Abstract
Protein ubiquitination and deubiquitination are two reversible processes catalyzed by ubiquitin ligases and deubiquitinating enzymes, respectively. In Arabidopsis, lots of substrates of ubiquitin ligases were found, whereas only a few targets of deubiquitinating enzymes were identified. Recently, we reported that a functional UBIQUITIN-SPECIFIC PROTEASE16 (UBP16) was involved in salt tolerance through positively regulating plasma membrane Na+/H+ antiport activity and at least partially modulating SERINE HYDROXYMETHYLTRANSFERASE1 (SHM1) stability and activity. Here, we report that UBP16 interacts with HEAVY METAL ASSOCIATED ISOPRENYLATED PLANT PROTEIN27 (HIPP27), a metallochaperone containing a predicted heavy-metal-associated domain, which has been reported to play an important role in cadmium detoxification. Meanwhile, the ubp16 mutant showed more sensitive to cadmium than wild-type. Taken together, HIPP27 may be another target of UBP16 in cadmium response.
Keywords: UBIQUITIN-SPECIFIC PROTEASE, HEAVY METAL ASSOCIATED ISOPRENYLATED PLANT PROTEIN, cadmium detoxification, ubiquitination, deubiquitination
Modification of protein by ubiquitin was widely distributed in eukaryotes, which was sequentially catalyzed by E1 ubiquitin-activating enzyme, E2 ubiquitin-conjugating enzyme and E3 ubiquitin-protein ligase.1,2 In Arabidopsis, more than 1 400 ubiquitin ligases were identified, which target specific substrates.3 Deubiquitinating enzymes (DUBs) are proteases that reverse the modification of proteins by ubiquitin. DUBs are divided into two general groups, ubiquitin C-terminal hydrolases (UCHs) and ubiquitin-specific proteases (UBPs) based on their amino acid sequence and substrate specificity.4,5 Estimated in a recent phylogenetic analysis, there are at least 64 UCHs and 27 UBPs in Arabidosis.6 The number of DUBs is significantly less than that of ubiquitin ligase, suggesting that one DUB may target different substrates under variant conditions.
Recently, though systemic analysis of UBPs family members responsive to salt tolerance, we found the ubp16 mutant was more sensitive to salt than wild-type and other UBPs mutants. UBP16 positively regulates plasma membrane Na+/H+ antiport activity and the ubp16 mutant accumulated more sodium. Through yeast two-hybrid assay, we identified a putative target of UBP16, SHM1. The stability and activity of SHM1 were changed in the ubp16 mutant, resulting in accelerated cell death and accumulation of reactive oxygen species. Taken together, UBP16 was involved in salt tolerance in Arabidopsis by regulating Na+/H+ antiport activity and repressing cell death partially through regulating SHM1 stability and activity.7 Besides SHM1, we obtained another positive clone from Arabidopsis cDNA library through yeast two-hybrid (Fig. 1). Sequence analysis revealed that this clone harbored the full-length coding sequence of HIPP27, which belongs to the metallochaperone-like proteins that traffic metal ion within cells and sequester metals in cell compartment.8
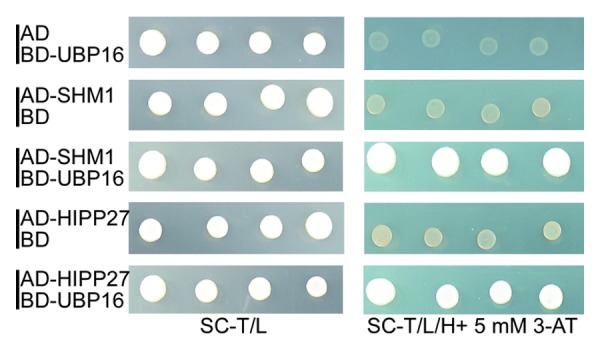
Figure 1. UBP16 interacts with HIPP27 through yeast two-hybrid. Yeast strains containing AD-HIPP27 and BD-UBP16 grew better than that harboring AD and BD-UBP16 or AD-HIPP27 and BD vector on synthetic complete (SC) medium without Trp, Ler and His, 5 mM 3-AT plus. SHM1 as positive control.
Cadmium (Cd) is a widespread non-essential toxic heavy metal and considered as a serious environmental pollutant. Cadmium has adverse effects on plant development such as inhibition of root growth and induction of chlorosis in leaves.9 In Arabidopsis, it was reported that HIPP20, HIPP22, HIPP26, and HIPP27 played an important role in Cd-detoxification.8 In order to determine whether UBP16 is involved in Cadmium tolerance, we transferred 7-d-old seedlings grown on MS medium to MS medium with or without 30 μM CdCl2 for 12 d. In the absence of CdCl2, there was no significant difference in shoot growth and anthocynanin contents between wild-type and the ubp16 mutant seedlings. However, in the presence of 30 μM CdCl2, shoot growth of the ubp16 mutant was more severely impaired than wild-type (Fig. 2), and the anthocynanin contents of the ubp16 mutant was significant higher than that of wild-type, suggesting that the ubp16 mutant is more sensitive to CdCl2. In summary, these evidences implied that HIPP27 may be another target of UBP16 and their interaction modulates Cadmium tolerance, although other evidences are needed.
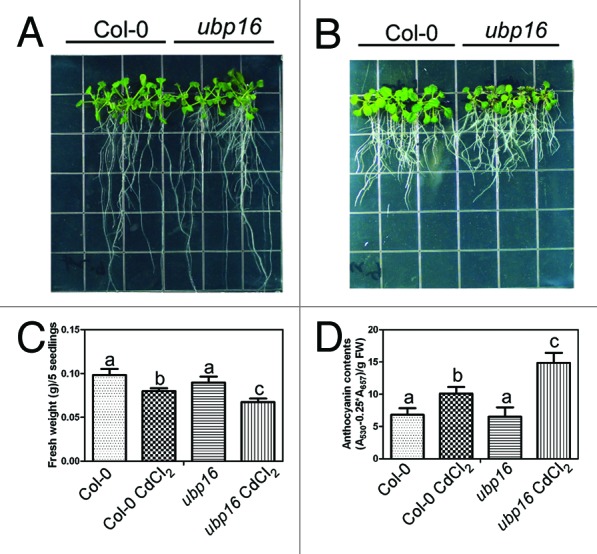
Figure 2. Analysis of Cadmium sensitivity in wild-type (Col-0) and the ubp16 mutant seedlings. (A) and (B) Col-0 and the ubp16 seedlings grown on MS medium with or without 30 μM CdCl2. Photograph were taken 12 d after transfer. (C) Fresh weight. (D) Anthocynanin contents. Error bars represent SD (n > 6). Statistical significance was determined by the Student t-test.
Acknowledgment
This research was supported by the National Natural Science Foundation of China (Grant 31170246).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1.Scheffner M, Nuber U, Huibregtse JM. Protein ubiquitination involving an E1-E2-E3 enzyme ubiquitin thioester cascade. Nature. 1995;373:81–3. doi: 10.1038/373081a0. [DOI] [PubMed] [Google Scholar]
- 2.Cui F, Liu LJ, Zhao QZ, Zhang ZH, Li QL, Lin BY, et al. Arabidopsis ubiquitin conjugase UBC32 is an ERAD component that functions in brassinosteroid-mediated salt stress tolerance. Plant Cell. 2012;24:233–44. doi: 10.1105/tpc.111.093062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vierstra RD. The ubiquitin-26S proteasome system at the nexus of plant biology. Nat Rev Mol Cell Biol. 2009;10:385–97. doi: 10.1038/nrm2688. [DOI] [PubMed] [Google Scholar]
- 4.Wilkinson KD. Regulation of ubiquitin-dependent processes by deubiquitinating enzymes. FASEB J. 1997;11:1245–56. doi: 10.1096/fasebj.11.14.9409543. [DOI] [PubMed] [Google Scholar]
- 5.Yan N, Doelling JH, Falbel TG, Durski AM, Vierstra RD. The ubiquitin-specific protease family from Arabidopsis. AtUBP1 and 2 are required for the resistance to the amino acid analog canavanine. Plant Physiol. 2000;124:1828–43. doi: 10.1104/pp.124.4.1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu YF, Wang F, Zhang HY, He H, Ma LG, Deng XW. Functional characterization of the Arabidopsis ubiquitin-specific protease gene family reveals specific role and redundancy of individual members in development. Plant J. 2008;55:844–56. doi: 10.1111/j.1365-313X.2008.03557.x. [DOI] [PubMed] [Google Scholar]
- 7.Zhou HP, Zhao JF, Yang YQ, Chen CX, Liu YF, Jin XH, et al. Ubiquitin-specific protease16 modulates salt tolerance in Arabidopsis by regulating Na(+)/H(+) antiport activity and serine hydroxymethyltransferase stability. Plant Cell. 2012;24:5106–22. doi: 10.1105/tpc.112.106393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tehseen M, Cairns N, Sherson S, Cobbett CS. Metallochaperone-like genes in Arabidopsis thaliana. Metallomics. 2010;2:556–64. doi: 10.1039/c003484c. [DOI] [PubMed] [Google Scholar]
- 9.Liu XM, Kim KE, Kim KC, Nguyen XC, Han HJ, Jung MS, et al. Cadmium activates Arabidopsis MPK3 and MPK6 via accumulation of reactive oxygen species. Phytochemistry. 2010;71:614–8. doi: 10.1016/j.phytochem.2010.01.005. [DOI] [PubMed] [Google Scholar]


