Abstract
MYC2 is an important transcription factor, which modulates transcription by directly binding to Z-, G- and E-box elements present in the promoters of light and different stress responsive genes.1–3 Very recently, we have shown that MYC2 plays a role in the regulation of Z- and/or G-box containing promoters during both seedling and adult plant growth.4 Although, MYC2 does not bind to the GATA box light responsive element (LRE) in vitro as shown in DNA binding assays1, its involvement in the regualtion of GATA-box containing promoter in planta, if any, is not known. Here, we report that the promoter activity of GATA-NOS101 in atmyc2 mutant was found to be similar to wild-type in BL and dark grown seedlings, whereas it was found to be lower compared with wild-type as revealed from GUS staining results. Further, we will discuss the consequences of MYC2 regulating GATA-box containing promoter in combination with G-box containing promoters.
Keywords: Arabidopsis, light responsive elements, MYC2, GATA-box, transcription factors
The spatio-temporal expression of genes are determined by the cis-acting elements present in the particular promoter, which confer responsiveness to particular type of stimuli such as biotic, abiotic, hormonal or nutritional signals. Light is one of the very important abiotic factor that regulates gene expression primarily at the transcription level.5 Light is known to affect the expression of nearly one-third of the total genes present in Arabidopsis. Analysis of promoters of light inducible genes, such as RBCS, CAB and CHS have identified a number of cis-acting light responsive elements (LREs) such as G-, GATA-, GT1- ,and Z-box that are essential for the light mediated gene expression.6 From the literature it is understood that cis-acting elements differ greatly from one promoter to another.7 Furthermore, it was suggested that combinatorial interplay of more than one cis-acting elements are required to confer proper light responsiveness of light-regulated promoters in plants.8-12
GATA-box (core motif: ATGATAAGG) has been reported to present in many LHC and RBCS genes in a number of different species.13 In plants, GATA LRE motifs have been implicated in light-dependent and nitrate-dependent control of transcription.6,14 Indeed, presence of core GATA motif in the set of relative modules in the promoters of many light regulated genes has been reported. Also, GATA motifs are known to often associated with other LREs including G-boxes.7 For example, the combination of G-box with GATA element has been shown to be critical for the promoter activation in response to signals from multiple photoreceptors as well as for repression by the COP/DET system.10 Mutation in either G- or GATA-box affects the expression of RBCS-1A promoter, suggesting both the LREs are necessary for the proper expression.8
Most of the light signaling transcription factors identified so far bind to the G- and Z-box LREs.1,5,15,16, Although the trans-acting factors, which bind to GATA-box LRE have been reported, only few have been shown to play role in light signaling. GATA-box binding transcription factors such as GATA2, GATA4, GATA21 (GNC) and GATA22 are some of the key players, which are thought to play role in light signaling. GATA2 and GATA4 show higher expression levels in dark grown seedlings, and their expression goes down in seedlings treated with different wavelengths of light, but not UV-A. Unlike, GATA2 and GATA4, GATA21 (GNC) and GATA22 show strong upregulation in light grown seedlings.17 GATA2 has been reported to cross-talk between light and brassinosteriod signaling pathways, and acts as a positive regulator of photomorphogenesis.18 GATA2 directly regulates both light and BR responsive genes, whereas BZR1 has been shown to directly repress GATA2 transcription in a BR dependent manner. Additionally, COP1 has been shown to degrade GATA2 protein in dark, whereas light stabilizes the GATA2 protein, likely by inhibiting a COP1-dependent degradation process.18 Recently, 2 GATA transcription factors, GNC and CGA1 have been shown to be involved in chloroplast development and hypocotyl growth in (Arabidopsis thaliana). Both GNC and CGA1 are highly expressed in green tissues in a cytokinin dependent manner.19
MYC2 Differentially Regulates the Expression of GATA-Box Containing Promoters
MYC2 has been identified as a Z-box binding specific transcription factor, and reported to regulate Z-box containing promoters.1,4 It has been shown through DNA-binding assays that MYC2 does not bind to GATA box LRE. However, it is not known whether MYC2 is involved in the regulation of GATA box containing promoter in planta. To address this question, we introduced GATA/NOS101-GUS promoter construct into atmyc2 mutant background from wild-type (WT) through genetic cross, and then homozygous mutant transgenic lines were generated and used for further analysis. Seedlings grown in constant dark and blue light (BL) for 6-days were subjected to GUS staining analysis, and the results suggest that the expression of GATA/NOS101-GUS was found to be at the similar levels in wild type and atmyc2 mutants (Fig. 1), suggesting that MYC2 does not regulate the expression of GATA/NOS101 promoter in both dark and BL conditions. However, examination of 6-days-old red light (RL) and far-red light (FL) grown seedlings suggest that GATA/NOS101 promoter activity was reduced in atmyc2 (more strikingly in the hypocotyl region) mutant than wild-type, suggesting that MYC2 is positively influencing the expression of GATA/NOS101 promoter in RL and FR (Fig. 1). The fact that MYC2 does not bind to GATA-box LRE indicates that the regulation of GATA/NOS101 by MYC2 in RL and FL is likely to be indirect. Although the exact mechanism of GATA/NOS101 regulation by MYC2 is not known, it is possible that MYC2 might be modulating the expression of some of the GATA binding factors. Similar to MYC2, HY5 has been shown to bind and regulate G-box containing promoters, however does not regulate GATA/NOS101-GUS promoter in constant dark and light grown seedlings.11
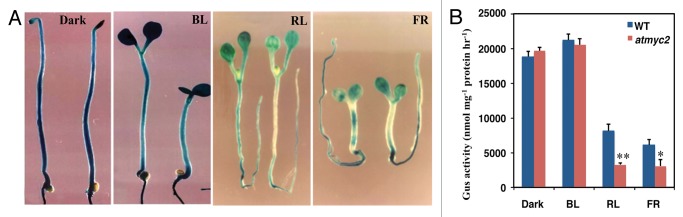
Figure 1. MYC2/ZBF1 differentially regulates the expression of GATA-box containing promoter. In each panel of A, WT and atmyc2/zbf1 mutant seedlings carrying the transgene are shown on the left and right, respectively. Six-day-old WT and atmyc2 seedlings were stained for the similar length of time. (A) GUS staining patterns of 4GATA/NOS1O1-GUS transgenic seedlings grown under constant dark, BL (30 µmol m−2 s−1), RL (90 µmol m−2 s−1), and FR (60 µmol m−2 s−1). (B) GUS activities of 6-d-old wild-type and atmyc2/zbf1 seedlings carrying 4G/NOS101-GUS transgene grown in constant dark, BL (30 µmol m−2 s−1), RL (90 µmol m−2 s−1), and FR (60 µmol m−2 s−1). Error bars represent SD (n = 3). *P ≤ 0.05 and ** P ≤ 0.01 for values significantly differ from WT in respective light conditions. The experiment was performed at least twice with similar results.
MYC2 has been shown to bind to Z- and G-box, and negatively regulates the expression of Z/NOS101 and 4G/NOS101 promoter irrespective of wavelength of light and in dark during seedling development.1,4 However, when G- or Z-box LRE is fused to GATA LRE, the resulting combinatorial promoters (G-GATA/NOS101, Z-GATA/NOS) behaves differently with respect to tissue specific expression and light responsiveness.11,12 Whereas G/NOS101, Z/NOS101 and GATA/NOS101 promoters expresses in all most all parts of the seedlings, the promoter activity in G-GATA/NOS101 and Z-GATA/NOS101 is only limited to tip of the cotyledons.11,12 Although, MYC2 does not seem to regulate GATA/NOS101 in both dark and BL, it negatively regulates the G-GATA/NOS101 promoter activity.4 HY5 does not regulate the GATA/NOS101 promoter in light and dark grown seedlings, whereas it positively regulates the expression of G-GATA/NOS101 and Z-GATA/NOS101 promoters in a light dependent manner.11,12 Thereby, it could be envisioned that interplay of at least 2 transcription factors is necessary for the proper regulation of G-GATA and Z-GATA promoters, this could be in combination with either G- or Z-box binding factors (MYC2/ZBF1, GBF2/ZBF2, HY5, PIFs [Phytochrome Interacting Factor]) or an unknown GATA-box binding factor. However, we can rule out the possibility of GATA factors such as GATA4 and GATA22, as GATA22 expression has been shown to be strongly correlated with the expression of some of the light signaling genes such as HY5 and HYH, suggesting that possible role of GATA22 in light signaling.17 Furthermore, the co-expression of GATA4 with light signaling transcription factors such as PIL5/PIF1, PIF3, SPT, and HFR1 leads to the speculation that GATA4 has overlapping functions with light signaling transcription factors.17
Light signaling molecules such as FHY3/FAR1, which contain 2 domains that are found in GATA factors, have been shown to functionally interact with HY5. HY5 has been reported to act antagonistically to FHY3 and suppresses FHY1/FHL expression by interfering with FHY3/FAR1 binding to the FHY1/FHL promoters,20 whereas it co-operatively function with FHY3 for the regulation of COP1 gene expression in a UV-B dependent manner.21 In spite of many examples leading to the speculation that GATA box binding transcription factors could be the potential functional partners of G- and/or Z-box box transcription factors, no such evidence is available at this point of time. Hence, future research addressing the combinatorial interaction of cis-acting elements by discrete LRE binding transcription factors will give us some insight into the combinatorial regulation of light regulated promoters.
Acknowledgments
This work is financially supported by Department of Science and Technology (JC Bose National Fellowship Grant), Government of India to SC.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1.Yadav V, Mallappa C, Gangappa SN, Bhatia S, Chattopadhyay S. A basic helix-loop-helix transcription factor in Arabidopsis, MYC2, acts as a repressor of blue light-mediated photomorphogenic growth. Plant Cell. 2005;17:1953–66. doi: 10.1105/tpc.105.032060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gangappa SN, Prasad VB, Chattopadhyay S. Functional interconnection of MYC2 and SPA1 in the photomorphogenic seedling development of Arabidopsis. Plant Physiol. 2010;154:1210–9. doi: 10.1104/pp.110.163717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kazan K, Manners JM. MYC2: the master in action. Mol Plant. 2013;6:686–703. doi: 10.1093/mp/sss128. [DOI] [PubMed] [Google Scholar]
- 4.Gangappa SN, Maurya JP, Yadav V, Chattopadhyay S. The regulation of the Z- and G-box containing promoters by light signaling components, SPA1 and MYC2, in Arabidopsis. PLoS One. 2013;8:e62194. doi: 10.1371/journal.pone.0062194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jiao Y, Lau OS, Deng XW. Light-regulated transcriptional networks in higher plants. Nat Rev Genet. 2007;8:217–30. doi: 10.1038/nrg2049. [DOI] [PubMed] [Google Scholar]
- 6.Terzaghi WB, Cashmore AR. Light-regulated transcription. Annu Rev Plant Physiol Plant Mol Biol. 1995;46:445–74. doi: 10.1146/annurev.pp.46.060195.002305. [DOI] [Google Scholar]
- 7.Arguello-Astorga G, Herrera-Estrella L. Evolution of light-regulated plant promoters. Annu Rev Plant Physiol Plant Mol Biol. 1998;49:525–55. doi: 10.1146/annurev.arplant.49.1.525. [DOI] [PubMed] [Google Scholar]
- 8.Donald RGK, Cashmore AR. Mutation of either G box or I box sequences profoundly affects expression from the Arabidopsis rbcS-1A promoter. EMBO J. 1990;9:1717–26. doi: 10.1002/j.1460-2075.1990.tb08295.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Puente P, Wei N, Deng XW. Combinatorial interplay of promoter elements constitutes the minimal determinants for light and developmental control of gene expression in Arabidopsis. EMBO J. 1996;15:3732–43. [PMC free article] [PubMed] [Google Scholar]
- 10.Chattopadhyay S, Puente P, Deng XW, Wei N. Combinatorial interaction of light-responsive elements plays a critical role in determining the response characteristics of light-regulated promoters in Arabidopsis. Plant J. 1998;15:69–77. doi: 10.1046/j.1365-313X.1998.00180.x. a. [DOI] [PubMed] [Google Scholar]
- 11.Chattopadhyay S, Ang LH, Puente P, Deng XW, Wei N. Arabidopsis bZIP protein HY5 directly interacts with light-responsive promoters in mediating light control of gene expression. Plant Cell. 1998;10:673–83. doi: 10.1105/tpc.10.5.673. b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yadav V, Kundu S, Chattopadhyay D, Negi P, Wei N, Deng XW, et al. Light regulated modulation of Z-box containing promoters by photoreceptors and downstream regulatory components, COP1 and HY5, in Arabidopsis. Plant J. 2002;31:741–53. doi: 10.1046/j.1365-313X.2002.01395.x. [DOI] [PubMed] [Google Scholar]
- 13.Grob U, Stüber K. Discrimination of phytochrome dependent light inducible from non-light inducible plant genes. Prediction of a common light-responsive element (LRE) in phytochrome dependent light inducible plant genes. Nucleic Acids Res. 1987;15:9957–73. doi: 10.1093/nar/15.23.9957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reyes JC, Muro-Pastor MI, Florencio FJ. The GATA family of transcription factors in Arabidopsis and rice. Plant Physiol. 2004;134:1718–32. doi: 10.1104/pp.103.037788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mallappa C, Yadav V, Negi P, Chattopadhyay S. A bzip transcription factor, GBF1, regulates blue light mediated photomor- phogenic growth in Arabidopsis. J Biol Chem. 2006;281:22190–9. doi: 10.1074/jbc.M601172200. [DOI] [PubMed] [Google Scholar]
- 16.Kushwaha R, Singh A, Chattopadhyay S. Calmodulin7 plays an important role as transcriptional regulator in Arabidopsis seedling development. Plant Cell. 2008;20:1747–59. doi: 10.1105/tpc.107.057612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Manfield IW, Devlin PF, Jen CH, Westhead DR, Gilmartin PM. Conservation, convergence, and divergence of light-responsive, circadian-regulated, and tissue-specific expression patterns during evolution of the Arabidopsis GATA gene family. Plant Physiol. 2006;143:941–58. doi: 10.1104/pp.106.090761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Luo XM, Lin WH, Zhu S, Zhu JY, Sun Y, Fan XY, et al. Integration of light- and brassinosteroid-signaling pathways by a GATA transcription factor in Arabidopsis. Dev Cell. 2010;19:872–83. doi: 10.1016/j.devcel.2010.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chiang YH, Zubo YO, Tapken W, Kim HJ, Lavanway AM, Howard L, et al. Functional characterization of the GATA transcription factors GNC and CGA1 reveals their key role in chloroplast development, growth, and division in Arabidopsis. Plant Physiol. 2012;160:332–48. doi: 10.1104/pp.112.198705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li J, Li G, Gao S, Martinez C, He G, Zhou Z, et al. Arabidopsis transcription factor ELONGATED HYPOCOTYL5 plays a role in the feedback regulation of phytochrome A signaling. Plant Cell. 2010;22:3634–49. doi: 10.1105/tpc.110.075788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang X, Ouyang X, Yang P, Lau OS, Li G, Li J, et al. Arabidopsis FHY3 and HY5 positively mediate induction of COP1 transcription in response to photomorphogenic UV-B light. Plant Cell. 2012;24:4590–606. doi: 10.1105/tpc.112.103994. [DOI] [PMC free article] [PubMed] [Google Scholar]


