Abstract
We recently showed that—in Arabidopsis thaliana suspension cells—phosphoinositide dependent-phospholipase C (PI-PLC) and diacylglycerol kinase (DGK) negatively regulated the basal expression of most DREB2 genes. DREB2 genes encode transcription factors that bind to Drought Responsive Elements (DRE). Those elements are also bound by DREB1 factors. While DREB2 factors are mostly involved in drought and heat responses, DREB1s are induced in the response to chilling. We here show that the pharmacological inhibition of PI-PLC or DGK leads to the basal induction of DREB1 genes. However, the induction is much less marked for the DREB1 genes than that of DREB2A, a member of the DREB2 family. This illustrates that DREB1 and DREB2 genes, while having the same targets, are not submitted to the same transcription regulation, and that lipid signaling might in part explain these differences in the regulation of the DREB genes.
Keywords: Phospholipase C, Diacylglycerol kinase, U73122, R59022, DREB1/CBF, DREB2, Arabidopsis
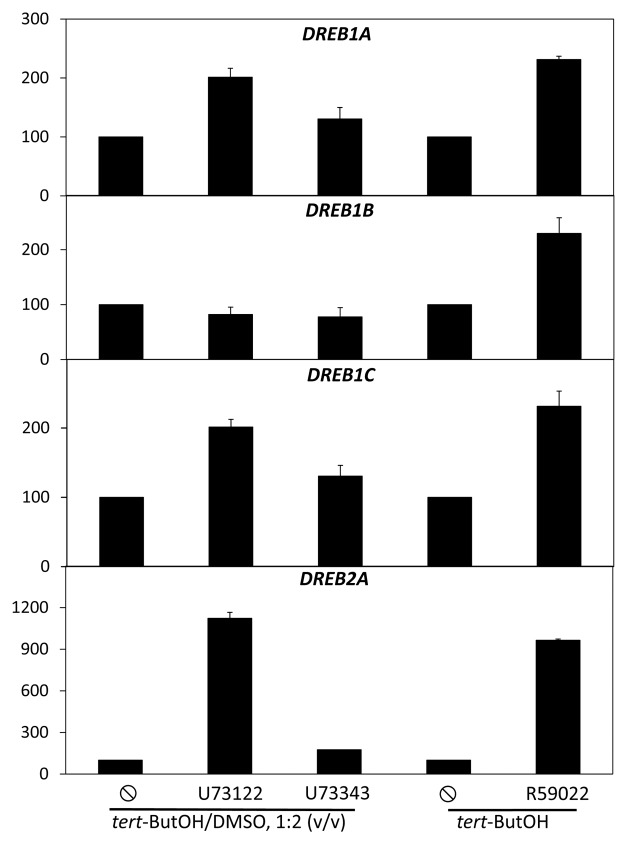
Plants are always submitted to changes in their environment. They can be submitted to abiotic stresses, such as drought, exposure to heat, or to low temperatures, changes of light intensity or quality. They can also be submitted to biotic stresses. Because plants are sessile organisms, they need, in order to survive, to be able to cope with these stresses. A major process of the resistance of plants to stresses is transcriptome remodelling. This occurs through the early induction of transcription factors. DREB2A and DREB2B transcription factors are highly induced by drought, NaCl, or heat, while poor induction is seen in response to cold or abscissic acid.1,2 The encoded protein binds to drought responsive elements (DRE)/C-repeat (CRT) on the promoters of genes, thus inducting their transcription. Interestingly, the same DRE/CRT elements can be bound by DREB1 proteins.3 These factors, also named CRT-binding factors (CBF), are induced by cold, but not by drought.4 DREB1 and DREB2 proteins share identity in the DNA binding domain, but are very little identical in the rest of the protein.5 Because the DREB1 and DREB2 proteins are key regulators of the response to major abiotic stress, it is of high importance to understand how they are regulated. The regulation of DREB gene expression occurs in response to a stress, but also in basal conditions. Among the signaling pathways active in control conditions are the ones that generate or consume bioactive phosphoglycerolipids. Phosphoinositide-dependent phospholipase C (PI-PLC) hydrolyses phosphatidylinositol-4,5-bisphosphate into inositol triphosphate and diacylglycerol. This lipid can be phosphorylated into phosphatidic acid by diacylglycerol-kinases (DGK).6 The PI-PLC/DGK pathway is active in non-stimulated Arabidopsis cells or plants.7,8 We recently showed that this basal activity negatively regulated the expression of DREB2A, DREB2B, DREB2C, DREB2E, and DREB2H genes. Indeed, when inhibiting the activity of PI-PLC by edelfosine or U73122, or when inhibiting the activity of DGK by R59022 or R59949, the expression of these genes was upregulated in Arabidopsis suspension cells.7 We wanted to know if this was also the case for the DREB1 genes. Cells were treated by U73122 or its inactive analog U73343. Cells were also separately inhibited with R59022. Four hours later, cells were harvested and transcripts isolated. The level of DREB1A-C were quantified by real-time PCR and compared with that of DREB2A. Inhibiting basal PI-PLC or DGK led to the induction of DREB1 genes. However, the induction is much less marked than that of DREB2A. U73122 led to a 10-fold increase of DREB2A expression when compared with the effect of U7343; on the contrary, U73122 had no effect on the expression of DREB1B, and only slight inducing effects on that of DREB1A or DREB1C. R59022 led to a 10-fold increase of DREB2A expression, while it led only to a 2.3-fold increase of that of DREB1A-C genes (Fig. 1). This illustrates that DREB1 and DREB2 genes are not submitted to the same transcription regulation. This is true in response to stresses, since they are not induced by the same stresses. But it also true in basal conditions. The way those genes are regulated is still poorly understood. It was shown that ABRE-BINDING PROTEIN 1, ABRE-BINDING PROTEIN 2, and ABRE-BINDING FACTOR 3 transcription factors can bind to and activate the DREB2A promoter in an ABRE-dependent manner. Concerning the DREB1 genes, ICE1—that encodes a MYC-like transcriptional activator— binds specifically to the MYC recognition sequences in the DREB1A promoter. ICE1 overexpression in wild-type plants enhances the expression of the CBF regulon in the cold.9 The CALMODULIN BINDING TRANSCRIPTION ACTIVATOR 3 is a positive regulator of DREB1C expression.10 However, to what extent those transcription factors and cis-elements are important for the control of basal expression is not known. A challenging task would be now to identify which part of the DREB2A promoter is responsible for the regulation by basal PI-PLC/DGK activity. Finally, it has to be reminded that not all DREB2 genes are dependent on the PI-PLC/DGK for their basal regulation. DREB2D expression is not stimulated by the inhibitors of these enzymes, while DREB2F and DREB2G transcripts could never be detected in any of the conditions tested.7
Figure 1. Effects of inhibitors of PI-PLC or DGK on the expression levels of DREB2A and DREB1 genes. U73122 and U73343 were used at final concentration 60 µM. R59022 was used at final concentration 100 µM. Transcript levels were quantified by qPCR, and expressed as % of solvent-treated cells. Cells were treated with inhibitors for 4 h, at 22°C, before harvesting. tert-ButOH is for tertiary butanol.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Glossary
Abbreviations:
- C-repeat
CRT
- CRT-binding factors
CBF
- diacylglycerol-kinase
DGK
- drought responsive elements
DRE
- Phosphoinositide-dependent phospholipase C
PI-PLC
References
- 1.Nakashima K, Shinwari ZK, Sakuma Y, Seki M, Miura S, Shinozaki K, Yamaguchi-Shinozaki K. Organization and expression of two Arabidopsis DREB2 genes encoding DRE-binding proteins involved in dehydration- and high-salinity-responsive gene expression. Plant Mol Biol. 2000;42:657–65. doi: 10.1023/A:1006321900483. [DOI] [PubMed] [Google Scholar]
- 2.Sakuma Y, Maruyama K, Qin F, Osakabe Y, Shinozaki K, Yamaguchi-Shinozaki K. Dual function of an Arabidopsis transcription factor DREB2A in water-stress-responsive and heat-stress-responsive gene expression. Proc Natl Acad Sci U S A. 2006;103:18822–7. doi: 10.1073/pnas.0605639103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sakuma Y, Liu Q, Dubouzet JG, Abe H, Shinozaki K, Yamaguchi-Shinozaki K. DNA-binding specificity of the ERF/AP2 domain of Arabidopsis DREBs, transcription factors involved in dehydration- and cold-inducible gene expression. Biochem Biophys Res Commun. 2002;290:998–1009. doi: 10.1006/bbrc.2001.6299. [DOI] [PubMed] [Google Scholar]
- 4.Zarka DG, Vogel JT, Cook D, Thomashow MF. Cold induction of Arabidopsis CBF genes involves multiple ICE (inducer of CBF expression) promoter elements and a cold-regulatory circuit that is desensitized by low temperature. Plant Physiol. 2003;133:910–8. doi: 10.1104/pp.103.027169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ruelland E, Vaultier M, Zachowski A, Hurry V. Cold Signalling and Cold Acclimation in Plants. In: Advances in Botanical Research. Elsevier: 2009, 35-150. [Google Scholar]
- 6.Pokotylo I, Kolesnikov Y, Kravets V, Zachowski A, Ruelland E. Plant phosphoinositide-dependent phospholipases C: Variations around a canonical theme. Biochimie. 2014 doi: 10.1016/j.biochi.2013.07.004. In press. [DOI] [PubMed] [Google Scholar]
- 7.Djafi N, Vergnolle C, Cantrel C, Wietrzyñski W, Delage E, Cochet F, Puyaubert J, Soubigou-Taconnat L, Gey D, Collin S, et al. The Arabidopsis DREB2 genetic pathway is constitutively repressed by basal phosphoinositide-dependent phospholipase C coupled to diacylglycerol kinase. Front Plant Sci. 2013;4:307. doi: 10.3389/fpls.2013.00307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Perera IY, Hung C-Y, Moore CD, Stevenson-Paulik J, Boss WF. Transgenic Arabidopsis plants expressing the type 1 inositol 5-phosphatase exhibit increased drought tolerance and altered abscisic acid signaling. Plant Cell. 2008;20:2876–93. doi: 10.1105/tpc.108.061374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chinnusamy V, Ohta M, Kanrar S, Lee B-H, Hong X, Agarwal M, Zhu J-K. ICE1: a regulator of cold-induced transcriptome and freezing tolerance in Arabidopsis. Genes Dev. 2003;17:1043–54. doi: 10.1101/gad.1077503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Doherty CJ, Van Buskirk HA, Myers SJ, Thomashow MF. Roles for Arabidopsis CAMTA transcription factors in cold-regulated gene expression and freezing tolerance. Plant Cell. 2009;21:972–84. doi: 10.1105/tpc.108.063958. [DOI] [PMC free article] [PubMed] [Google Scholar]