Abstract
Three O-methyltransferases (BX10a, b, c) catalyze the conversion of 2,4-dihydroxy-7-methoxy-1,4-benzoxazin-3-one glucoside (DIMBOA-Glc) to 2-hydroxy-4,7-dimethoxy-1,4-benzoxazin-3-one glucoside (HDMBOA-Glc) in maize (Zea mays). Variation in benzoxazinoid accumulation and resistance to Rhopalosiphum maidis (corn leaf aphid) was attributed to a natural CACTA family transposon insertion that inactivates Bx10c. Whereas maize inbred line B73 has this transposon insertion, line CML277 does not. To characterize the phenotypic effects of DIMBOA-Glc methyltransferase activity, we created near-isogenic lines derived from B73 and CML277 that do or do not contain the transposon insertion. Bx10c inactivation causes high DIMBOA-Glc, low HDMBOA-Glc, and decreased aphid reproduction relative to near-isogenic lines that have a functional Bx10c gene. These results confirm the importance of this locus in maize aphid resistance. The availability of Bx10c near-isogenic lines will facilitate further research on the function of different benzoxazinoids and DIMBOA-Glc methyltransferase activity in maize defense against herbivores and pathogens.
Keywords: Rhopalosiphum maidis, Benzoxazinoid, HDMBOA, DIMBOA, Zea mays
Maize (Zea mays), one of the world’s most productive crops, is used for food, feed, and biofuel production.1 More than 90 insect species are known to attack maize, resulting in losses of 6% to 19% in overall maize productivity.2 Therefore, extensive research has been conducted to identify factors associated with maize herbivore resistance and susceptibility.3
A recently developed maize nested association mapping (NAM) population was generated by crossing a diverse population of 25 maize inbred lines to the sequenced reference line B73.4-6 This set of ~5000 recombinant inbred lines (RILs) has been used to genetically map numerous maize traits, including resistance to the corn leaf aphid (Rhopalosiphum maidis) and benzoxazinoid accumulation.7 Benzoxazinoids, a class of secondary metabolites found primarily in grasses, including maize, wheat, and rye,8,9 have been demonstrated to inhibit growth of fungi, insect herbivores, and even competing plants.10,11 Nine genes (Bx1-Bx9) catalyze successive steps in the pathway of 2,4-dihydroxy-7-methoxy-1,4-benzoxazin-3-one glucoside (DIMBOA-Glc) from indole-3-glycerol phosphate.10,12 Maize predominantly produces 2 benzoxazinoids, DIMBOA-Glc and 2-hydroxy-4,7-dimethoxy-1,4-benzoxazin-3-one glucoside (HDMBOA-Glc),12,13 with significant variation among different inbred lines.7
In a recent publication,7 we mapped an aphid resistance quantitative trait locus (QTL) using RILs derived from maize inbred lines B73 and CML277, identifying 3 genes (Bx10a, b, c) encoding O-methyltransferases that convert DIMBOA-Glc to HDMBOA-Glc. In comparison to B73, CML277 has constitutively elevated HDMBOA-Glc content. This phenotype was attributed to a natural CACTA family transposon14 insertion that inactivates Bx10c (GRMZM2G023325) in B73 and other maize lines with low constitutive HDMBOA-Glc accumulation. R. maidis progeny production is negatively correlated with DIMBOA-Glc and positively correlated with HDMBOA-Glc production. In the current study, we use near-isogenic lines (NILs) derived from B73 and CML277 to further characterize the effects of Bx10c inactivation on benzoxazinoid accumulation and aphid resistance in maize.
Genotyped RILs of the NAM population are on average 3.6% heterozygous,4 making it possible to find individual lines that are still heterozygous for almost any desired genomic interval. Such heterozygotes can be self-pollinated and genotyped to create heterogeneous inbred families (HIFs).15 Five RILs derived from a B73 × CML277 cross, Z005E0109, Z005E0153, Z005E0174, Z005E0176, and Z005E018, were predicted to be heterozygous in a region containing the Bx10a,b, c genes (Fig. 1). To genotype the Bx10c alleles, genomic DNA was amplified by PCR using the following 3 primers: forward, 5′- AGCACGGCAA CAACCTTGG-3′; reverse for the transposon insertion allele 5′- CGCGGTGGTG AGAACCGTTT -3′; and reverse for the functional gene, 5′-AAGTGCACGT TGCCATCAGA TGGAG -3′, as described previously..6 Heterozygosity at the Bx10c gene could be verified only for Z005E0109 and Z005E0174, and the latter was chosen for further experiments. As Z005E0174 was predicted to be heterozygous for regions of chromosome 1, 2, 3, and 4 (Fig. 1A), it was selfed for 3 generations, prior to selecting sibling lines with the B73 or CML277 alleles, respectively, at the Bx10c locus. Single nucleotide polymorphism (SNP) markers in the other previously heterozygous regions of chromosomes 1–4 (Fig. 1A) were genotyped by PCR amplification and Sanger sequencing (Table 1) to confirm that these regions of the genome are homozygous for the B73 or CML277 alleles in the NILs (illustrated graphically in Figure 1B). In addition to the predicted SNPs from www.panzea.org, our DNA sequencing identified 5 other SNPs in the amplified regions that are polymorphic between B73 and CML322 (Table 1). The CML277 allele of one SNP was found to be different from that which was predicted at www.panzea.org.
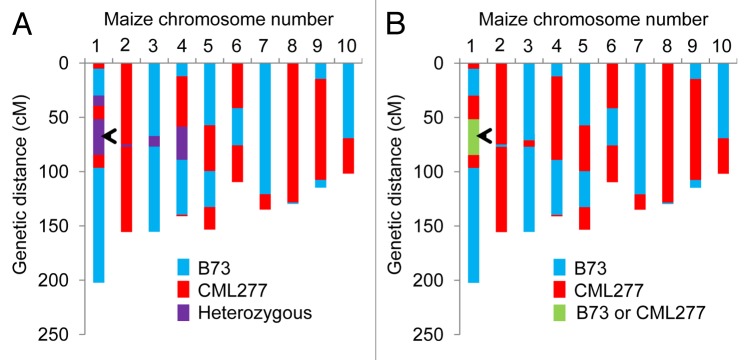
Figure 1. Generation of near-isogenic lines for the Bx10a, b, c locus. (A) Genotype data for Z005E0174, a recombinant inbred line derived from B73 × CML277, are shown (marker data were downloaded from www.panzea.org). (B) Genotype of Bx10a, b, c near-isogenic lines derived from Z005E0174, with the area of the genome that differs between the 2 near-isogenic lines, Bx10cNIL-B73 and Bx10cNIL-CML277, marked in green. The approximate Bx10a, b, c position on chromosome 1 is marked with an arrow.
Table 1. Genotyping SNPs of near-isogenic lines derived from Z005E0174 on chromosome 1 to 4.
| SNP Name1 | SNP location2 | PCR amplification primer sequence (5’ – 3’)3 | B73 | CML277 | Bx10cNIL-B73 | Bx10cNIL-CML277 | |
|---|---|---|---|---|---|---|---|
| PZA01271.1 | Chr1: 16,581,396 | AGCCATCGATAGTCGAAACG | C | T | T | T | |
| TAGGCACGAATTTGCTTGAA | |||||||
| PZA02032.1 | Chr1: 17,746,383 | TCCAAGCGTATGCTCAAGAA | T | C | C | C | |
| GCACAATAGCGAGATGGTCA | |||||||
| Chr2: 93,103,729 | TAATCCTGGCGATGAGATCC | T | C | T | T | ||
| TTTGCCAAGCTCCTCCATAC | |||||||
| Chr3: 156,790,281 | GAACCTCGCATGGTTGAGAT | A | T | A | A | ||
| AGGGCACTTGGTTCCAGATA | |||||||
| PZA00828.2 | Chr3: 158,896,370 | GCTCATCAGGCTTTTGGAAG | T | A | T | T | |
| GCACAAGCCACCCACTATTT | |||||||
| Chr3: 158,896,399 | GCTCATCAGGCTTTTGGAAG | A | T | A | A | ||
| GCACAAGCCACCCACTATTT | |||||||
| PZA01396.1 | Chr3: 166,377,572 | CCACGGTTACAGCACTAGCA | A | G | G | G | |
| GTCCAACCCGAAACTGAAAA | |||||||
| Chr3: 166,377,629 | CCACGGTTACAGCACTAGCA | A | T | T | T | ||
| GTCCAACCCGAAACTGAAAA | |||||||
| PZA03459.1 | Chr4: 135,157,951 | CAACACGCGTTATTGGACAC | C | T 4 | T | T | |
| GCCAGAACTGGACGAGTAGC | |||||||
| PZA02114.1 | Chr4: 155,907,930 | CCAAACCAAGCACCGTCTAT | A | G | G | G | |
| CCAGAAGGAGAGGGAGAACC | |||||||
| Chr4: 155,908,028 | CCAAACCAAGCACCGTCTAT | T | C | C | C | ||
| CCAGAAGGAGAGGGAGAACC | |||||||
Marker names from www.panzea.org, if previously described; 2Base pair position in maize genome assembly (AGP version 2); 3PCR conditions that were applied for all reactions: 95 °C for 3 min followed by 34 cycles of amplification (95 °C for 30 s, 0 °C for 30 s, and 72 °C for 30 s) and 72 °C for 10 min; 4CML277 allele at this locus is different from information provided at www.panzea.org
To measure benzoxazinoid content of the selected NILs, the tip (approximately 5 cm) of the third leaf was collected, extracted, and analyzed by HPLC as described previously,7 with minor modification of the extraction solvent (500 μL methanol/water/formic acid, 30:69.5:0.5, v/v). The levels of DIMBOA-Glc (Fig. 2A) and HDMBOA-Glc (Fig. 2B) were quantified using authentic standards. DIMBOA-Glc was observed in Bx10cNIL-B73, but not in Bx10cNIL-CML277. HDMBOA-Glc was present in both lines, but was significantly less abundant in the Bx10cNIL-B73.This is similar to the benzoxazinoid levels observed in the B73 and CML277 NAM parental lines,7 indicating that the Bx10a, b, c locus is the major determinant of the DIMBO-Glc:HDMBOA-Glc ratio in these maize lines.
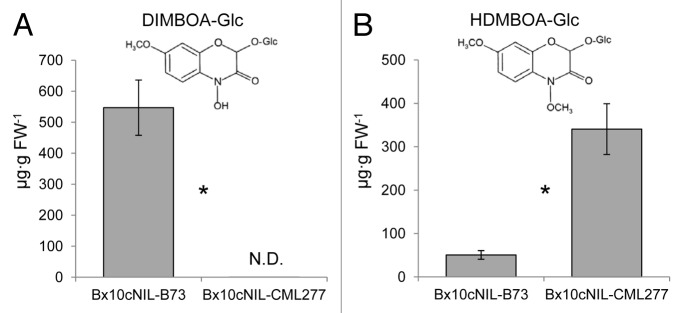
Figure 2. Benzoxazinoid content in Bx10c NILs. Concentration of (A) DIMBOA-Glc and (B) HDMBOA-Glc, as detected by HPLC. Mean ± SE; n = 10–11; *P < 0.05, Wilcoxon rank-sum test; N.D. = not detected; FW = fresh weight.
We examined the effect of the Bx10a, b, c locus on R. maidis progeny production. Plants were used for aphid bioassays at the age of 2 weeks (V2-V3 stage). Ten adult aphids were confined on each of the seedlings using microperforated polypropylene bags (15.25 cm × 61 cm; PJP Marketplace). After one week, Bx10cNIL-CML277 had significantly higher numbers of aphid progeny than Bx10cNIL-B73 (Fig. 3).
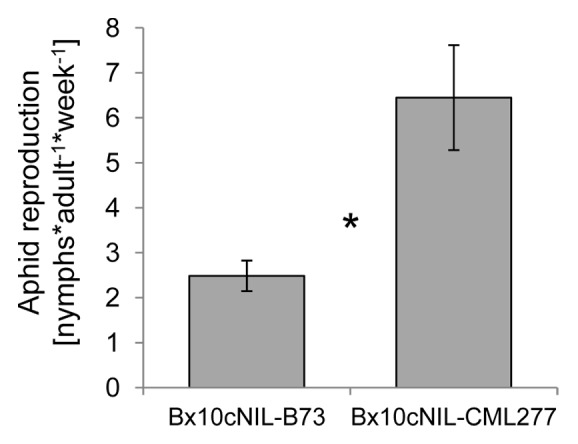
Figure 3. Aphid progeny production on Bx10c NILs. The number of R. maidis progeny produced per adult over one week is shown. Mean ± SE; n = 15, Bx10cNIL-B73; n = 12, Bx10cNIL-CML277; *P < 0.05, Wilcoxon rank-sum test.
As suggested by our prior genetic mapping,7 the current maize NIL experiments demonstrate a large effect of the Bx10a, b, c locus on R. maidis reproduction and benzoxazinoid content. Although it is likely that differences in the methylation of DIMBOA-Glc to make HDMBOA-Glc are caused by the presence or absence of a Bx10c transposon insertion, we cannot rule out the possibility that the phenotype is caused by as yet unknown variation in the function of BX10a and/or BX10b, which also have DIMBOA-Glc methyltransferase activity in vitro.7 Further crosses to select genetic recombination between these 3 genes will be necessary to determine their respective in vivo functions.
The large differences in benzoxazinoid content among inbred lines,7 as well as the herbivory-induced methylation of DIMBOA-Glc to make HDMBOA-Glc,16 suggest divergent functions of these metabolites in maize defense. However, in the absence of mutations specifically affecting DIMBOA-Glc to HDMBOA-Glc conversion, it has been difficult to assess the relative roles of these benzoxazinoids in maize defense. Thus, the NILs described here provide an important new resource for studying the relative importance of constitutive DIMBOA-Glc and HDMBOA-Glc levels in maize defense against a wide variety of herbivores and pathogens in the laboratory or in the field.
Although the relatively low level of inbreeding in the NAM population could be a liability for genetic mapping projects, it facilitates the rapid generation of near-isogenic lines. We have taken advantage of natural variation in the Bx10c locus to create NILs that can be used to study maize-insect interactions. The function of other maize defense-related genes could be studied in a similar manner without the use of targeted mutagenesis or the complications associated with doing field studies with transgenic maize plants.
Acknowledgments
This research was funded by US National Science Foundation Research Experience for Undergraduates grant DBI-1061199 for Mijares V, US National Science Foundation award IOS-1139329 to Jander G, United States Department of Agriculture award 2011–67012–30675 to Meihls LN, and Vaadia-BARD Postdoctoral Fellowship Award FI-471–2012 to Tzin V.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1.Searchinger T, Heimlich R, Houghton RA, Dong F, Elobeid A, Fabiosa J, Tokgoz S, Hayes D, Yu TH. Use of U.S. croplands for biofuels increases greenhouse gases through emissions from land-use change. Science. 2008;319:1238–40. doi: 10.1126/science.1151861. [DOI] [PubMed] [Google Scholar]
- 2.Oerke EC. Crop losses to pests. J Agric Sci. 2006;144:31–43. doi: 10.1017/S0021859605005708. [DOI] [Google Scholar]
- 3.Meihls LN, Kaur H, Jander G. Natural variation in maize defense against insect herbivores. Cold Spring Harb Symp Quant Biol. 2012;77:269–83. doi: 10.1101/sqb.2012.77.014662. [DOI] [PubMed] [Google Scholar]
- 4.McMullen MD, Kresovich S, Villeda HS, Bradbury P, Li H, Sun Q, Flint-Garcia S, Thornsberry J, Acharya C, Bottoms C, et al. Genetic properties of the maize nested association mapping population. Science. 2009;325:737–40. doi: 10.1126/science.1174320. [DOI] [PubMed] [Google Scholar]
- 5.Yu J, Holland JB, McMullen MD, Buckler ES. Genetic design and statistical power of nested association mapping in maize. Genetics. 2008;178:539–51. doi: 10.1534/genetics.107.074245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Flint-Garcia SA, Thuillet AC, Yu J, Pressoir G, Romero SM, Mitchell SE, Doebley J, Kresovich S, Goodman MM, Buckler ES. Maize association population: a high-resolution platform for quantitative trait locus dissection. Plant J. 2005;44:1054–64. doi: 10.1111/j.1365-313X.2005.02591.x. [DOI] [PubMed] [Google Scholar]
- 7.Meihls LN, Handrick V, Glauser G, Barbier H, Kaur H, Haribal MM, Lipka AE, Gershenzon J, Buckler ES, Erb M, et al. Natural variation in maize aphid resistance is associated with DIMBOA-Glc methyltransferase activity. Plant Cell. 2013 doi: 10.1105/tpc.113.112409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zúñiga GE, Argandoña VH, Niemeyer HM, Corcuera LJ. Hydroxamic acid content in wild and cultivated Gramineae. Phytochemistry. 1983;22:2665–8. doi: 10.1016/S0031-9422(00)97669-6. [DOI] [Google Scholar]
- 9.Jonczyk R, Schmidt H, Osterrieder A, Fiesselmann A, Schullehner K, Haslbeck M, Sicker D, Hofmann D, Yalpani N, Simmons C, et al. Elucidation of the final reactions of DIMBOA-glucoside biosynthesis in maize: characterization of Bx6 and Bx7. Plant Physiol. 2008;146:1053–63. doi: 10.1104/pp.107.111237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Niemeyer HM. Hydroxamic acids derived from 2-hydroxy-2H-1,4-benzoxazin-3(4H)-one: key defense chemicals of cereals. J Agric Food Chem. 2009;57:1677–96. doi: 10.1021/jf8034034. [DOI] [PubMed] [Google Scholar]
- 11.Ahmad S, Veyrat N, Gordon-Weeks R, Zhang YH, Martin J, Smart L, Glauser G, Erb M, Flors V, Frey M, et al. Benzoxazinoid metabolites regulate innate immunity against aphids and fungi in maize. Plant Physiol. 2011;157:317–27. doi: 10.1104/pp.111.180224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Frey M, Schullehner K, Dick R, Fiesselmann A, Gierl A. Benzoxazinoid biosynthesis, a model for evolution of secondary metabolic pathways in plants. Phytochemistry. 2009;70:1645–51. doi: 10.1016/j.phytochem.2009.05.012. [DOI] [PubMed] [Google Scholar]
- 13.Frey M, Chomet P, Glawischnig E, Stettner C, Grün S, Winklmair A, Eisenreich W, Bacher A, Meeley RB, Briggs SP, et al. Analysis of a chemical plant defense mechanism in grasses. Science. 1997;277:696–9. doi: 10.1126/science.277.5326.696. [DOI] [PubMed] [Google Scholar]
- 14.Bercury SD, Panavas T, Irenze K, Walker EL. Molecular analysis of the Doppia transposable element of maize. Plant Mol Biol. 2001;47:341–51. doi: 10.1023/A:1011606529513. [DOI] [PubMed] [Google Scholar]
- 15.Tuinstra MR, Ejeta G, Goldsbrough PB. Heterogeneous inbred family (HIF) analysis: a method for developing near-isogenic lines that differ at quantitative trait loci. Theor Appl Genet. 1997;95:1005–11. doi: 10.1007/s001220050654. [DOI] [Google Scholar]
- 16.Oikawa A, Ishihara A, Tanaka C, Mori N, Tsuda M, Iwamura H. Accumulation of HDMBOA-Glc is induced by biotic stresses prior to the release of MBOA in maize leaves. Phytochemistry. 2004;65:2995–3001. doi: 10.1016/j.phytochem.2004.09.006. [DOI] [PubMed] [Google Scholar]


