with the planktonic bacteria. These results showed similar levels across the 2-log challenge, as well as instances where the highest or lowest challenge dose resulted in the greatest levels of individual cytokines/chemokines. Previous studies have identified that challenge of epithelial cell cultures with planktonic bacteria, irrespective of the MOI, results in a very disjointed host–bacterial interaction at the individual cell level. Thus, even though these are proposed to be identical cells in culture, microscopic evaluation demonstrated some cells with tens to hundreds of bacteria coating their surface, while the juxtaposed neighboring cells have no bacteria attached to the host cell surface. Thus, population based evaluation of cellular responses can be affected by how the individual bacteria interact with the targeted host cells, as well as the relatively broad capacity of Toll-like receptors to interact with bacterial structures [52]. However, the novel inclusion of monospecies oral bacterial biofilms demonstrated response patterns, some of which at the bacterial species and/or analyte levels were parallel between the biofilms and planktonic bacteria. However, other bacterial-host interactions clearly showed that the biofilms stimulated unique patterns of responses, and in some cases as a biofilm the bacteria were significantly inhibitory to basal mediator production by the epithelial cells. An additional example of these variations was noted with different patterns of responses across the oral streptococcal species that were evaluated. While the oral viridans streptococci have been historically considered similar in their physiology and ecological niches within the commensal microbiota, recent genomic and functional studies have provided clear evidence the not only do these bacteria have some unique metabolic and structural characteristics, but also integrate into complex multispecies biofilms in unique ways [53–56]. Thus, variations in responses of the epithelial cells to these individual species should not be unexpected.
An interesting aspect of these studies was that the majority were conducted under standard aerobic tissue culture conditions. However, the literature supports that at the site of a periodontal disease lesion the ecology reflects an anaerobic microenvironment for the bacteria to interact with the host. A single recent study has been reported examining the variation in cellular response to oral bacterial challenge in aerobic versus anaerobic conditions [57]. This report showed that under reduced oxygen tension (i.e. 2% oxygen) selected oral bacteria, e.g. Tannerella forsythia, P. gingivalis, and Prevotella intermedia elicited elevated levels of IL-8 and TNFα with the highest levels in the low oxygen environment. Thus, with the variations in the subgingival microbial environment, including oxygen tension [58], presence of volatile sulfur compounds [59], and elevated pH [60], results are lacking regarding the impact of these environmental changes on the host–bacterial interactions related to periodontal disease. The model reported in this study will enable us to combine investigations of oral microbial biofilms, both monospecies and multispecies, to directly address the environmental control of host cell responses that could occur in the oral cavity.
While the mediators examined in this study do not cover the extent of response capacity of the oral epithelium, the variation even with two pro-inflammatory cytokines (IL-1α and IL-6) indicates that data is clearly needed to document the characteristics of responses of epithelial cells to the format in which oral bacteria exist in situ in the oral cavity. As importantly, these studies were limited in directly evaluating monospecies biofilms as a basis to understand the capabilities of individual bacteria, recognizing the ultimate need to delineate how these bacteria in complex multispecies biofilms would interact with and stimulate these host cells.
Acknowledgments
We thank Michelle J. Steffen and Jason Stevens for their excellent technical assistance. This work was supported by U.S.P.H.S. grant R21 DE018177 from the National Institute for Dental and Craniofacial Research.
References
- 1.Madianos PN, Bobetsis YA, Kinane DF. Generation of inflammatory stimuli: how bacteria set up inflammatory responses in the gingiva. J Clin Periodontol. 2005;32:57–71. doi: 10.1111/j.1600-051X.2005.00821.x. [DOI] [PubMed] [Google Scholar]
- 2.Bozkurt FY, Yetkin Ay Z, Berker E, Tepe E, Akkus S. Anti-inflammatory cytokines in gingival crevicular fluid in patients with periodontitis and rheumatoid arthritis: a preliminary report. Cytokine. 2006;35:180–5. doi: 10.1016/j.cyto.2006.07.020. [DOI] [PubMed] [Google Scholar]
- 3.Bascones-Martinez A, Munoz-Corcuera M, Noronha S, Mota P, Bascones-Ilundain C, Campo-Trapero J. Host defence mechanisms against bacterial aggression in periodontal disease: basic mechanisms. Med Oral Patol Oral Cir Bucal. 2009:e680–5. doi: 10.4317/medoral.14.e680. [DOI] [PubMed] [Google Scholar]
- 4.Huang C, Altimova Y, Strange S, Ebersole J. Polybacterial challenge effects on cytokine/chemokine production by macrophages and dendritic cells. Inflamm Res. 2011;60:119–25. doi: 10.1007/s00011-010-0242-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kornman KS. Mapping the pathogenesis of periodontitis: a new look. J Periodontol. 2008;79:1560–8. doi: 10.1902/jop.2008.080213. [DOI] [PubMed] [Google Scholar]
- 6.Stathopoulou PG, Benakanakere MR, Galicia JC, Kinane DF. Epithelial cell pro-inflammatory cytokine response differs across dental plaque bacterial species. J Clin Periodontol. 2010;37:24–9. doi: 10.1111/j.1600-051X.2009.01505.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ji S, Kim Y, Min BM, Han SH, Choi Y. Innate immune responses of gingival epithelial cells to nonperiodontopathic and periodontopathic bacteria. J Periodontal Res. 2007;42:503–10. doi: 10.1111/j.1600-0765.2007.00974.x. [DOI] [PubMed] [Google Scholar]
- 8.Hasegawa Y, Mans JJ, Mao S, Lopez MC, Baker HV, Handfield M, et al. Gingival epithelial cell transcriptional responses to commensal and opportunistic oral microbial species. Infect Immun. 2007;75:2540–7. doi: 10.1128/IAI.01957-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mikolajczyk-Pawlinska J, Travis J, Potempa J. Modulation of interleukin-8 activity by gingipains from Porphyromonas gingivalis: implications for pathogenicity of periodontal disease. FEBS Lett. 1998;440:282–6. doi: 10.1016/s0014-5793(98)01461-6. [DOI] [PubMed] [Google Scholar]
- 10.Eskan MA, Benakanakere MR, Rose BG, Zhang P, Zhao J, Stathopoulou P, et al. Interleukin-1{beta} modulates proinflammatory cytokine production in human epithelial cells. Infect Immun. 2008;76:2080–9. doi: 10.1128/IAI.01428-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Asai Y, Hashimoto M, Fletcher HM, Miyake K, Akira S, Ogawa T. Lipopolysaccharide preparation extracted from Porphyromonas gingivalis lipoprotein-deficient mutant shows a marked decrease in toll-like receptor 2-mediated signaling. Infect Immun. 2005;73:2157–63. doi: 10.1128/IAI.73.4.2157-2163.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Krauss JL, Potempa J, Lambris JD, Hajishengallis G. Complementary Tolls in the periodontium: how periodontal bacteria modify complement and Toll-like receptor responses to prevail in the host. Periodontology 2000. 2010;52:141–62. doi: 10.1111/j.1600-0757.2009.00324.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Taylor JJ. Cytokine regulation of immune responses to Porphyromonas gingivalis. Periodontology 2000. 2010;54:160–94. doi: 10.1111/j.1600-0757.2009.00344.x. [DOI] [PubMed] [Google Scholar]
- 14.Ren L, Jiang ZQ, Fu Y, Leung WK, Jin L. The interplay of lipopolysaccharide-binding protein and cytokines in periodontal health and disease. J Clin Periodontol. 2009;36:619–26. doi: 10.1111/j.1600-051X.2009.01436.x. [DOI] [PubMed] [Google Scholar]
- 15.Jin L. An update on innate defense molecules of human gingiva. Periodontology 2000. 2011;56:125–42. doi: 10.1111/j.1600-0757.2010.00364.x. [DOI] [PubMed] [Google Scholar]
- 16.Sbordone L, Bortolaia C. Oral microbial biofilms and plaque-related diseases: microbial communities and their role in the shift from oral health to disease. Clin Oral Investig. 2003;7:181–8. doi: 10.1007/s00784-003-0236-1. [DOI] [PubMed] [Google Scholar]
- 17.Silva N, Dutzan N, Hernandez M, Dezerega A, Rivera O, Aguillon JC, et al. Characterization of progressive periodontal lesions in chronic periodontitis patients: levels of chemokines, cytokines, matrix metalloproteinase-13, periodontal pathogens and inflammatory cells. J Clin Periodontol. 2008;35:206–14. doi: 10.1111/j.1600-051X.2007.01190.x. [DOI] [PubMed] [Google Scholar]
- 18.Talbert J, Elter J, Jared HL, Offenbacher S, Southerland J, Wilder RS. The effect of periodontal therapy on TNF-, IL-6 and metabolic control in type 2 diabetics. J Dent Hyg. 2006;80:7. [PubMed] [Google Scholar]
- 19.Socransky SS, Haffajee AD, Cugini MA, Smith C, Kent RL. Microbial complexes in subgingival plaque. J Clin Periodontol. 1998;25:134–44. doi: 10.1111/j.1600-051x.1998.tb02419.x. [DOI] [PubMed] [Google Scholar]
- 20.Ter Steeg PF, Van Der Hoeven JS. Development of periodontal microflora on human serum. Microb Ecol Health Dis. 1989;2:1–10. [Google Scholar]
- 21.Bradshaw DJ, Homer KA, Marsh PD, Beighton D. Metabolic cooperation in oral microbial communities during growth on mucin. Microbiology. 1994;140:3407–12. doi: 10.1099/13500872-140-12-3407. [DOI] [PubMed] [Google Scholar]
- 22.Kolenbrander PE, Palmer RJ, Periasamy S, Jakubovics NS. Oral multispecies biofilm development and the key role of cell–cell distance. Nat Rev Microbiol. 2010;8:471–80. doi: 10.1038/nrmicro2381. [DOI] [PubMed] [Google Scholar]
- 23.Marsh PD, Devine DA. How is the development of dental biofilms influenced by the host? J Clin Periodontol. 2011;38:28–35. doi: 10.1111/j.1600-051X.2010.01673.x. [DOI] [PubMed] [Google Scholar]
- 24.Guggenheim B, Gmur R, Galicia J, Stathopoulou P, Benakanakere M, Meier A, et al. In vitro modeling of host-parasite interactions: the ‘subgingival’ biofilm challenge of primary human epithelial cells. BMC Microbiol. 2009;9:280. doi: 10.1186/1471-2180-9-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Belibasakis GN, Meier A, Guggenheim B, Bostanci N. Oral biofilm challenge regulates the RANKL–OPG system in periodontal ligament and dental pulp cells. Microb Pathog. 2011;50:6–11. doi: 10.1016/j.micpath.2010.11.002. [DOI] [PubMed] [Google Scholar]
- 26.Peyyala R, Kirakodu SS, Ebersole JL, Novak KF. Characterization of a novel model for multispecies biofilms using rigid gas permeable lenses. Appl Environ Microbiol. 2011;77:3413–21. doi: 10.1128/AEM.00039-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Peyyala R, Kirakodu S, Novak KF, Ebersole JL. Epithelial interleukin-8 responses to oral bacterial biofilms. Clin Vaccine Immunol. 2011;18:1770–2. doi: 10.1128/CVI.05162-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Könönen E. Development of oral bacterial flora in young children. Ann Med. 2000;32:107–12. doi: 10.3109/07853890009011759. [DOI] [PubMed] [Google Scholar]
- 29.Marsh PD. Dental plaque as a microbial biofilm. Caries Res. 2004;38:204–11. doi: 10.1159/000077756. [DOI] [PubMed] [Google Scholar]
- 30.Marsh PD. Microbiology of dental plaque biofilms and their role in oral health and caries. Dent Clin North Am. 2010;54:441–54. doi: 10.1016/j.cden.2010.03.002. [DOI] [PubMed] [Google Scholar]
- 31.Kebschull M, Papapanou PN. Periodontal microbial complexes associated with specific cell and tissue responses. J Clin Periodontol. 2011;38:17–27. doi: 10.1111/j.1600-051X.2010.01668.x. [DOI] [PubMed] [Google Scholar]
- 32.Scheres N, Laine ML, De Vries TJ, Everts V, Van Winkelhoff AJ. Gingival and periodontal ligament fibroblasts differ in their inflammatory response to viable Porphyromonas gingivalis. J Periodontal Res. 2010;45:262–70. doi: 10.1111/j.1600-0765.2009.01229.x. [DOI] [PubMed] [Google Scholar]
- 33.Huang GT-J, Kim D, Lee JK-H, Kuramitsu HK, Haake SK. Interleukin-8 and intercellular adhesion molecule 1 regulation in oral epithelial cells by selected periodontal bacteria: multiple effects of Porphyromonas gingivalis via antagonistic mechanisms. Infect Immun. 2001;69:1364–72. doi: 10.1128/IAI.69.3.1364-1372.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Uehara A, Naito M, Imamura T, Potempa J, Travis J, Nakayama K, et al. Dual regulation of interleukin-8 production in human oral epithelial cells upon stimulation with gingipains from Porphyromonas gingivalis. J Med Microbiol. 2008;57:500–7. doi: 10.1099/jmm.0.47679-0. [DOI] [PubMed] [Google Scholar]
- 35.Sliepen I, Van Damme J, Van Essche M, Loozen G, Quirynen M, Teughels W. Microbial interactions influence inflammatory host cell responses. J Dent Res. 2009;88:1026–30. doi: 10.1177/0022034509347296. [DOI] [PubMed] [Google Scholar]
- 36.Marsh PD, Devine DA. How is the development of dental biofilms influenced by the host? J Clin Periodontol. 2011;38(Suppl 11):28–35. doi: 10.1111/j.1600-051X.2010.01673.x. [DOI] [PubMed] [Google Scholar]
- 37.Kuramitsu HK, Wang BY. The whole is greater than the sum of its parts: dental plaque bacterial interactions can affect the virulence properties of cariogenic Streptococcus mutans. Am J Dent. 2011;24:153–4. [PMC free article] [PubMed] [Google Scholar]
- 38.Kolenbrander PE. Multispecies communities: interspecies interactions influence growth on saliva as sole nutritional source. Int J Oral Sci. 2011;3:49–54. doi: 10.4248/IJOS11025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kebschull M, Papapanou PN. Periodontal microbial complexes associated with specific cell and tissue responses. J Clin Periodontol. 2011;38(Suppl 11):17–27. doi: 10.1111/j.1600-051X.2010.01668.x. [DOI] [PubMed] [Google Scholar]
- 40.Beikler T, Flemmig TF. Oral biofilm-associated diseases: trends and implications for quality of life, systemic health and expenditures. Periodontology 2000. 2011;55:87–103. doi: 10.1111/j.1600-0757.2010.00360.x. [DOI] [PubMed] [Google Scholar]
- 41.Silva TA, Garlet GP, Fukada SY, Silva JS, Cunha FQ. Chemokines in oral inflammatory diseases: apical periodontitis and periodontal disease. J Dent Res. 2007;86:306–19. doi: 10.1177/154405910708600403. [DOI] [PubMed] [Google Scholar]
- 42.Liu Y-CG, Lerner UH, Teng Y-TA. Cytokine responses against periodontal infection: protective and destructive roles. Periodontology 2000. 2010;52:163–206. doi: 10.1111/j.1600-0757.2009.00321.x. [DOI] [PubMed] [Google Scholar]
- 43.Brocker C, Thompson D, Matsumoto A, Nebert DW, Vasiliou V. Evolutionary divergence and functions of the human interleukin (IL) gene family. Hum Genomics. 2010;5:30–55. doi: 10.1186/1479-7364-5-1-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xing Z, Gauldie J, Cox G, Baumann H, Jordana M, Lei XF, et al. IL-6 is an antiinflammatory cytokine required for controlling local or systemic acute inflammatory responses. J Clin Invest. 1998;101:311–20. doi: 10.1172/JCI1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rodero MP, Khosrotehrani K. Skin wound healing modulation by macrophages. Int J Clin Exp Pathol. 2010;3:643–53. [PMC free article] [PubMed] [Google Scholar]
- 46.Gainet J, Chollet-Martin S, Brion M, Hakim J, Gougerot-Pocidalo MA, Elbim C. Interleukin-8 production by polymorphonuclear neutrophils in patients with rapidly progressive periodontitis: an amplifying loop of polymorphonuclear neutrophil activation. Lab Invest. 1998;78:755–62. [PubMed] [Google Scholar]
- 47.Bechara C, Chai H, Lin PH, Yao Q, Chen C. Growth related oncogene-alpha (GRO-alpha): roles in atherosclerosis, angiogenesis and other inflammatory conditions. Med Sci Monit. 2007;13:RA87–90. [PubMed] [Google Scholar]
- 48.Fevang B, Yndestad A, Damås JK, Bjerkeli V, Ueland T, Holm AM, et al. Chemokines and common variable immunodeficiency; possible contribution of the fractalkine system (CX3CL1/CX3CR1) to chronic inflammation. Clin Immunol. 2009;130:151–61. doi: 10.1016/j.clim.2008.09.002. [DOI] [PubMed] [Google Scholar]
- 49.Taub DD, Lloyd AR, Conlon K, Wang JM, Ortaldo JR, Harada A, et al. Recombinant human interferon-inducible protein 10 is a chemoattractant for human monocytes and T lymphocytes and promotes T cell adhesion to endothelial cells. J Exp Med. 1993;177:1809–14. doi: 10.1084/jem.177.6.1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gaga M, Ong YE, Benyahia F, Aizen M, Barkans J, Kay AB. Skin reactivity and local cell recruitment in human atopic and nonatopic subjects by CCL2/MCP-1 and CCL3/MIP-1α. Allergy. 2008;63:703–11. doi: 10.1111/j.1398-9995.2007.01578.x. [DOI] [PubMed] [Google Scholar]
- 51.Cook D, Beck M, Coffman T, Kirby S, Sheridan J, Pragnell I, et al. Requirement of MIP-1 alpha for an inflammatory response to viral infection. Science. 1995;269:1583–5. doi: 10.1126/science.7667639. [DOI] [PubMed] [Google Scholar]
- 52.Brown J, Wang H, Hajishengallis GN, Martin M. TLR-signaling networks: an integration of adaptor molecules, kinases, and cross-talk. J Dent Res. 2011;90:417–27. doi: 10.1177/0022034510381264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Peyyala R, Kirakodu SS, Ebersole JL, Novak KF. Novel model for multispecies biofilms that uses rigid gas-permeable lenses. Appl Environ Microbiol. 2011;77:3413–21. doi: 10.1128/AEM.00039-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Whitmore SE, Lamont RJ. The pathogenic persona of community-associated oral streptococci. Mol Microbiol. 2011;81:305–14. doi: 10.1111/j.1365-2958.2011.07707.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Burton JP, Wescombe PA, Cadieux PA, Tagg JR. Beneficial microbes for the oral cavity: time to harness the oral streptococci? Benef Microbes. 2011;2:93–101. doi: 10.3920/BM2011.0002. [DOI] [PubMed] [Google Scholar]
- 56.Nobbs AH, Lamont RJ, Jenkinson HF. Streptococcus adherence and colonization. Microbiol Mol Biol Rev. 2009;73:407–50. doi: 10.1128/MMBR.00014-09. (Table of Contents) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Grant MM, Kolamunne RT, Lock FE, Matthews JB, Chapple ILC, Griffiths HR. Oxygen tension modulates the cytokine response of oral epithelium to periodontal bacteria. J Clin Periodontol. 2010;37:1039–48. doi: 10.1111/j.1600-051X.2010.01622.x. [DOI] [PubMed] [Google Scholar]
- 58.Mettraux GR, Gusberti FA, Graf H. Oxygen tension (pO2) in untreated human periodontal pockets. J Periodontol. 1984;55:516–21. doi: 10.1902/jop.1984.55.9.516. [DOI] [PubMed] [Google Scholar]
- 59.Torresyap G, Haffajee AD, Uzel NG, Socransky SS. Relationship between periodontal pocket sulfide levels and subgingival species Beziehung zwischen Sulfid-Niveau der parodontalen Tasche und subgingivalen Spezies Relation entre les niveaux de sulfate des poches parodontales et les espèces sousgingivales. J Clin Periodontol. 2003;30:1003–10. doi: 10.1034/j.1600-051x.2003.00377.x. [DOI] [PubMed] [Google Scholar]
- 60.Eggert FM, Drewell L, Bigelow JA, Speck JE, Goldner M. The pH of gingival crevices and periodontal pockets in children, teenagers and adults. Arch Oral Biol. 1991;36:233–8. doi: 10.1016/0003-9969(91)90091-8. [DOI] [PubMed] [Google Scholar]