Abstract
RAR1 is identified as a critical protein involved in plant innate immunity. We investigated the role of RAR1 in Agrobacterium-mediated plant transformation based on the previous findings that accessory proteins associated with the E3 ligase complex such as SGT1, which tightly interacts with RAR1, play a role in the transformation process. RAR1 gene silencing in Nicotiana benthamiana and Arabidopsis rar1 mutant analysis suggested that RAR1 is required for early stages of Agrobacterium-mediated plant transformation. This finding further illustrates that RAR1, along with SGT1, that serve as a HSP90 co-chaperone is important for Agrobacterium-mediated plant transformation.
Keywords: Agrobacterium, F-box proteins, plant transformation, SCF-E3 ligase, VIGS
Introduction
One of the critical steps in Agrobacterium-mediated genetic transformation involves the uncoating of the transferred DNA (T-DNA) from its cognate protein complex before integration. This step in the transformation process is not clearly understood, even though the involvement of the host ubiquitin-proteosome system (UPS) has been suggested.1 One of the most extensively studied UPS in plants includes the SKP1/Cullin/ F-box (SCF) E3 ubiquitin ligase complex which is induced upon Agrobacterium tumefaciens infection2 and mediates degradation of the cognate proteins either through the induction of a plant specific F-box protein VBF3 or by the Agrobacterium F-box protein VirF, which is exported into the host cell.4
The plant SCF complex mediates polyubiquitination of host and foreign proteins for degradation, thus controlling a plethora of biological process within the cell. Recent studies have shown differential expression of the UPS-associated genes upon Agrobacterium inoculation,2 some of which likely affects Agrobacterium infection.2,5 In addition, accessory proteins such as SGT1 (suppressor of the G2 allele of skp1) which loosely associate with SCF E3 ligases but tightly interacts with RAR1, a protein required for R-gene resistance to powdery mildew in barley,6,7 also affect Agrobacterium infection.2 Both RAR1 and SGT1 were identified as components of plant innate immunity.8-12 Here we report the role of RAR1 in early stages of Agrobacterium-mediated plant transformation.
Results and Discussion
Gene silencing of RAR1 attenuates Agrobacterium-mediated plant transformation
Using a virus-induced gene silencing (VIGS)-based approach, we previously showed2 that NbRAR1 silenced Nicotiana benthamiana plants produced significantly smaller and fewer tumors when compared with non-silenced control plants upon inoculation with an oncogenic strain, A. tumefaciens A348.13 For further characterizing the role of RAR1 in plant transformation, we performed additional experiments. Relying on the transient expression assay of β-glucuronidase (GUS) gene described by Anand et al.,14 we observed moderate reduction in 5-bromo-4-chloro-3-indolyl β-D-glucuronide (X-Gluc) staining and GUS activity in the leaf disks derived from NbRAR1 silenced N. benthamiana plants as compared with leaf disks derived from control (TRV::GFP inoculated plants) or wild-type plants at early stages of transformation (4 and 7 dpi; Figure 1A). Based on the above data we suggest that gene silencing of RAR1 results in reduced transient T-DNA gene expression. In addition to transient transformation, stable transformation experiments were performed to select glufosinate ammonium (GF)-resistant calli as described.14 Silencing of NbRAR1 produced fewer GF-resistant calli (70.6%), when compared with wild-type or controls (85.6% and 91.1%, respectively, Figure 1B). When cultured on a non-selective callus-inducing medium, leaf disks from NbRAR1 silenced and control N. benthamiana plants produced calli at equal efficiency (data not shown). These data suggest that RAR1 gene silencing did not affect the cell division machinery in phytohormone-rich medium. The above observations lead us to conclude that RAR1 might be involved at the early stages of transformation.
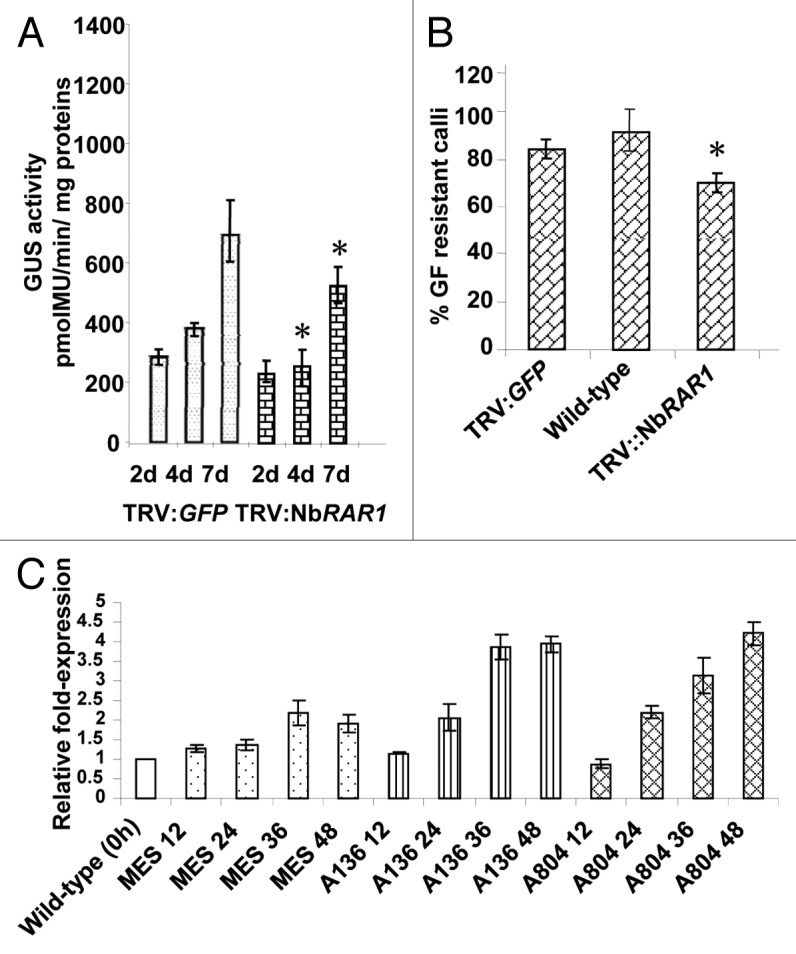
Figure 1. Transformation assays on RAR1 gene silenced plants of N. benthamiana and induction of RAR1 in response to Agrobacterium infection. (A) Transient transformation assay. Leaf disks of the gene silenced plants and control plants were inoculated with non-tumorigenic strain A. tumefaciens GV2260 carrying pBISN1 (At804; has the uidA-intron gene within the T-DNA). Leaf disks were collected periodically and were used for measuring the fluorescence of 4-methylumbelliferone (4-MU). (B) Leaf disk stable transformation assay. Leaf disks of silenced and control plants were inoculated with a non-tumorigenic strain, A. tumefaciens GV2260, harboring the binary vector pCAS1 (contains a bar gene within the T-DNA) and were incubated on callus inducing medium (CIM) with glufosinate ammonium (GF). Data represent the mean of 3 experiments with a minimum of 150 leaf disks each per treatment with their SE values shown as error bars. (C) Differential gene expression of RAR1 upon Agrobacterium infection. Individual leaves of a minimum of 3 N. benthamiana plants were syringe (needleless) infiltrated with either: buffer (10 mM MES, dotted bars); an avirulent strain A. tumefaciens A136 (lacks Ti plasmid – cannot transfer T-DNA, line bars); or a T-DNA transfer-competent strain, A. tumefaciens At804 (checkered bars). Asterisks denote significant difference compared with controls using Fisher's least significant difference test at P = 0.05.
RAR1 is induced in response to Agrobacterium inoculation
The expression of RAR1 was analyzed to check whether the gene is responsive to Agrobacterium inoculation. To characterize RAR1 gene expression following Agrobacterium inoculation, the leaves of N. benthamiana plants were separately infiltrated with a transfer competent strain, A. tumefaciens At804, a transfer deficient strain, A. tumefaciens A136, and the buffer control as described.14 RAR1 showed differential expression following both Agrobacterium and buffer inoculations. RAR1 gene induction was greater (1.5- to 2-fold) in the leaves infected with Agrobacterium at 24–48 h post-infection as compared with buffer inoculated plants (Fig. 1C). These results suggest that RAR1 is induced upon both general stress and also upon Agrobacterium infection. This finding complements the work we previously published involving microarray analysis on uninfected and Agrobacterium infected Arabidopsis (Col-0) leaves.15 AtRAR1 induction (~2.5-fold) was observed at 48hpi, while a relatively lower expression (< 1.5-fold) was observed at 72 hpi.15 We further reconfirmed the above data through quantitative RT-PCR (qRT-PCR) analyses on Agrobacterium infected and uninfected leaf samples (Table 1) and the results were consistent with the microarray data.
Table1. qRT-PCR results showing the differential gene expression of Arabidopsis RAR1 following Agrobacterium infection.
| Gene ID | Gene Symbol | Infected (48h) | Mock (48h) | Infected (72h) | Mock (72h) |
|---|---|---|---|---|---|
| At5g51700 | Rar1 | 4.6 ± 0.5 | 1.9 ± 0.18 | 2.9 ± 0.4 | 1.2 ± 0.6 |
Arabidopsis rar1 mutant is recalcitrant to Agrobacterium-mediated root transformation
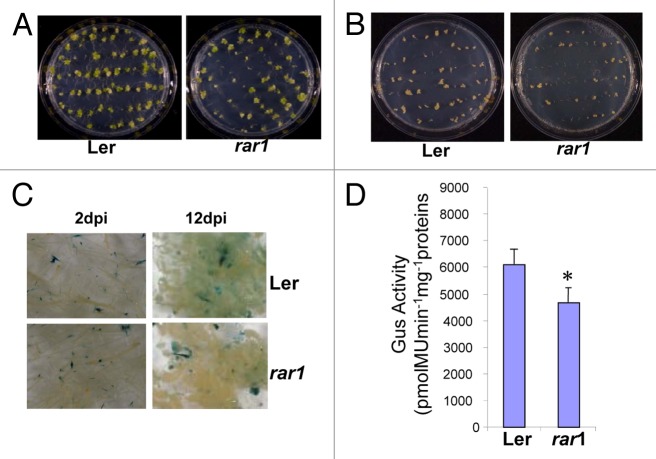
Using Arabidopsis rar1 (Ler-rar1)16 mutant, the function of RAR1 protein in Agrobacterium-mediated plant transformation in another plant species was confirmed. Figure 2 summarizes the root transformation assay results15 for the mutant along with its corresponding wild-type. A significant reduction in tumor formation was observed in the rar1 mutant as compared with the wild-type plant upon inoculation with tumorigenic strain A. tumefaciens, A208 (Table 2, Figure 2A). The rar1 mutant was also evaluated for stable transformation phenotype by scoring for phosphinothricin (ppt)-resistant calli following infection with disarmed strain Agrobacterium At872 (T-DNA carries the bar gene) (Fig. 2B). Consistent with the tumor assay (Fig. 2A), we observed significantly lower stable transformation of rar1 mutant when infected with a disarmed Agrobacterium strain (Table 2). Consistent with the NbRAR1 gene silenced data, the rar1 mutant was also found to be slightly defective in transient gene expression, as seen through the reduced GUS activity following infection with Agrobacterium strain containing the uidA-intron in the T-DNA of the binary vector (Fig. 2C and D). Taken together, the data from both gene silencing in N. benthamiana and mutant analysis in Arabidopsis imply the role of RAR1 in the early stages of Agrobacterium-mediated plant transformation.
Figure 2. Root transformation assays in the Arabidopsis rar1 mutant. (A) Root tumorigenesis assays. Roots of wild-type (Ler) and rar1 mutant plants were infected with a tumorigenic strain A. tumefaciens A208 (nopaline strain), and tumors incited on the roots were visualized and scored 4 weeks after infection. (B) Stable root transformation assays. The root segments of the mutant and the wild-type (Ler) plants were inoculated with a disarmed strain, A. tumefaciens At872, containing the bar gene within the T-DNA and were incubated on callus-inducing medium (CIM) with glufosinate ammonium (GF). Photographs were taken 4 weeks after inoculation. (C) Roots of the wild-type and mutant plants were inoculated with a disarmed strain A. tumefaciens GV3101 carrying pBISN1. The inoculated roots were periodically collected at 2 and 12 dpi and stained with X-Gluc. (D) Quantification of GUS activity. The root segments were collected at 12 dpi (C) and were used for measuring the fluorescence of 4-methylumbelliferone (4-MU). All the experiments were repeated 2 times and obtained similar results. Asterisk denotes significant difference compared with controls using Fisher's least significant difference test at P = 0.05.
Table2: Root transformation assays in Arabidopsis wild-type and rar1 mutant Root tumorigenesis assay.
| Genotype | % Tumor (total # roots) 0.D600 (0.5) |
|---|---|
| Ler | 79.3 ± 6.2 (670) a |
| rar1 | 62.3 ± 9.2 (670) b |
| Callus assay (GF-resistant calli) |
| Genotype | %GF-resistant calli (total # roots) 0.D600 (0.5) |
|---|---|
| Ler | 75.3 ± 7.7 (404)a |
| rar1 | 62.3 ± 7.7 (404)b |
In conclusion, we show that RAR1 silenced N. benthamiana plants and rar1 Arabidopsis mutant are partially recalcitrant to Agrobacterium-mediated plant transformation. The RAR1 gene silenced plants and the rar1 mutant showed reduced transient and stable transformation and is similar to the previously published data on SGT1 silenced plants and Arabidopsis sgt1 mutant.2 The differential expression of the RAR1 gene upon Agrobacterium infection and its known association with SGT1 might suggest a role in plant defenses against Agrobacterium-infection. Current research suggests that SGT1 and RAR1 associate as co-chaperones with HSP90 and with a possible role in resistance (R) protein activation.17 Furthermore, SGT1 and RAR1 are thought to function in disease resistance by forming multiple protein complexes influencing the conformation of R protein complexes and targeting proteins for degradation.17 The data from this manuscript and the recently published data2 suggest that E3 ubiquitin-ligases are also involved in Agrobacterium-mediated plant transformation. The definite role of these genes in Agrobacterium-mediated plant transformation warrants further research.
Acknowledgments
This work was supported by the Samuel Roberts Noble Foundation and in part by a grant from the National Science Foundation (grant IOB 0445799)
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1.Magori S, Citovsky V. The role of the ubiquitin-proteasome system in Agrobacterium tumefaciens-mediated genetic transformation of plants. Plant Physiol. 2012;160:65–71. doi: 10.1104/pp.112.200949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anand A, Rojas CM, Tang Y, Mysore KS. Several components of SKP1/Cullin/F-box E3 ubiquitin ligase complex and associated factors play a role in Agrobacterium-mediated plant transformation. New Phytol. 2012;195:203–16. doi: 10.1111/j.1469-8137.2012.04133.x. [DOI] [PubMed] [Google Scholar]
- 3.Zaltsman A, Lacroix B, Gafni Y, Citovsky V. Disassembly of synthetic Agrobacterium T-DNA-protein complexes via the host SCF(VBF) ubiquitin-ligase complex pathway. Proc Natl Acad Sci U S A. 2013;110:169–74. doi: 10.1073/pnas.1210921110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tzfira T, Vaidya M, Citovsky V. Involvement of targeted proteolysis in plant genetic transformation by Agrobacterium. Nature. 2004;431:87–92. doi: 10.1038/nature02857. [DOI] [PubMed] [Google Scholar]
- 5.Zaltsman A, Krichevsky A, Loyter A, Citovsky V. Agrobacterium induces expression of a host F-box protein required for tumorigenicity. Cell Host Microbe. 2010;7:197–209. doi: 10.1016/j.chom.2010.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu Y, Burch-Smith TM, Schiff M, Feng S, Dinesh-Kumar SP. Molecular chaperone Hsp90 associates with resistance protein N and its signaling proteins SGT1 and Rar1 to modulate an innate immune response in plants. J Biol Chem. 2004;279:2101–8. doi: 10.1074/jbc.M310029200. [DOI] [PubMed] [Google Scholar]
- 7.Takahashi A, Casais C, Ichimura K, Shirasu K. HSP90 interacts with RAR1 and SGT1 and is essential for RPS2-mediated disease resistance in Arabidopsis. Proc Natl Acad Sci U S A. 2003;100:11777–82. doi: 10.1073/pnas.2033934100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Austin MJ, Muskett P, Kahn K, Feys BJ, Jones JDG, Parker JE. Regulatory role of SGT1 in early R gene-mediated plant defenses. Science. 2002;295:2077–80. doi: 10.1126/science.1067747. [DOI] [PubMed] [Google Scholar]
- 9.Azevedo C, Sadanandom A, Kitagawa K, Freialdenhoven A, Shirasu K, Schulze-Lefert P. The RAR1 interactor SGT1, an essential component of R gene-triggered disease resistance. Science. 2002;295:2073–6. doi: 10.1126/science.1067554. [DOI] [PubMed] [Google Scholar]
- 10.Azevedo C, Betsuyaku S, Peart J, Takahashi A, Noël L, Sadanandom A, Casais C, Parker J, Shirasu K. Role of SGT1 in resistance protein accumulation in plant immunity. EMBO J. 2006;25:2007–16. doi: 10.1038/sj.emboj.7601084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu F, Ni W, Griffith ME, Huang Z, Chang C, Peng W, Ma H, Xie D. The ASK1 and ASK2 genes are essential for Arabidopsis early development. Plant Cell. 2004;16:5–20. doi: 10.1105/tpc.017772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shirasu K, Lahaye T, Tan M-W, Zhou F, Azevedo C, Schulze-Lefert P. A novel class of eukaryotic zinc-binding proteins is required for disease resistance signaling in barley and development in C. elegans. Cell. 1999;99:355–66. doi: 10.1016/S0092-8674(00)81522-6. [DOI] [PubMed] [Google Scholar]
- 13.Garfinkel DJ, Simpson RB, Ream LW, White FF, Gordon MP, Nester EW. Genetic analysis of crown gall: fine structure map of the T-DNA by site-directed mutagenesis. Cell. 1981;27:143–53. doi: 10.1016/0092-8674(81)90368-8. [DOI] [PubMed] [Google Scholar]
- 14.Anand A, Vaghchhipawala Z, Ryu CM, Kang L, Wang K, del-Pozo O, Martin GB, Mysore KS. Identification and characterization of plant genes involved in Agrobacterium-mediated plant transformation by virus-induced gene silencing. Mol Plant Microbe Interact. 2007;20:41–52. doi: 10.1094/MPMI-20-0041. [DOI] [PubMed] [Google Scholar]
- 15.Anand A, Krichevsky A, Schornack S, Lahaye T, Tzfira T, Tang Y, Citovsky V, Mysore KS. Arabidopsis VIRE2 INTERACTING PROTEIN2 is required for Agrobacterium T-DNA integration in plants. Plant Cell. 2007;19:1695–708. doi: 10.1105/tpc.106.042903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Muskett PR, Kahn K, Austin MJ, Moisan LJ, Sadanandom A, Shirasu K, Jones JDG, Parker JE. Arabidopsis RAR1 exerts rate-limiting control of R gene-mediated defenses against multiple pathogens. Plant Cell. 2002;14:979–92. doi: 10.1105/tpc.001040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shirasu K, Schulze-Lefert P. Complex formation, promiscuity and multi-functionality: protein interactions in disease-resistance pathways. Trends Plant Sci. 2003;8:252–8. doi: 10.1016/S1360-1385(03)00104-3. [DOI] [PubMed] [Google Scholar]