Abstract
Aux/IAA genes play a pivotal role in auxin transcriptional regulation. Their functions were mainly studied in Arabidopsis through analysis of gain-of-function mutants. In the tomato, the Solanaceae reference species, different studies on Sl-IAA down-regulated lines showed specific role for Sl-IAA genes. Our recent work revealed that the Sl-IAA27 gene displays a distinct behavior compared with most Aux/IAA genes, being down-regulated by auxin. Interestingly, the silencing of Sl-IAA27 leads to altered chlorophyll accumulation in leaves, reduced fertilization, altered fruit development and altered root formation. Here we report that IAA27 could be a key auxin signaling gene involved in AM in tomato and also in Medicago model plant. Indeed both Sl-IAA27 and its closest homolog in Medicago truncatula, Mt-IAA27, are overexpressed in mycorrhized roots. These data are in line with the putative role of auxin in arbuscular mycorrhization.
Keywords: auxin, Aux/IAA genes, root, tomato, arbuscular mycorrhizal fungus
The plant hormone auxin has an essential role in plant growth and development processes. Auxin is involved in the regulation of cell growth affecting both cell division and cell elongation and also in specific differentiation events such as embryogenesis, root development, vascular differentiation, apical dominance, gravitropic and phototropic response, fruit set, and fruit development.1,2 The auxin signaling pathway is modulated by the interaction of 3 main multigenes family coding for the TIR1/AFB receptors, the Aux/IAA repressors and the auxin response factor (ARF) transcription factors. The binding of auxin to the TIR1/AFB nuclear receptors leads to the degradation of the Aux/IAA proteins via the ubiquitin-proteasome pathway and allows the release of the ARF activators that could activate or repress their target auxin responsive genes.3-6
In the tomato, a reference species for Solanaceae plants, 25 Aux/IAA genes were identified and the understanding of their function in planta was achieved for 4 of them through the characterization of down-regulated plants.7-14 Our recent publication aims with the functional characterization of a specific tomato Aux/IAA gene, the Sl-IAA27 gene.8 Indeed, the Sl-IAA27 protein harbors the 4 typical Aux/IAA domains but we also identified another motif in the protein that is conserved among putative orthologous IAA27 proteins in monocot and dicot species. Moreover, the Sl-IAA27 auxin expression is atypical being downregulated by auxin treatment whereas the expression of Aux/IAA genes is generally overexpressed by auxin. Interestingly, our functional analysis of Sl-IAA27 by reverse genetic revealed an implication of this gene in various plant developmental processes. First, the level of chlorophyll content was reduced in leaves of Sl-IAA27 downregulated lines and was correlated with downregulation of many genes involved in chlorophyll synthesis. Second, the reproductive development of the Sl-IAA27 RNAi lines was altered at different levels. Indeed, the size of the Sl-IAA27 RNAi fruits was smaller with an enlarged placenta and the fertility of both ovule and pollen were dramatically reduced compared with wild-type plants. Moreover, the transgenic lines displayed an alteration of root development with an increased primary root length and more lateral roots. Interestingly, Sl-IAA27 overexpressing plants displayed a reduced primary root growth and no lateral root formation at the opposite of Sl-IAA27 RNAi lines.8
Previous studies have shown that AM symbiosis positively affects tomato plant productivity and tomato phenology. Indeed, mycorrhization accelerates flowering and fruit development, increases fruit yield and enhances the nutritional and nutraceutical value of the fruit.15-17 Following inoculation with AM fungi, a significant increase in the level of free auxin was observed in different species.18 However, even though such an increase in auxin levels has not been reported yet in tomato, analysis of mycorrhization in 2 auxin tomato mutants revealed that auxin signaling is required for normal fungal infection.19 Partial transcriptomic analysis using the TOM2 microarray showed that several genes involved in auxin metabolism, such as GH3-like protein, are differentially expressed in mycorrhized tomato plants compared with the controls.20 All these data suggest a role for auxin during the process of tomato mycorrhization. Since our previous work indicated that modulation of Sl-IAA27 expression impacts root development, and considering that AM symbiosis greatly impacts root development, we investigated the potential effect of mycorrhization on Sl-IAA27 expression. Transcript accumulation of Sl-IAA27 assessed by qRT-PCR revealed a slight but significant upregulation of Sl-IAA27 in tomato mycorrhized roots with a level of expression 1.7 fold higher than in uninoculated roots (Fig. 1A). To uncover whether this induction of Sl-IAA27 expression is a common feature during AM symbiosis, we also assessed the expression of the putative ortholog of Sl-IAA27 in Medicago truncatula, a plant species widely used as a model for the study of the mycorrhization process. We performed TBLASTN analysis on the M. truncatula genome using Sl-IAA27 protein sequence (http://www.plantgdb.org/MtGDB/). This in silico search identified Medtr2g122570.1, here named Mt-IAA27, as the closest homolog Sl-IAA27 in M. truncatula. The Mt-IAA27 protein displays 67% of amino acid identity with Sl-IAA27 and presents the specific domain (YxGLS) of tomato Aux/IAA clade B comprising Sl-IAA27 (Fig. 2).8 Interestingly, Mt-IAA27 transcript accumulation is significantly induced in mycorrhized roots with a level more than 5-fold higher than in uninoculated roots (Fig. 1B). The upregulation of IAA27 expression in mycorrhized roots uncovered in this study in 2 different plant species, strongly suggests a role of this gene during AM symbiosis. These data add putative new functions to Aux/IAA members and open novel opportunities to study auxin signaling during AM symbiosis. Future work will focus on the response of the IAA27 gene to the Myc-LCOs and COs and its role during AM symbiosis.21,22
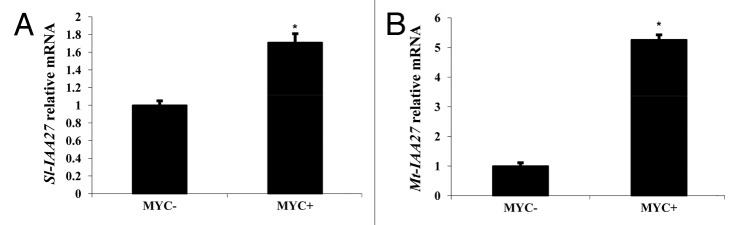
Figure 1. Accumulation of Sl-IAA27 (A) and Mt-IAA27 (B) transcripts in uninoculated roots (MYC−) and in mycorrhized roots (MYC+) of 12-week-old tomato plants and 8-week-old Medicago truncatula plants, respectively. qRT-PCR analyses were performed according to Lauressergues et al.,23 with n = 6, error bars represent SEM. Stars indicate significant differences when compared with corresponding control, according to the Kruskal–Wallis test (P< 0.05).
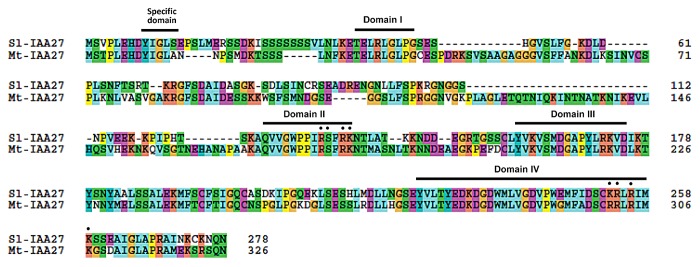
Figure 2. Sequence alignment of Sl-IAA27 protein and its closest homolog in Medicago truncatula, Mt-IAA27 obtained with ClustalX and manual correction. Amino acid residues, part of nuclear localization signal, are indicated by stars. The amino acid position is given on the right of each sequence.
Acknowledgments
This work was supported by the Laboratoire d’Excellence (LABEX, TULIP ANR-10-LABX-41), the French government (MESR grant for CB) and the Iranian plant research company, Zistfanafarin Co (grant for ME).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1.Gillaspy G, Ben-David H, Gruissem W. Fruits: A Developmental Perspective. Plant Cell. 1993;5:1439–51. doi: 10.1105/tpc.5.10.1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vanneste S, Friml J. Auxin: a trigger for change in plant development. Cell. 2009;136:1005–16. doi: 10.1016/j.cell.2009.03.001. [DOI] [PubMed] [Google Scholar]
- 3.Chapman EJ, Estelle M. Mechanism of auxin-regulated gene expression in plants. Annu Rev Genet. 2009;43:265–85. doi: 10.1146/annurev-genet-102108-134148. [DOI] [PubMed] [Google Scholar]
- 4.Kepinski S, Leyser O. The Arabidopsis F-box protein TIR1 is an auxin receptor. Nature. 2005;435:446–51. doi: 10.1038/nature03542. [DOI] [PubMed] [Google Scholar]
- 5.Dharmasiri N, Dharmasiri S, Estelle M. The F-box protein TIR1 is an auxin receptor. Nature. 2005;435:441–5. doi: 10.1038/nature03543. [DOI] [PubMed] [Google Scholar]
- 6.Tan X, Calderon-Villalobos LI, Sharon M, Zheng C, Robinson CV, Estelle M, et al. Mechanism of auxin perception by the TIR1 ubiquitin ligase. Nature. 2007;446:640–5. doi: 10.1038/nature05731. [DOI] [PubMed] [Google Scholar]
- 7.Audran-Delalande C, Bassa C, Mila I, Regad F, Zouine M, Bouzayen M. Genome-wide identification, functional analysis and expression profiling of the Aux/IAA gene family in tomato. Plant Cell Physiol. 2012;53:659–72. doi: 10.1093/pcp/pcs022. [DOI] [PubMed] [Google Scholar]
- 8.Bassa C, Mila I, Bouzayen M, Audran-Delalande C. Phenotypes associated with down-regulation of Sl-IAA27 support functional diversity among Aux/IAA family members in tomato. Plant Cell Physiol. 2012;53:1583–95. doi: 10.1093/pcp/pcs101. [DOI] [PubMed] [Google Scholar]
- 9.Chaabouni S, Jones B, Delalande C, Wang H, Li Z, Mila I, et al. Sl-IAA3, a tomato Aux/IAA at the crossroads of auxin and ethylene signalling involved in differential growth. J Exp Bot. 2009;60:1349–62. doi: 10.1093/jxb/erp009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Salma C, Alain L, Claude PJ, Mondher B. Tomato Aux/IAA3 and HOOKLESS are important actors of the interplay between auxin and ethylene during apical hook formation. Plant Signal Behav. 2009;4:559–60. doi: 10.4161/psb.4.6.8748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deng W, Yan F, Liu M, Wang X, Li Z. Down-regulation of SlIAA15 in tomato altered stem xylem development and production of volatile compounds in leaf exudates. Plant Signal Behav. 2012;7:911–3. doi: 10.4161/psb.20723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deng W, Yang Y, Ren Z, Audran-Delalande C, Mila I, Wang X, et al. The tomato SlIAA15 is involved in trichome formation and axillary shoot development. New Phytol. 2012;194:379–90. doi: 10.1111/j.1469-8137.2012.04053.x. [DOI] [PubMed] [Google Scholar]
- 13.Wang H, Jones B, Li Z, Frasse P, Delalande C, Regad F, et al. The tomato Aux/IAA transcription factor IAA9 is involved in fruit development and leaf morphogenesis. Plant Cell. 2005;17:2676–92. doi: 10.1105/tpc.105.033415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang H, Schauer N, Usadel B, Frasse P, Zouine M, Hernould M, et al. Regulatory features underlying pollination-dependent and -independent tomato fruit set revealed by transcript and primary metabolite profiling. Plant Cell. 2009;21:1428–52. doi: 10.1105/tpc.108.060830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Giovannetti M, Avio L, Barale R, Ceccarelli N, Cristofani R, Iezzi A, et al. Nutraceutical value and safety of tomato fruits produced by mycorrhizal plants. Br J Nutr. 2012;107:242–51. doi: 10.1017/S000711451100290X. [DOI] [PubMed] [Google Scholar]
- 16.Salvioli A, Zouari I, Chalot M, Bonfante P. The arbuscular mycorrhizal status has an impact on the transcriptome profile and amino acid composition of tomato fruit. BMC Plant Biol. 2012;12:44. doi: 10.1186/1471-2229-12-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Subramanian KS, Santhanakrishnan P, Balasubramanian P. Responses of field grown tomato plants to arbuscular mycorrhizal fungal colonization under varying intensities of drought stress. Sci Hortic (Amsterdam) 2005;107:245–53. doi: 10.1016/j.scienta.2005.07.006. [DOI] [Google Scholar]
- 18.Ludwig-Müller J, Güther M. Auxins as signals in arbuscular mycorrhiza formation. Plant Signal Behav. 2007;2:194–6. doi: 10.4161/psb.2.3.4152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hanlon MT, Coenen C. Genetic evidence for auxin involvement in arbuscular mycorrhiza initiation. New Phytol. 2011;189:701–9. doi: 10.1111/j.1469-8137.2010.03567.x. [DOI] [PubMed] [Google Scholar]
- 20.Fiorilli V, Catoni M, Miozzi L, Novero M, Accotto GP, Lanfranco L. Global and cell-type gene expression profiles in tomato plants colonized by an arbuscular mycorrhizal fungus. New Phytol. 2009;184:975–87. doi: 10.1111/j.1469-8137.2009.03031.x. [DOI] [PubMed] [Google Scholar]
- 21.Genre A, Chabaud M, Balzergue C, Puech-Pagès V, Novero M, Rey T, et al. Short-chain chitin oligomers from arbuscular mycorrhizal fungi trigger nuclear Ca2+ spiking in Medicago truncatula roots and their production is enhanced by strigolactone. New Phytol. 2013;198:190–202. doi: 10.1111/nph.12146. [DOI] [PubMed] [Google Scholar]
- 22.Maillet F, Poinsot V, André O, Puech-Pagès V, Haouy A, Gueunier M, et al. Fungal lipochitooligosaccharide symbiotic signals in arbuscular mycorrhiza. Nature. 2011;469:58–63. doi: 10.1038/nature09622. [DOI] [PubMed] [Google Scholar]
- 23.Lauressergues D, et al. The microRNA miR171h modulates arbuscular mycorrhizal colonization of Medicago truncatula by targeting NSP2. Plant J. Cell Mol Biol. 2012;72:512–22. doi: 10.1111/j.1365-313X.2012.05099.x. [DOI] [PubMed] [Google Scholar]


