Abstract
Regulated proteolysis by the ubiquitin/26S proteasome system (UPS) has emerged as a major posttranslational control mechanism regulating transcription factor (TF) activity in plants. Anthocyanin biosynthesis in Arabidopsis is regulated by a ternary complex comprised of basic helix-loop-helix (bHLH), R2R3MYB and WD-repeat (WDR) proteins. The bHLH TF, TRANSPARENT TESTA 8 (TT8), and the WDR protein, TRANSPARENT TESTA GLABRA1 (TTG1), are essential for expression of late flavonoid biosynthesis genes. Previous studies have demonstrated that the turnover of several anthocyanin pathway regulators is controlled by the UPS. Here, we showthatTT8 and TTG1areshort-lived and targeted by the UPS for degradation. Our findings further extend our understanding of the role of the UPS in the regulation of anthocyanin biosynthesis in plants.
Keywords: anthocyanins, flavonoids, 26S proteasome, TT8, TTG1
The anthocyanin biosynthesis pathway is one of the most extensively studied metabolic pathways in plants. Biosynthesis and accumulation of anthocyanins is dependent upon several factors including light, phytohormones and nutrients.1-3 Upon perception of developmental or environmental signals, regulatory proteins begin accumulating in the cell, and subsequently, trigger the expression of structural genes leading to biosynthesis of anthocyanins. In plants studied to date, a ternary transcription factor (TF) complex, comprised of basic helix-loop-helix (bHLH), R2R3MYBs and WD-repeat (WDR) proteins, regulates the transcription of structural genes in the pathway.4,5 Recent studies suggest that, in addition to transcriptional control, the anthocyanin pathway is also regulated by posttranscriptional and posttranslational control mechanisms.6-10 To maintain cellular homeostasis, cells control the transcriptional machinery either through the expression of negative regulators, small RNAs, or by selectively degrading the proteins involved in metabolic pathways. In Arabidopsis, the single repeat MYBs, CAPRICE (CPC) and MYBL2, act as negative regulators of the anthocyanin biosynthesis pathway.11-13They compete with the R2R3MYBs, PAP1/PAP2, to interact with bHLH factors to form an inactive/repressor complex thereby affecting the transcription of structural genes. The small RNAs have also been shown to play a critical role in the anthocyanin pathway by regulating expression of the regulators such asPAP1.6,8
Both TFs and enzymatic proteins have an intrinsic half-life depending on their function and cellular requirement. Metabolic enzymes are generally more stable than TFs and the rapid turnover of TFs is considered to be a part of the regulatory mechanism.14 The ubiquitin/26S proteasome system (UPS) has emerged as the most prevalent posttranslational control mechanism dictating the activities of TFs in plants. One hypothesis for the rapid turnover of regulatory proteins is that timely degradation of the “spent” regulatory proteins allows promoter clearance for new rounds of transcription.15 In Arabidopsis, loss of 26S proteasome function results in higher accumulation of anthocyanin pigments in vegetative tissues indicating the involvement of UPS in anthocyanin regulation.16,17 This hypothesis is strengthened by recent reports demonstrating that the activities of TFs involved in the regulation of anthocyanin biosynthesis in plants are regulated by UPS. The R2R3MYBs, PAP1 and PAP2in Arabidopsis, and MdMYB1 in apple, are degraded in the dark by the UPS suggesting that the light-mediated anthocyanin accumulation in Arabidopsis and apple skin is due, at least in part, to the stabilization of these factors in light.7,9 We have recently shown that the bHLH factors, GL3 and EGL3 that control anthocyanin biosynthesis and epidermal cell differentiation in Arabidopsis, are short-lived and targeted by the UPS for degradation.10
In Arabidopsis, the bHLH TF TT8 serves as a component of the regulatory complex that positively regulates the expression of most of the late biosynthesis genes in the flavonoid pathway leading to the accumulation of anthocyanins and proanthocyanidins.18 The WD-repeat protein TTG1 works coordinately with the bHLH and R2R3MYB TFs to regulate flavonoid biosynthesis and trichome development in Arabidopsis.19 Here, we demonstrate that TT8 and TTG1 are also short-lived and are targeted by UPS for proteolytic degradation.
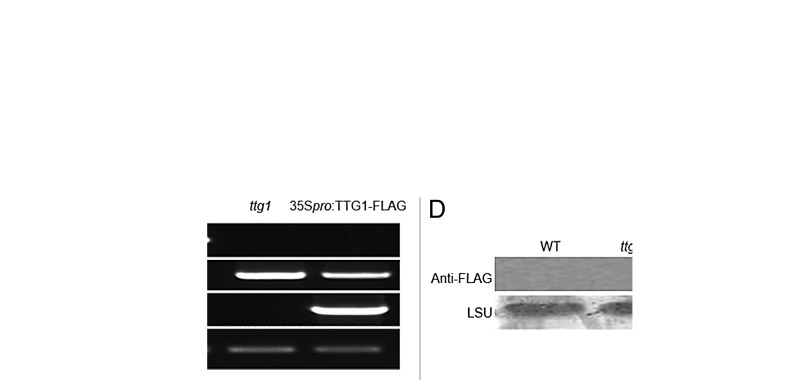
To test the stability of TT8 and TTG1 we ectopically expressed both TFs in stable transgenic lines in their respective homozygous tt8-1 (SALK_030966) and ttg1 (Salk_104152) mutant backgrounds. Homozygosity of the mutants was confirmed by PCR (Fig.1A and C). Modified pCAMBIA1300 vector carrying full-length cDNA of TT8 or TTG1 were used for plant transformation. For efficient immunodetection, both TFs were C-terminally fused to a 3×FLAG tag. Homozygous transgenic plants were verified for accumulation of the fusion proteins by Western blot (Fig.1B and D). Ectopic expression of TT8 or TTG1did not show any adverse effects on the overall growth and development of homozygous transgenic lines.
Figure 1. Genotyping and Western blot analysis of homozygous tt8-1 and ttg1 plants expressing FLAG- tagged TT8 and TTG1. (A) DNA was isolated from 2-week-old plants. Homozygousity was confirmed by PCR. TT8-FLAG transcripts were PCR amplified from total RNA isolated from 2-week-old seedlings using TT8-F and FLAG-R primers. Actin 2 was used as a control. (B) Total protein was isolated from 2-week-old seedlings and the presence of TT8-FLAG fusion protein was checked by Western blot using Anti-FLAG M2 antibody. Ponceau S-stained membrane with LSU is shown as a loading control. (C) and (D) Genotyping, RT-PCR, and Western blot analysis were performed for the plants expressing TTG1-FLAG as in (A) and (B).
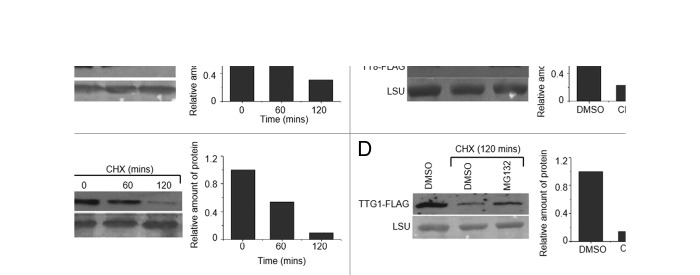
We then independently investigated whether TT8 and TTG1 are targeted for degraded by UPS. Two-week-old seedlings of 35Spro:TT8-FLAG or 35Spro:TTG1-FLAG transgenic plants were treated with 200 µM cycloheximide (CHX) in half-strength MS medium at room temperature for different time intervals and the depletion rates of TT8-FLAG and TTG1-FLAG proteins were monitored by Western blot analysis using anti-FLAG–M2 monoclonal antibody (Sigma). The amount ofTT8 or TTG1 fusion protein was depleted significantly after 120 minutes of incubation of the seedlings in CHX (Fig.2A and C). The transcript levels of TT8-FLAG/ TTG1-FLAG remained unchanged after CHX treatment for 60 and 120 minutes, indicating that the depletion of the TT8 and TTG1 is posttranslational and the TF proteins are unstable and degraded in the plant cell (Fig.3A and C). Next, in order to test whether degradation of TT8 and TTG1 is dependent on 26S proteasome, 2-week-old seedlings of the transgenic lines were treated with either CHX alone or in combination with MG132, an inhibitor of proteasome activity, for 120 minutes. DMSO-only treated plants served as control. Degradation of both TT8 and TTG1 proteins were inhibited significantly by MG132 (Fig. 2B and D). Similar to CHX treatment, the combined CHX and MG132 treatment did not affect the transgene transcriptional level (Fig. 3B and D). Taken together, our results demonstrate that, in Arabidopsis, TT8 and TTG1 proteins are short-lived and targeted for proteasomal degradation.
Figure 2. (A) Stability assay for TT8. Two-week-old tt8-1seedlings expressing a 35Spro:TT8-FLAG (TT8-FLAGox) transgene were treated with 200 μM cycloheximide (CHX) for the indicated time period and used for Western blot analyses with anti-FLAG antibodies. Ponceau S-stained membrane with LSU is shown as a loading control. The relative amount of protein in each sample is normalized against the corresponding LSU. (B) Stabilization of TT8by MG132. Two-week-old tt8-1 seedlings expressing a 35Spro:TT8-FLAG were treated with CHX, alone or in combination with MG132 (200 µM), for 120 minutes and used for Western blot analyses with anti-FLAG antibodies. DMSO-only treated plants served as control. The relative amount of protein in each sample is normalized against the corresponding LSU. (C) and (D) TTG1stability and MG132 stabilization assays were performed using two week-old ttg1 plants expressing a 35Spro:TTG1-FLAG (TTG1-FLAGox) as in (A) and (B).
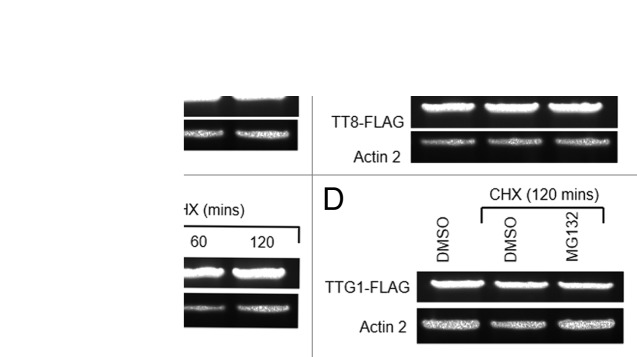
Figure 3. TT8-FLAG transcript levels in the seedlings, either untreated or treated with 200µM CHX, were examined using RT-PCR. Actin 2 was used as a control. TT8-FLAG transcript levels in the seedlings, treated with either DMSO or 200µM CHX (± 200µM MG132) for 120 minutes, were detected using RT-PCR. Actin 2 was used as a control. (A) and (D) TTG1 transcript levels were detected in 2 week-old ttg1 plants expressing35Spro:TTG1-FLAG (TTG1-FLAGox) as in (A) and (B).
Transcriptional regulation is considered to be the major mechanism dictating metabolic pathway gene expression in plants. However, recent studies on the regulation of flavonoid biosynthesis pathways suggest that posttranscriptional and posttranslational mechanisms also play significant roles in the regulation of metabolic pathways.6-10 The UPS has been demonstrated to regulate the activities of key regulatory proteins, including GL3, EGL3, PAP1 and PAP2, in the Arabidopsis flavonoid pathway.9,10 Here we show that two additional TFs, TT8 and TTG1, in the pathway are also targeted for proteasomal degradation, suggesting that the activities of most, if not all, TFs in the ternary complex that regulate the flavonoid pathway in Arabidopsis are regulated by UPS. UPS mediated degradation is a multistep process that involves several enzymatic reactions. E3 ligases interact with target proteins to facilitate their degradation through UPS. Proteasomal degradation of PAP1 and PAP2 is mediated by the E3 ligase, COP1/SPA.9 We recently demonstrated that the HECT domain-containing E3 ligase, Ubiquitin Protein Ligase 3 (UPL3), interacts with GL3 and EGL3 and mediates their degradation through the UPS.10 The E3 ligase protein that mediates the degradation of TT8 and TTG1 has yet to be identified. Collectively, these findings highlight the importance of UPS in flavonoid pathway regulation in plants.
Materials and Methods
Plant materials and growth conditions
All mutant and transgenic lines were in Arabidopsis thaliana ecotype Col‑0. The T‑DNA insertion mutant lines, tt8-1 (SALK_030966) and ttg1 (Salk_104152), were obtained from The Arabidopsis Biological Resource Center (ABRC). For germination, seeds were sterilized using 75% ethanol and 3% commercial bleach, and plated on half-strength MS medium (Caisson Labs). Seeds were stratified for 2 days, and plants were grown in a controlled environment chamber (16-h-light at 22‑24 °C and 8-h-dark at 17‑19 ºC). DNA was isolated from 2-week-old plants. Homozygousity was confirmed by PCR using gene specific LP and RP primers, as well as by using LBb1.3 and gene specific RP primers.
To generate TT8 and TTG1 overexpression lines, full-length cDNAs were cloned into a modified pCAMBIA1300 vector containing the CaMV35S promoter and rbcS terminator. Three tandem repeats encoding the FLAG epitope (3×GACTACAAAG ACGATGACGA CAAA) were fused in‑frame to the C-terminal end of TT8 and TTG1 cDNA. The resulting constructs, 35Spro:TT8-FLAG or 35Spro:TTG1-FLAG were introduced into Agrobacterium tumefaciens strain GV3850 and used for transformation of tt8-1 and ttg1 plants by the floral dip method.20 To check for the TT8-FLAG or TTG1-FLAG transcripts, total RNA was isolated from 2-week-old seedlings and used for first strand cDNA synthesis. TT8-FLAG or TTG1-FLAG transcripts were PCR amplified using forward primerTT8-F (5′-ATGGATGAAT CAAGTATTAT TCC-3′) or TTG1-F (5′-ATGGATAATT CAGCTCCAGA TT-3′) in combination with reverse primer FLAG-R (5′-TTTGTCGTCA TCGCTTTTGT AGTC-3′). Actin 2 was amplified using Actin 2 F (5′-AACCCAAAGG CCAACAGAGA-3′) and Actin 2 R (5′- AAGGTCACGT CCAGCAAGGT-3′) primers and used as experimental control.
Treatments
For protein stability and proteasome inhibition assays, 2-week-old seedlings were incubated with either 200 µM cycloheximide (CHX) alone or in combination with 200 µM MG132. The CHX and MG132 stocks were prepared in water and DMSO, respectively, and all control experiments contained an equal amount of solvent.
Immunoblotting analysis
Total protein extracts were prepared as described previously,21 separated by SDS-PAGE, and transferred to nitrocellulose membranes (Bio-Rad). Membranes were stained with Ponceau S to ensure equal loading, blocked with 3% BSA, probed with the anti-FLAG antibody (clone M2, Sigma) followed by the HRP-conjugated secondary antibody (Thermo Scientific). Signal was detected using the Super Signal West Pico Chemiluminescent Substrate (Thermo Scientific). The amount of proteins present relative to the untreated or only solvent treated plants were determined densitometrically using ImageJ (http://rsb.info.nih.gov/ij/) software.
Acknowledgement
This work is supported by a grant from the Kentucky Tobacco Research and Development Center (to LY). We thank KA Shen for critical comments of the manuscript.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1.Loreti E, Povero G, Novi G, Solfanelli C, Alpi A, Perata P. Gibberellins, jasmonate and abscisic acid modulate the sucrose-induced expression of anthocyanin biosynthetic genes in Arabidopsis. New Phytol. 2008;179:1004–16. doi: 10.1111/j.1469-8137.2008.02511.x. [DOI] [PubMed] [Google Scholar]
- 2.Cominelli E, Gusmaroli G, Allegra D, Galbiati M, Wade HK, Jenkins GI, et al. Expression analysis of anthocyanin regulatory genes in response to different light qualities in Arabidopsis thaliana. J Plant Physiol. 2008;165:886–94. doi: 10.1016/j.jplph.2007.06.010. [DOI] [PubMed] [Google Scholar]
- 3.Lillo C, Lea US, Ruoff P. Nutrient depletion as a key factor for manipulating gene expression and product formation in different branches of the flavonoid pathway. Plant Cell Environ. 2008;31:587–601. doi: 10.1111/j.1365-3040.2007.01748.x. [DOI] [PubMed] [Google Scholar]
- 4.Broun P. Transcriptional control of flavonoid biosynthesis: a complex network of conserved regulators involved in multiple aspects of differentiation in Arabidopsis. Curr Opin Plant Biol. 2005;8:272–9. doi: 10.1016/j.pbi.2005.03.006. [DOI] [PubMed] [Google Scholar]
- 5.Ramsay NA, Glover BJ. MYB-bHLH-WD40 protein complex and the evolution of cellular diversity. Trends Plant Sci. 2005;10:63–70. doi: 10.1016/j.tplants.2004.12.011. [DOI] [PubMed] [Google Scholar]
- 6.Hsieh LC, Lin SI, Shih AC, Chen JW, Lin WY, Tseng CY, et al. Uncovering small RNA-mediated responses to phosphate deficiency in Arabidopsis by deep sequencing. Plant Physiol. 2009;151:2120–32. doi: 10.1104/pp.109.147280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li YY, Mao K, Zhao C, Zhao XY, Zhang HL, Shu HR, et al. MdCOP1 ubiquitin E3 ligases interact with MdMYB1 to regulate light-induced anthocyanin biosynthesis and red fruit coloration in apple. Plant Physiol. 2012;160:1011–22. doi: 10.1104/pp.112.199703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Luo QJ, Mittal A, Jia F, Rock CD. An autoregulatory feedback loop involving PAP1 and TAS4 in response to sugars in Arabidopsis. Plant Mol Biol. 2012;80:117–29. doi: 10.1007/s11103-011-9778-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maier A, Schrader A, Kokkelink L, Falke C, Welter B, Iniesto E, et al. Light and the E3 ubiquitin ligase COP1/SPA control the protein stability of the MYB transcription factors PAP1 and PAP2 involved in anthocyanin accumulation in Arabidopsis. Plant J. 2013;74:638–51. doi: 10.1111/tpj.12153. [DOI] [PubMed] [Google Scholar]
- 10.Patra B, Pattanaik S, Yuan L. Ubiquitin protein ligase 3 mediates the proteasomal degradation of GLABROUS 3 and ENHANCER OF GLABROUS 3, regulators of trichome development and flavonoid biosynthesis in Arabidopsis. Plant J. 2013;74:435–47. doi: 10.1111/tpj.12132. [DOI] [PubMed] [Google Scholar]
- 11.Matsui K, Umemura Y, Ohme-Takagi M. AtMYBL2, a protein with a single MYB domain, acts as a negative regulator of anthocyanin biosynthesis in Arabidopsis. Plant J. 2008;55:954–67. doi: 10.1111/j.1365-313X.2008.03565.x. [DOI] [PubMed] [Google Scholar]
- 12.Dubos C, Le Gourrierec J, Baudry A, Huep G, Lanet E, Debeaujon I, et al. MYBL2 is a new regulator of flavonoid biosynthesis in Arabidopsis thaliana. Plant J. 2008;55:940–53. doi: 10.1111/j.1365-313X.2008.03564.x. [DOI] [PubMed] [Google Scholar]
- 13.Zhu HF, Fitzsimmons K, Khandelwal A, Kranz RG. CPC, a single-repeat R3 MYB, is a negative regulator of anthocyanin biosynthesis in Arabidopsis. Mol Plant. 2009;2:790–802. doi: 10.1093/mp/ssp030. [DOI] [PubMed] [Google Scholar]
- 14.Henriques R, Jang IC, Chua NH. Regulated proteolysis in light-related signaling pathways. Curr Opin Plant Biol. 2009;12:49–56. doi: 10.1016/j.pbi.2008.10.009. [DOI] [PubMed] [Google Scholar]
- 15.Lipford JR, Smith GT, Chi Y, Deshaies RJ. A putative stimulatory role for activator turnover in gene expression. Nature. 2005;438:113–6. doi: 10.1038/nature04098. [DOI] [PubMed] [Google Scholar]
- 16.Fatimababy AS, Lin YL, Usharani R, Radjacommare R, Wang HT, Tsai HL, et al. Cross-species divergence of the major recognition pathways of ubiquitylated substrates for ubiquitin/26S proteasome-mediated proteolysis. FEBS J. 2010;277:796–816. doi: 10.1111/j.1742-4658.2009.07531.x. [DOI] [PubMed] [Google Scholar]
- 17.Smalle J, Kurepa J, Yang P, Babiychuk E, Kushnir S, Durski A, et al. Cytokinin growth responses in Arabidopsis involve the 26S proteasome subunit RPN12. Plant Cell. 2002;14:17–32. doi: 10.1105/tpc.010381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nesi N, Debeaujon I, Jond C, Pelletier G, Caboche M, Lepiniec L. The TT8 gene encodes a basic helix-loop-helix domain protein required for expression of DFR and BAN genes in Arabidopsis siliques. Plant Cell. 2000;12:1863–78. doi: 10.1105/tpc.12.10.1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Walker AR, Davison PA, Bolognesi-Winfield AC, James CM, Srinivasan N, Blundell TL, et al. The TRANSPARENT TESTA GLABRA1 locus, which regulates trichome differentiation and anthocyanin biosynthesis in Arabidopsis, encodes a WD40 repeat protein. Plant Cell. 1999;11:1337–50. doi: 10.1105/tpc.11.7.1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–43. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- 21.Kurepa J, Smalle JA. Assaying transcription factor stability. Methods Mol Biol. 2011;754:219–34. doi: 10.1007/978-1-61779-154-3_12. [DOI] [PubMed] [Google Scholar]