Abstract
Ourmia melon virus (OuMV), Epirus cherry virus (EpCV) and Cassava virus C (CsVC) are three species placed in the genus Ourmiavirus. We cloned and sequenced their RNA genomes. The sizes of the three genomic RNAs of OuMV, the type member of the genus, were 2814, 1064 and 974 nt and each had one open reading frame. RNA1 potentially encoded a 97.5 kDa protein carrying the GDD motif typical of RNA-dependent RNA polymerases (RdRps). The putative RdRps of ourmiaviruses are distantly related to known viral RdRps, with the closest similarity and phylogenetic affinity observed with fungal viruses of the genus Narnaviridae. RNA2 encoded a 31.6 kDa protein which, expressed in bacteria as a His-tag fusion protein and in plants through agroinfiltration, reacted specifically with antibodies made against tubular structures found in the cytoplasm. The ORF2 product is significantly similar to movement proteins of the genus Tombusviridae, and phylogenetic analysis supported this evolutionary relationship. The product of OuMV ORF3 is a 23.8 kDa protein. This protein was also expressed in bacteria and plants, and reacted specifically with antisera against the OuMV coat protein. The sequence of the ORF3 protein showed limited but significant similarity to capsid proteins of several plant and animal viruses, although phylogenetic analysis failed to reveal its most likely origin. Taken together, these results indicate that ourmiaviruses comprise a unique group of plant viruses that might have evolved by reassortment of genomic segments of RNA viruses infecting hosts belonging to different eukaryotic kingdoms, in particular, fungi and plants.
Introduction
Ourmia melonvirus (OuMV) is the cause of a mosaic disease of melons (Cucumis melo) in the north-western Azerbaijan district of Iran. Field-infected plants have leaves showing round, light green spots, grow poorly and produce smaller fruits. The virus is easily mechanically transmitted to a rather wide range of dicotyledonous plants (34 species in 14 families), including several plants of economic importance; it has been purified and a rabbit antiserum has been produced (Lisa et al., 1988). A recent survey in the Guilan region of northern Iran has confirmed the importance of this virus for melon crops in Iran (Gholamalizadeh et al., 2008). OuMV has unique morphological characteristics: the bacilliform virions are composed of a series of particles with conical ends (apparently hemi-icosahedra) and cylindrical bodies 18 nm in diameter, composed of two, three, four or six double discs (Lisa et al., 1988; Matthews, 1991; Milne, 2005; Accotto & Milne, 2008). The particles consist of three linear single-stranded positive-sense RNAs and one protein with an estimated molecular weight of about 25 kDa (Lisa et al., 1988; Marzachi et al., 1992; Accotto et al., 1997). Cassava virus C (CsVC) and Epirus cherry virus (EpCV), the other Ourmiavirus species isolated from equatorial Africa and Greece, respectively (Fauquet et al., 2005), have similar particles (Aiton et al., 1988; Avgelis et al., 1989) and EpCV is known to have three RNAs and a single coat protein (CP), each slightly smaller than those of OuMV (Accotto et al., 1997).
In this paper, we describe the genome organization of one isolate of each of OuMV, EpCV and CsVC. We provide data on the roles of the three proteins encoded by RNAs 1, 2 and 3 of OuMV, and discuss the phylogenetic relationships deduced for each of the RNAs and the implications for the evolutionary significance and taxonomic position of the genus Ourmiavirus.
Methods
Virus isolates and their maintenance.
All virus isolates were maintained and mechanically propagated in Nicotiana benthamiana plants in the greenhouse in previously described conditions (Turina et al., 2006). The sources of the OuMV and EpCV isolates have been described by Avgelis et al. (1989) and Lisa et al. (1988). CsVC isolates (Aiton et al., 1988) from the Ivory Coast (IC) and from Malawi (MV) were kindly provided by M. M. Aiton (now Mrs M. Swanson, Scottish Crop Research Institute, Invergowrie, Dundee, UK). Of these, we used the IC isolate.
Virus particle and tubule purification.
One hundred grams of N. benthamiana systemically infected with OuMV, EpCV and CsVC was used for purification following the method described by Lisa et al. (1988).
To purify the tubular structures found in the cytoplasm of OuMV-infected cells (see Results and Fig. 1), 100 g infected N. benthamiana (local lesions) was homogenized in 3 volumes of 0.5 M potassium phosphate buffer, pH 7, containing 5 mM EDTA, 20 mM Na2SO3 and 1 % Triton X-100. The homogenate was centrifuged at 1464 g for 5 min and the pellet was resuspended in 50 ml 0.2 M potassium phosphate buffer, pH 7. After centrifugation at 164 g for 4 min, the supernatant was retained. Pellet resuspension and centrifugation were repeated twice and all the supernatants were retained and pooled. These were incubated overnight with anti-CP and anti-healthy plant antisera, as described by Luisoni et al. (1995). Samples were centrifuged at low speed to remove the precipitate, and the supernatant was ultracentrifuged at 265 000 g for 15 min. The pellet was resuspended in 0.6 ml 0.2 M potassium phosphate buffer, pH 7. Negative staining and electron microscopy were used to monitor the presence of tubules in the various fractions.
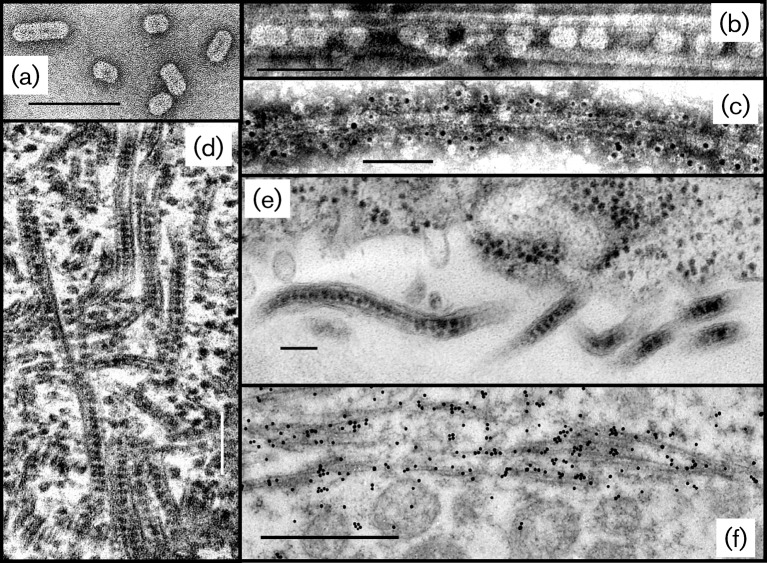
Fig. 1.
Transmission electron micrographs of structures related to OuMV. Bars, 100 nm unless otherwise stated. (a) Purified preparation of virions negatively stained in 0.5 % uranyl acetate. (b) Crude sap extract of a 7 day systemic infection in N. benthamiana showing a tubule containing virus particles; uranyl acetate negative stain. (c) A tubule shown in (b) immunolabelled with tubule-specific rabbit antibody followed by goat anti-rabbit IgG coupled to 5 nm gold particles; uranyl acetate negative stain. (d) Thin section of leaf tissue from a 4 week systemic infection in Nicotiana clevelandii showing cytoplasm with tubules containing virus particles; aldehyde-osmium fixation, epoxy resin embedding. Bar, 200 nm. (e) Thin section of a leaf parenchyma cell from an 8 day systemic infection in N. clevelandii, showing plasmodesmata that contain tubules, in turn containing virus particles, within the cell wall (lower half of the image); preparation as in (d). (f) Thin section of a leaf parenchyma cell from a 7 day systemic infection in N. benthamiana, showing tubules (but not mitochondria) immunolabelled with anti-tubule rabbit serum followed by goat anti-rabbit IgG conjugated to 15 nm gold particles. Bar, 500 nm.
Antisera already existed against OuMV CP (Lisa et al., 1988). An antiserum to the OuMV tubular structures was prepared as previously described (Luisoni et al., 1995).
Electron microscopy.
The procedures employed were those described by Milne (1993), Milne et al. (1995) and Vaira et al. (1997). We used 400-mesh grids bearing Formvar-carbon films, and a Philips CM 10 transmission electron microscope (Eindhoven, The Netherlands) operated at 60 kV. For negative staining, we used 0.5 % aqueous uranyl acetate; immunolabelling was carried out with rabbit primary antisera and goat anti-rabbit IgG conjugated with either 5 or 15 nm gold particles (Amersham Life Sciences); thin sections were cut using an Ultracut E microtome (Reichert-Jung) and Diatome diamond knives (Taab Laboratories) from blocks fixed either in glutaraldehyde and osmium tetroxide and embedded in Epon equivalent (Taab) or, for immunolabelling, in formaldehyde–glutaraldehyde and embedded in LR Gold acrylic resin (London Resin). In order to be sure of the appearance of OuMV particles in thin sections of infected plants, we mixed equal volumes of a pellet of purified virus and 6 % low-melting-point agarose (Sigma) held at 45 °C. After solidification, the mixture was cut into small cubes, fixed in glutaraldehyde–osmium and embedded in Epon.
Western blot analysis of leaf extracts.
Total leaf protein extracts, SDS-PAGE and Western blots were carried out as described by Turina et al. (2000). Antisera were used at a final dilution of 1/1000. Secondary horseradish peroxidase (Sigma) antibodies were used at a final dilution of 1/4000.
cDNA libraries and Northern blot analysis.
RNA was extracted from the purified viruses and from healthy and infected plants using two different methods: Triazol and PureLink RNA Reagent (Invitrogen). RNA was separated on 1 % agarose TAE gels and bands of genomic RNA were extracted from the gel and purified using RNaid W/Spin kit (Bio 101).
A cDNA library was assembled from purified genomic RNA as described by Turina et al. (2006). The inserts were sequenced using the dideoxy chain-termination method (Sanger et al., 1977). In order to check the viral origin of the cloned cDNA, we carried out Northern blots using specific clones as probes. Total RNA from infected N. benthamiana leaves was prepared through homogenization and PureLink extraction from leaves as suggested by the manufacturer (Invitrogen). RNA samples were separated in denaturing conditions (glyoxal method) as previously detailed, but using HEPES–EDTA buffer (Sambrook et al., 1989; Turina et al., 2003).
Northern blots were carried out using a Dig-labelled RNA probe prepared from transcripts from linearized purified plasmids containing cDNA clones in both orientations, following the manufacturer’s protocols and reagents (Roche). Once cDNA clones were identified for each genomic RNA, we performed RT-PCR using specific oligonucleotides designed from the sequences already available from cDNA cloning in order to cover gaps among the various sequences of the cDNA library.
To determine the 5′ ends of the three genomic RNAs, we used the method described by Hirzmann et al. (1993). dGTP or, in some cases, dATP was used to synthesize a polyG or polyA tail using rTdT terminal deoxynucleotidyl transferase (Promega). The final PCR step was carried out using oligo-dC (Eco-bam-dc12) or oligo dT-V, each time using a specific primer for each RNA (oligonucleotides OuRNA1-140R, OuMV-2-180R and OuMV-3-120R, respectively; Supplementary Table S1, available in JGV Online). PCR products were cloned and sequenced as above.
The 3′ ends of genomic RNAs were determined by 3′ RACE, after polyadenylation of each genomic RNA with polyA polymerase and ATP (Ambion). Reverse transcription and PCR were carried out as described above using oligo dT-V for reverse transcription and oligo dT-V and a specific forward primer upstream the putative 3′ end of each RNA (OuRNA1-2600F, OuMV-2-900F and OuMV-3-400F, respectively; Supplementary Table S1).
Overlapping sequences were assembled and ORF analysis was carried out on each sequence using the Vector NTI 10 (Invitrogen). Cloning and sequence analysis of the EpCV and CsVC genomes were carried out as described for OuMV.
Determining the polarity of OuMV genomic RNAs.
To determine the polarity of the RNA species that accumulate in the plant during OuMV replication and those that are encapsidated in particles, full-length RNA1, RNA2 and RNA3 cDNAs were prepared through RT-PCR using the oligonucleotides OuMV-RNA1-1F, OuMV-RNA1-3′Rev, OuRNA2-5′F, OuRNA2-3′R, OuRNA3-5′F and OuRNA3-3′R (Supplementary Table S1). The PCR products were cloned using Zero Blunt Topo PCR Cloning kit (Invitrogen) following the manufacturer’s instructions. EcoICRI and the T7 promoter, and PstI and the SP6 promoter were used to synthesize minus and plus strand synthetic RNA2 and RNA3 transcripts using MaxiScript kit (Ambion). For RNA1, HindIII was used instead of EcoICRI in order to linearize the plasmid before transcription. Minus and plus strand Dig-labelled probes were also prepared following the manufacturer’s instructions (Roche). Total RNA from infected and healthy N. benthamiana leaves was extracted with PureLink Plant RNA reagent using 2.5 mm zirconia/silica beads (Biospec Products) following the manufacturer’s instructions (Invitrogen). This method was also used to extract RNA from purified virus. Northern blot analyses were carried out as described above.
Expression of polyprotein fragments encoded by OuMV RNA2 and RNA3 in Escherichia coli and in N. benthamiana leaves through agroinfiltration.
DNA fragments to be ligated into the pRSET-A expression vector (Invitrogen) were obtained through RT-PCR, using total RNA from infected plants as template, in the conditions described above, using the following primers (see Supplementary Table S1): RNA1-Bam-F and RNA1-1560-R for RNA1, RNA2-Bam-F and RNA2-Bam-R for RNA2, and OuMV-3BamF and RNA3wh-BamR for RNA3. RT-PCR products were first cloned in pGEM-T easy vector. Plasmids with the expected insert were selected for digestion with BamHI and ligation into the digested and dephosphorylated pRSET-A vector (Invitrogen). Expression cultures (1 ml) were processed as described by Turina et al. (2000). Western blot analysis of bacterial lysates was performed using the antisera 1 h after induction with IPTG (Turina et al., 2000).
Agrobacterium tumefaciens competent cells (strain C58C1) were transformed with recombinant plasmids derived from vector pBin61 (Bendahmane et al., 2002; Höfgen & Willmitzer 1988) containing ORF2 and ORF3 of OuMV. N. benthamiana plants were agroinfiltrated as described by Bendahmane et al. (1999). Leaf extracts of virus-infected or agroinfiltrated plants were prepared for protein separation and Western blot analysis as described by Margaria et al. (2007).
Sequence similarity search and phylogenetic analysis.
Sequence similarity searches were performed using psi-blast (Altschul et al. 1997) against the NCBI RefSeq database (Wheeler et al. 2008). Multiple alignments of protein sequences were constructed using the muscle (Edgar 2004) and HHalign (Soding 2005) programs. Alignments of capsid proteins were manually adjusted using vast (Gibrat et al., 1996) structural alignments as templates, where a 3D structure of at least one member of the respective protein family was available. The statistical support for the inter-family similarity was computed using the HHalign program (Soding, 2005). For phylogenetic tree reconstruction, alignment columns containing more than 50 % gap characters were removed. The alignments were also evaluated for the information content of individual columns by computing the homogeneity value as described previously (Yutin et al., 2008); positions with homogeneity values greater than 0.5 were considered to be confidently aligned. Preliminary maximum-likelihood (ML) trees were constructed using the phyml software (Guindon & Gascuel, 2003) with the Whelan and Goldman (WAG) evolutionary model (Whelan & Goldman, 2001) and gamma-distributed site rates. The trees obtain with phyml were then used to initialize Monte Carlo Markov Chain (MCMC) computations using the MrBayes program (Ronquist & Huelsenbeck, 2003) with mixed evolutionary model and gamma-distributed site rates. For two runs of four MCMCs, 1.1×106 generations were retained; the first 105 generations were discarded as burn-in, 1000 samples taken every 1000 generations from each of the runs were pooled together and consensus trees were constructed. The fractions of sampled trees with the given tree bipartition present were used as support values for selected clades. Additionally, ML trees were constructed using the TreeFinder (Jobb et al., 2004) and RAxML (Stamatakis et al., 2008) programs, with the evolutionary model preferred by Bayesian sampling and gamma-distributed site rates [blosum for polymerase and CP and WAG for the movement protein (MP)]. Each of these programs was also used to compute the statistical support for internal branches.
Results
Electron microscopy.
Fig. 1(a) shows a negatively stained purified preparation of OuMV particles. We confirmed that the particles of EpCV and CsVC both looked the same as those of OuMV (not shown). In crude preparations of OuMV-infected tissues, tubular structures often containing virions were frequently seen (Fig. 1b). The presence of such virion-containing tubules was also noted for CsVC (Aiton et al., 1988). The OuMV-related tubular structures could be detected with the antiserum raised against purified tubule preparations and absorbed to remove any reaction with the virions (Fig. 1c). In thin sections of pelleted virions, the virus particles appeared as dense, sometimes elongated objects of 16–25 nm in diameter (not shown). The same objects, distinct from ribosomes, were recognized as abundantly present in the cytoplasm of infected cells, often lined up within tubular structures (Fig. 1d) that were the homologues of those seen in Fig. 1(b and c). The tubules containing virus particles were also seen within plasmodesmata-like structures passing through cell walls (Fig. 1e). When tissues equivalent to those shown in Fig. 1(d) were aldehyde-fixed and embedded in acrylic resin, thin sections could be immunolabelled with the absorbed anti-tubule antiserum; label was concentrated over the tubules (Fig. 1f).
Apart from the presence of virions and tubules in the cytoplasm, no cytopathology specific to OuMV was observed.
Western blot analysis using antisera against OuMV CP and tubules.
Antiserum against OuMV particles specifically reacted with a band of approximately 25 kDa (Fig. 2a), the same size found for the purified CP. There was no cross-reaction with extracts of plants infected with EpCV or CsVC (not shown).
Fig. 2.
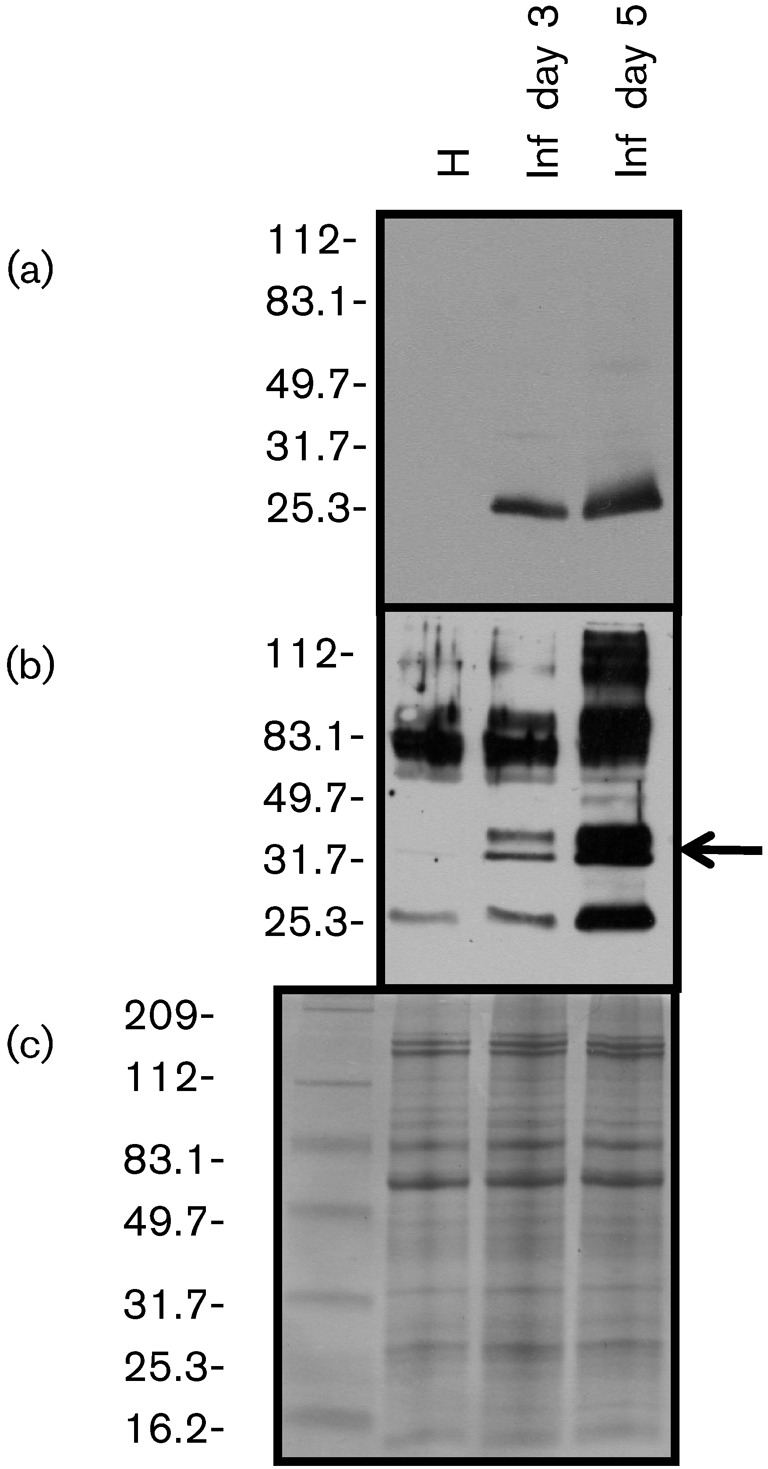
Western blot analysis using total RNA extracted from healthy N. benthamiana (H) and infected N. benthamiana 3 (Inf day 3) and 5 (Inf day 5) days after OuMV inoculation. Antiserum against OuMV CP (a) and tubule protein (b) was used. The arrow in (b) points to the OuMV-specific doublet band. (c) Coomassie blue staining of the same gel. Protein markers (kDa) are indicated to the left.
In preparations of infected crude sap, the absorbed tubule antiserum detected a doublet band running at about 30 kDa (Fig. 2b) specific to OuMV that was not detected in extracts of healthy or EpCV- and CsVC-infected plants (not shown).
Molecular characterization of OuMV, EpCV and CsVC.
RNA extracted from all three purified viruses and separated in a 1 % TAE gel showed two bands, the lower band present in greater abundance. The same samples run on 1.4 % agarose gels in HEPES buffer in denaturing conditions showed that the lower band resolved into two components of slightly different mobility (not shown). These results confirmed those of Marzachi et al. (1992) for OuMV.
The TAE gel-purified RNAs were used as templates for a cDNA library and we obtained a number of clones which hybridized specifically to viral RNAs. With OuMV, three cDNA probes (T2, B24 and B35) were used for Northern blot analysis and showed that we had cloned cDNAs specific for each of the three RNA segments (not shown). Assembly of contiguous cDNA clones allowed us to cover most of the three genome segments.
RT-PCR was carried out using an internal forward primer and the oligo-dT-V primer for reverse transcription. The negative result in this experiment confirmed the lack of a polyA tail at the 3′ ends, whereas, after artificial polyadenylation of the genomic segments, a band of the expected size was obtained in the same conditions (not shown).
The 5′ and 3′ ends of each segment were determined from the consensus of at least three clones for each RACE experiment. The same strategy was applied to purified RNA from EpCV and CsVC. Sequences of overlapping clones were assembled and deposited in GenBank. The main features derived from the full-length sequences of OuMV, EpCV and CsVC are shown in Table 1 and Fig. 3(a). The three viruses showed the same genome organization. Consistent coding capacity was present only in the positive sense of each of the three genomic RNAs, while a number of small ORFs were present in the negative sense sequences, but none of them was conserved in the three viruses (not shown).
Table 1. Comparison between the main genomic features of OuMV, EpCV and CsVC.
L, indicates the number of amino acids encoded by the respective ORFs; MW, predicted molecular mass (kDa); IP, predicted isoelectric point.
| Length of genomic RNA (bp) | ORF1 | ORF2 | ORF3 | |||||||||
| RNA1 | RNA2 | RNA3 | L | MW | IP | L | MW | IP | L | MW | IP | |
| OuMV | 2814 | 1064 | 974 | 860 | 97.5 | 8.9 | 288 | 31.6 | 8.9 | 210 | 23.8 | 9.7 |
| EpCV | 2769 | 1028 | 960 | 852 | 96.1 | 9.4 | 286 | 31.9 | 7.33 | 195 | 21.3 | 11.8 |
| CsVC | 2740 | 1137 | 958 | 870 | 98.5 | 9.3 | 289 | 31.5 | 8.23 | 214 | 23.6 | 10.5 |
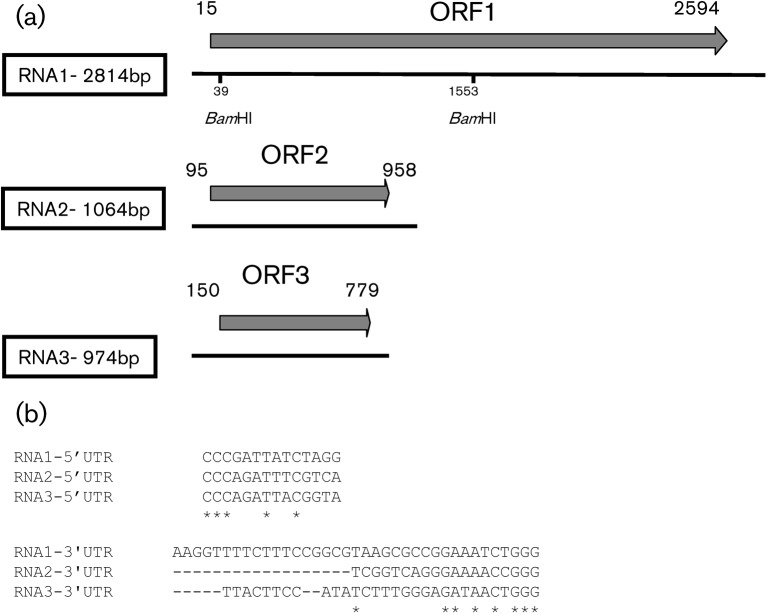
Fig. 3.
Main features of the ourmiaviruses genome organization derived from OuMV sequence analyses. (a) Diagram summarizing the OuMV genome organization. The filled arrows correspond to ORFs. The RNA lengths (bp) are shown in the box for each RNA. BamHI restriction enzyme sites are shown. (b) 5′ UTR and 3′ UTR conserved nucleotides (*) of the three OuMV RNAs.
The first three nucleotides at the 5′ end of each OuMV genome segment were the same (CCC) and the last three at the 3′ end were also the same (GGG), which implies that for the minus-strand synthesis, the same CCC sequences are also present at the polymerase initiation site. There were also conserved nucleotides at the 5′ untranslated region (UTR) and 3′UTR of OuMV (Fig. 3b). For EpCV, the 5′ and 3′ nucleotides in common to all three segments were CCCAG and TGGG, respectively. The three genomic segments of CsVC also had the same three nucleotides (CCC) at the 5′end and GGG at the 3′ end.
RNA1 of OuMV encoded a single protein of approximately 97.5 kDa with a putative palm domain of RNA-dependent RNA polymerase (RdRp) in the carboxy terminal part of the protein (Supplementary Data S1, available in JGV Online).
RNA2 and RNA3 of OuMV each carried one ORF in the positive sense potentially expressing proteins of 31.6 and 23.8 kDa, respectively. Through Western blot analysis of bacterially expressed recombinant proteins encoded by RNA2 and RNA3, we showed that the RNA2 ORF encodes the protein reacting with antibodies to the tubular structure, whereas RNA3 encodes the protein reacting with antibodies to the CP (not shown). Analogous results were obtained when the two proteins were agroinfiltrated from pBin61-derived clones: ORF3 accumulated abundantly, whereas ORF2 accumulated to a much lesser extent (Fig. 4).
Fig. 4.
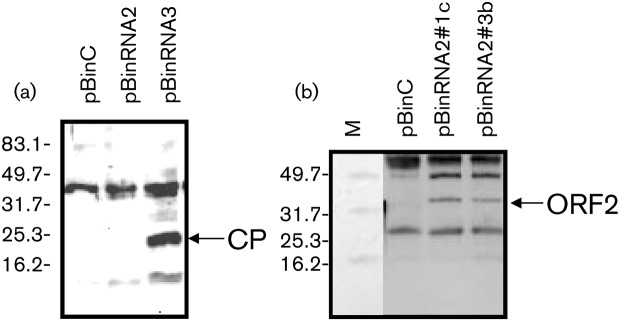
Western blot analysis for CP (a) and tubule structure protein (b) expressed through agroinfiltration in N. benthamiana 16C. pBinC was used as negative control. pBinRNA2#1c and pBinRNA2#3b are from two different agrobacterial colonies transformed with pBinRNA2. Antiserum against CP (a) and tubule structure protein (b) were used as probes. Arrows indicate the approximate positions of CP and ORF2-encoded protein (tubule structure). M, Protein molecular mass marker (kDa).
A number of commonly found motifs were present in the OuMV protein sequences (such as putative phosphorylation and N-myristoylation or N-glycosylation sites), but such putative sites were only conserved in the homologous gene products of EpCV and CsVC in two cases (protein kinase C phosphorylation sites in ORF2 protein and a low complexity region at the amino terminal of the ORF3 protein) (not shown).
Polarity of OuMV genomic RNAs.
Plus- and minus-sense RNA probes derived from full-length cDNA were used to test the polarity of the encapsidated RNA and that of the RNAs that accumulate during viral replication. Because viral RNA often displays extensive regions of internal pairing, we also included plus- and minus-strand full-length transcripts as controls. Fig. 5 shows a Northern blot carried out using RNA extracted from healthy plants, OuMV-infected plants, RNA extracted from purified OuMV particles and in vitro synthesized plus and minus strand OuMV full-length RNA2. We obtained a strong reaction between minus-strand probe and synthesized plus-strand RNA, RNA from purified virus and RNA from infected plants, whereas no reaction was obtained for the same minus-strand probe with in vitro synthesized minus-strand RNA (Fig. 5a). The plus-strand probe reacted with RNA extracted from infected plants and with minus-strand full-length transcripts, but not with plus-strand-synthesized OuMV RNA transcripts, or with RNA from purified OuMV (Fig. 5b). The same results were obtained when the same RNA extracts and the full-length RNA1 and RNA3 transcripts were hybridized with RNA1 and RNA3 plus- and minus-strand probes (not shown). We conclude that the RNAs encapsidated in OuMV particles are of positive sense, whereas a certain amount of minus-strand RNA accumulates during replication. No evidence of subgenomic RNA was observed in any of the Northern blots.
Fig. 5.
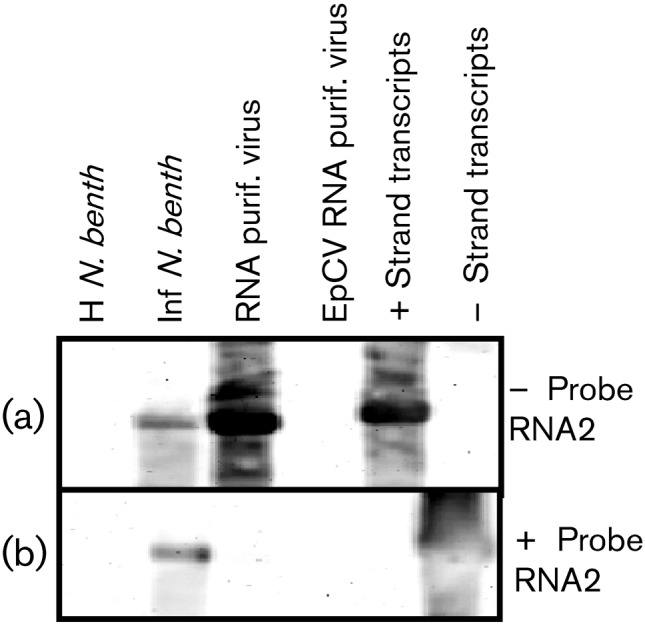
Northern blot analysis using 1.4 % agarose gels. Total RNA was extracted from healthy (H N. benth) and OuMV-infected (Inf N. benth) N. benthamiana and from purified OuMV (RNA purif. virus) and purified EpCV (EpCV RNA purif. virus). Minus- and plus-strand in vitro transcripts of RNA2 (− Strand transcripts and + Strand transcripts) were also run to evaluate self hybridization. Minus- (a) and plus- (b) strand probes for RNA2 were used to hybridize related RNAs.
Sequence similarity and phylogenetic analysis of ourmiavirus gene products.
The protein sequences of ourmiaviruses have no closely related homologues outside this group. Nevertheless, a detailed computational analysis of the proteins encoded by each of the three ourmiavirus genes revealed apparent evolutionary affinities with homologous proteins from distinct groups of RNA viruses. A search of the sequences of ssRNA viral proteins using the psi-blast program (Altschul et al., 1997) yielded a moderately significant similarity between the RNA1 product of ourmiaviruses and the RdRps of narnaviruses, viruses that infect fungi and encode the RdRp as the sole gene product (Buck et al., 2005). Specifically, the second iteration of psi-blast revealed a statistically significant alignment of the EpCV RNA 1 product with the RdRp of the Saccharomyces cerevisiae 20 S RNA narnavirus (expected value <10−4, 21 % identity). For the RNA2 product, limited but statistically significant similarity was detected with MPs of tombusviruses, in particular, those of the genus Aureusvirus (E-value <10−4 in the first psi-blast iteration, 24–26 % identity). Finally, for the RNA3 product, limited similarity was found with capsid proteins of several families of icosahedral plant and animal viruses including luteoviruses and nodaviruses. For instance, a search initiated with the CP sequence of EpCV as the query yielded alignment with E-values of approximately 0.03 with CP sequences of fish nodaviruses. A family-to-family profile alignment of capsid proteins shows that the capsid proteins of ourmiavirus exhibit approximately as much sequence similarity to those of picornaviruses and tombusviruses as those two groups of capsid proteins show to each other (Supplementary Table S2, available in JGV Online). The picornavirus and carmovirus (a member of the family Tombusviridae) capsid proteins are clearly homologous, with the vector alignment search tool (VAST) structural alignment spanning 152 positions at a root mean square distance of 3.0 Å (1HXS 3 vs 1OPO B). Thus, not unexpectedly, the structural similarity between capsid proteins of icosahedral viruses by far exceeds the sequence similarity so that, despite the relatively low sequence similarity, the capsid proteins of ourmiaviruses can be inferred to be homologous to those of other icosahedral positive-strand RNA viruses.
Overall, despite the high level of divergence, we conclude that the above results reflect bona fide homologous relationship between each of the three ourmiavirus genes and the corresponding genes of other viruses. Indeed, multiple alignments of the respective viral sequences were highly statistically significant for each ourmiavirus protein (Fig. 6), and contained all typical sequence motifs and structural elements of the RdRp, MP and CP (Supplementary Data S1–S3 and data not shown).
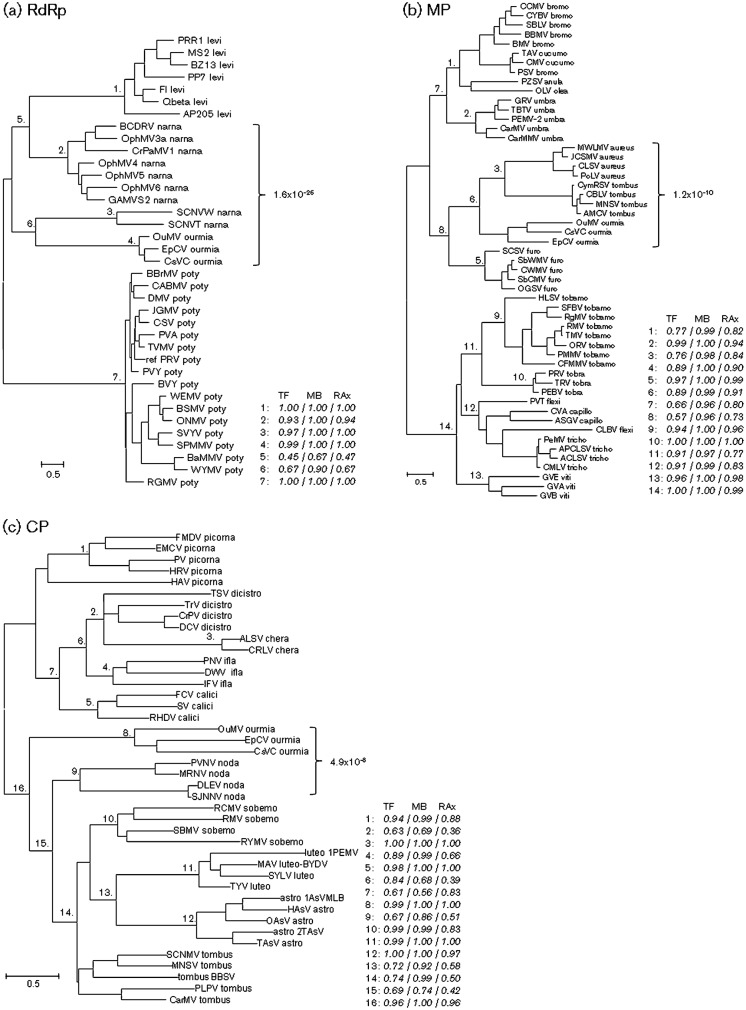
Fig. 6.
Phylogenies of ourmiavirus proteins RdRp (a), MP (b) and capsid protein (c). The ML trees were constructed as described in Methods. Selected internal branches are marked by numbers; the support for each of these nodes obtained using TreeFinder (TF), MrBayes (MB) and RAxML (RAx) is indicated. The P-value (calculated using HHalign) for the multiple alignment of ourmiavirus proteins with the most similar homologues from other viruses (denoted by a bracket) are shown. For each leaf of the tree, the abbreviated name of the virus and the family to which it belongs are indicated. Abbreviations are given in Supplementary Table S3, where the extended name of the virus species and the protein accession numbers are also given.
We used phylogenetic tree analysis in an attempt to reveal the specific phylogenetic affinities of the ourmiavirus genes (the GenBank accession numbers, viral species names and abbreviations of the sequences used for phylogenetic analysis are given in Supplementary Table S3, available in JGV Online). Phylogenetic analysis using three different ML methods and the Bayesian MCMC method consistently supported the affinity of the ourmiavirus RdRp with the RdRps of a subset of narnaviruses (Fig. 6a and Supplementary Fig. S1a), and the ourmiavirus MP with the MPs of tombusviruses (Fig. 6b and Supplementary Fig. S1b). The tree of the RdRps shown in Fig. 6(a) does not include polymerases of tombusviruses, so the possibility that the phylogenies of the RdRps and the MPs are congruent could not be formally rejected on the basis of this tree alone. Tombusvirus sequences were not part of the phylogenetic analysis of the RdRps because they are much less similar to the included sequences than the latter are to one another, which led to a considerable decrease in the number of confidently aligned positions under the employed criteria: there were 113 confidently aligned positions in the alignment used for the construction of the tree in Fig. 6(a), whereas, when tombusviruses were included, only 89 confidently aligned positions were obtained. Nevertheless, in a tree that included tombusvirus sequences, the affiliation of the ourmiaviruses with narnaviruses was retained (Supplementary Fig. S1a). The phylogeny of the CP is harder to interpret because, in this case, ourmiaviruses formed a deep branch that joined a diverse group including viruses that infect either plants (sobemo-, tombus- and luteoviruses) or animals (nodaviruses) (Fig. 6c and Supplementary Fig. S1c). It cannot be determined whether the CP gene of ourmiaviruses was derived from that of an icosahedral plant virus, such as a tombusvirus or a luteovirus, although the putative ancestral relationship is obscured by the length of the ourmiavirus branch, presumably resulting from the fast evolution of this gene in ourmiaviruses.
Discussion
OuMV, EpCV and CsVC isolated from different countries and hosts were found to possess very similar genome organizations, comprising three plus-strand RNAs each carrying one ORF. The genome seems to be minimal for viruses possessing an external phase, apparently encoding only an RdRp (ORF1 in RNA1), an MP (ORF2 in RNA2) and a CP (ORF 3 in RNA3). Among positive-sense ssRNA viruses, the combination appears unique in that the three genome segments each express one ORF, with no subgenomic RNAs. Sequence comparisons of the RNAs and proteins of the three viruses, together with data on their geographical locations, natural hosts and particle structures, indicate that they should be confirmed as three species in the same genus.
ORF2 encodes a protein that appears to be homologous to many MPs of plant viruses, with the most pronounced similarity and phylogenetic affinity with the MP of tombusviruses. This evidence is reinforced by the electron microscopy results showing that the product of OuMV ORF2 forms tubules that enclose the virions and pass through the cell walls via plasmodesmata. In this respect, ourmiaviruses resemble the families Bromoviridae, Comoviridae and Caulimoviridae, together with the genera Tospovirus and Trichovirus, in possessing an approximately 30 kDa MP and employing the tubule strategy to move from cell to cell (Hull, 2002). Plant virus MPs are often phosphorylated (Lee & Lucas, 2001) and the presence of a protein doublet reacting with tubule antibodies in Western blots of infected plant sap could be explained by a post-translational modification. In agreement with this hypothesis, there are some conserved putative protein kinase C phosphorylation sites in ORF2 (not shown).
The observed sequence conservation and phylogenetic affinities that were strongly statistically supported in the case of the RdRp and the MP (Fig. 6) suggest that ourmiaviruses possess chimeric genomes that might have evolved as a result of an association of RNA segments from at least two, but more likely three, distinct sources. The most parsimonious scenario for the origin of ourmiaviruses appears to be that a capsid-less narnavirus of a fungal plant pathogen or endophyte became a distinct plant virus as a result of the acquisition of the MP and CP genes from plant viruses. In the phylogenetic tree of the RdRps, the ourmiavirus branch is positioned within the narnavirus cluster, an observation that suggests a relatively late origin of the ourmiaviruses, postdating the divergence of the known narnaviruses. It seems most likely that the MP gene was acquired from a member of the Tombusviridae, whereas the CP gene might have been acquired from either the same or (more likely) a distinct virus.
Indications of distinct origins of different genes, which suggests that the origin of a viral genome is through recombination between different viruses, are common among plant viruses. In particular, the same family of MPs is involved in the systemic infection of plants by a wide range of viruses, including numerous families of RNA viruses and also the DNA-containing geminiviruses that are otherwise completely unrelated to RNA viruses (Koonin et al., 1991b; Melcher 2000). Thus, the extensive horizontal mobility of the MP gene that is a pre-requisite for systemic spread of a virus in the plant host (and therefore would be readily fixed in a viral genome that acquires it anew) is beyond doubt. Viruses in which the RdRp and CP genes are of distinct origins, indicating recombination during the evolution of the respective viral groups, are also known. For instance, potyviruses possess filamentous capsids, in contrast with other viruses of the broadly defined picorna-like viral superfamily that typically possess icosahedral capsids, indicating the role of an ancient recombination with a filamentous virus in the emergence of the family Potyviridae (Koonin et al., 2008).
However, the apparent chimerism seen in ourmiaviruses is so far unique in that each of the three segments of the viral genome was most probably derived from a separate source, implicating reassortment of genomic segments rather than recombination as the mechanism of evolution. The origin of ourmiaviruses might have involved the capture of subgenomic RNAs encoding the MP and CP from two distinct plant viruses or, possibly, two subgenomic RNAs from the same virus; these RNAs became genome segments of the ancestral ourmiavirus. Viral genomic segment reassortment between viruses belonging to the same genus or family was previously shown to play a role in the evolution of new species of plant viruses (White et al., 1995; Codoñer & Elena, 2008); however, reassortment between viruses with hosts from different kingdoms has not been reported so far. Furthermore, the ourmiavirus RdRp seems to be derived from a fungal virus. Evolutionary connections between plant and fungal viruses have been reported previously but they seem to differ from the ourmiavirus case in important aspects. The hypoviruses of plant-pathogenic fungi are related to the plant potyviruses (Koonin et al., 1991a); in this case, considering the evolutionary relationship between potyviruses and other picorna-like viruses, the direction of evolution seems to be from a plant pathogen to a fungal pathogen, with the CP gene being lost during the transition to the new host (Koonin et al., 2008). Another example of fungal viruses being phylogenetically affiliated with plant viruses is that of Botrytis cinerea virus F and Sclerotinia sclerotiorum debilitation-associated RNA virus, two viruses shown to belong to the family Flexiviridae, a plant virus taxon (Martelli et al., 2007), and hypothesized to be derived from a plant virus through loss of MP in one case, and of MP and CP in the second example. Southern tomato virus, a recently reported dsRNA virus isolated from tomato, was recently reported to be a link between the families Partitiviridae and Totiviridae (Sabanadzovic et al., 2009). The case of Cucurbit yellows associated virus (CYAV) RdRp that is highly similar to the RNA1 products of Agaricus bisporus virus 1 (Hong et al., 1998) is complicated by the fact that this RdRp was never clearly shown to come from a plant virus (Coffin & Coutts, 1995), and could be derived from a virus of a contaminant (or endophytic) fungus as is the case for another Totiviridae dsRNA extracted from diseased plants (Covelli et al., 2004; Kozlakidis et al., 2006). By contrast, in the case of the ourmiaviruses, there is no room for doubt that they really are plant viruses. Conversely, plant endornaviruses that replicate inside mitochondria seem to be derived from typical fungal, capsid-less, virus-like replicons (Fukuhara et al., 2006); essentially, endornaviruses retain the typical replication strategy of fungal RNA replicons even after the transfer to plants. The same fungal character was retained by the plant cryptoviruses (lacking an MP) in the family Partitiviridae (Fauquet et al., 2005; Rong et al., 2002). The ourmiaviruses are different from these cases in that a regular fungal virus appears to have evolved into a typical plant virus by capturing RNA segments encoding the proteins responsible for virion formation and systemic spread in plants.
The unique genome composition and the distant relationship between the RdRp, MP and CP genes of ourmiaviruses and the homologous genes from other plant viruses strongly suggest that ourmiaviruses should be classified in a novel viral family. This conclusion is reinforced by the unique particle morphology. The unique evolutionary route that led to the emergence of these viruses expands our ideas on the diversity of the paths of viral evolution.
AKNOWLEDGMENTS
We thank Maud Swanson (formerly Aiton), Scottish Crop Research Institute, for kindly providing isolates of CsVC, and Caterina Perrone for skilful technical assistance in the greenhouse. M. R. was recipient of a scholarship from the Iran Ministry of Science, Research and Technology, as part of her PhD project.
Footnotes
A supplementary figure, three tables and three sequence alignments (in FASTA format) are available with the online version of this paper.
References
- Accotto G. P., Milne R. G. (2008). Ourmiavirus. In Encyclopedia of Virology, pp. 500–501 Edited by Mahy B. W. J., Van Regenmortel M. H. V. London, UK: Elsevier [Google Scholar]
- Accotto G. P., Boccardo G., Riccioni L., Barba M. (1997). Comparison of some molecular properties of Ourmia melon and Epirus cherry viruses, two representatives of a proposed new virus group. J Plant Pathol 78, 87–91 [Google Scholar]
- Aiton M. M., Lennon A. M., Roberts I. M., Harrison B. D. (1988). Two new cassava viruses from Africa. In 5th International Congress of Plant Pathology, Kyoto, Japan, 1988, p. 43 [Google Scholar]
- Altschul S. F., Madden T. L., Schaffer A. A., Zhang J., Zhang Z., Miller W., Lipman D. J. (1997). Gapped blast and psi-blast: a new generation of protein database search programs. Nucleic Acids Res 25, 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avgelis A., Barba M., Rumbos I. (1989). Epirus cherry virus, an unusual virus isolated from cherry with rasp-leaf symptoms in Greece. J Phytopathol 126, 51–58 [Google Scholar]
- Bendahmane A., Kanyuka K., Baulcombe D. C. (1999). The Rx gene from potato controls separate virus resistance and cell death responses. Plant Cell 11, 781–791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendahmane A., Farnham G., Moffett P., Baulcombe D. C. (2002). Constitutive gain-of-function mutants in a nucleotide binding site – leucine rich repeat protein encoded at the Rx locus of potato. Plant J 32, 195–204 [DOI] [PubMed] [Google Scholar]
- Buck K. W., Esteban R., Hillman B. I. (2005). Family Narnaviridae. In Virus Taxonomy: Eighth Report of the International Committee for the Taxonomy of Viruses, pp. 751–756 Edited by Fauquet C. M., Mayo M. A., Maniloff J., Desselberger U., Ball L. A. San Diego, CA: Elsevier [Google Scholar]
- Codoñer F. M., Helena S. F. (2008). The promiscuous evolutionary history of the family Bromoviridae. J Gen Virol 89, 1739–1747 [DOI] [PubMed] [Google Scholar]
- Coffin R. S., Coutts R. H. A. (1995). Relationships among Trialeurodes vaporariorum-transmitted yellowing viruses from Europe and North America. J Phytopathol 143, 375–380 [Google Scholar]
- Covelli L., Coutts R. H. A., Di Serio F., Citir A., Açkgöz S., Hernández C., Ragozzino A., Flores R. (2004). Cherry chlorotic rusty spot and Amasya cherry diseases are associated with a complex pattern of mycoviral-like double-stranded RNAs. Characterization of a new species in the genus Chrysovirus. J Gen Virol 85, 3389–3397 [DOI] [PubMed] [Google Scholar]
- Edgar R. C. (2004). muscle: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32, 1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fauquet C. M., Mayo M. A., Maniloff J., Desselberger U., Ball L. A. (2005). Virus Taxonomy, Eighth Report of the International Committee on Taxonomy of Viruses. San Diego, CA: Elsevier [Google Scholar]
- Fukuhara T., Koga R., Aoki N., Yuki C., Yamamoto N., Oyama N., Udagawa T., Horiuchi H., Miyazaki S., et al. (2006). The wide distribution of endornaviruses, large double-stranded RNA replicons with plasmid-like properties. Arch Virol 151, 995–1002 [DOI] [PubMed] [Google Scholar]
- Gholamalizadeh R., Vahdat A., Hossein-Nia S. V., Elahinia A., Bananej K. (2008). Occurrence of Ourmia melon virus in the Guilan Province of Northern Iran. Plant Dis 92, 1135. [DOI] [PubMed] [Google Scholar]
- Gibrat J. F., Madej T., Bryant S. H. (1996). Surprising similarities in structure comparison. Curr Opin Struct Biol 6, 377–385 [DOI] [PubMed] [Google Scholar]
- Guindon S., Gascuel O. (2003). A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol 52, 696–704 [DOI] [PubMed] [Google Scholar]
- Hirzmann J., Luo D., Hahnen J., Hobom G. (1993). Determination of messenger RNA 5′- ends by reverse transcription of the cap structure. Nucleic Acids Res 21, 3597–3598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Höfgen R., Willmitzer L. (1988). Storage of competent cells for Agrobacterium transformation. Nucleic Acids Res 16, 9877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong Y., Cole T. E., Brzsier C. M., Buck K. W. (1998). Evolutionary relationships among putative RNA-dependent RNA polymerases encoded by a mitochondrial virus-like RNA in the Dutch elm disease fungus, Ophiostoma novo-ulmi, by other viruses and virus-like RNAs and by the Arabidopsis mitochondrial genome. Virology 246, 158–169 [DOI] [PubMed] [Google Scholar]
- Hull R. (2002). Matthews’ Plant Virology, 4th edn New York: Academic Press [Google Scholar]
- Jobb G., von Haeseler A., Strimmer K. (2004). treefinder: a powerful graphical analysis environment for molecular phylogenetics. BMC Evol Biol 4, 18. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Koonin E. V., Choi G. H., Nuss D. L., Shapira R., Carrington J. C. (1991a). Evidence for common ancestry of a chestnut blight hypovirulence-associated double-stranded RNA and a group of positive-strand RNA plant viruses. Proc Natl Acad Sci U S A 88, 10647–10651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin E. V., Mushegian A. R., Ryabov E. V., Dolja V. V. (1991b). Diverse groups of plant RNA and DNA viruses share related movement proteins that may possess chaperone-like activity. J Gen Virol 72, 2895–2903 [DOI] [PubMed] [Google Scholar]
- Koonin E. V., Wolf Y. I., Nagasaki K., Dolja V. V. (2008). The Big Bang of picorna-like virus evolution antedates the radiation of eukaryotic supergroups. Nat Rev Microbiol 6, 925–939 [DOI] [PubMed] [Google Scholar]
- Kozlakidis Z., Covelli L., Di Serio F., Citir A., Açkgöz S., Hernández C., Ragozzino A., Flores R., Coutts R. H. A. (2006). Molecular characterization of the largest mycoviral-like double-stranded RNAs associated with Amasya cherry disease, a disease of presumed fungal aetiology. J Gen Virol 87, 3113–3117 [DOI] [PubMed] [Google Scholar]
- Lee J.-Y., Lucas W. J. (2001). Phosphorylation of viral movement proteins – regulation of cell-to-cell trafficking. Trends Microbiol 9, 5–8 [DOI] [PubMed] [Google Scholar]
- Lisa V., Milne R. G., Accotto G. P., Boccardo G., Caciagli P., Parvizy R. (1988). Ourmia melon virus, a virus from Iran with novel properties. Ann Appl Biol 112, 291–302 [Google Scholar]
- Luisoni E., Milne R. G., Vecchiati M. (1995). Purification of Tomato yellow leaf curl geminivirus. New Microbiol 18, 253–260 [PubMed] [Google Scholar]
- Margaria P., Ciuffo M., Pacifico D., Turina M. (2007). Evidence that the nonstructural protein of Tomato spotted wilt virus is the avirulence determinant in the interaction with resistant pepper carrying the Tsw gene. Mol Plant Microbe Interact 20, 547–558 [DOI] [PubMed] [Google Scholar]
- Martelli G. P., Adams M. J., Kreuze J. F., Dolja V. V. (2007). Family Flexiviridae: a case study in virion and genome plasticity. Annu Rev Phytopathol 45, 73–100 [DOI] [PubMed] [Google Scholar]
- Marzachi C., Antoniazzi S., D’Aquilio M. A., Boccardo G. (1992). Molecular cloning of the segmented RNA genome of Ourmia melon virus. Riv Patol Veg S 2, 3–8 [Google Scholar]
- Matthews R. E. F. (1991). Plant Virology, 3rd edn San Diego, CA: Academic Press [Google Scholar]
- Melcher U. (2000). The ‘30K’ superfamily of viral movement proteins. J Gen Virol 81, 257–266 [DOI] [PubMed] [Google Scholar]
- Milne R. G. (1993). Electron microscopy of in vitro preparations. In Diagnosis of Plant Virus Diseases, pp. 215–251 Edited by Matthews R. E. F. Boca Raton: CRC Press [Google Scholar]
- Milne R. G. (2005). Genus Ourmiavirus. In Virus Taxonomy, Eighth Report of the International Committee on Taxonomy of Viruses, pp. 1059–1061 Edited by Fauquet C. M., Mayo M. A., Maniloff J., Desselberger U., Ball L. A. San Diego, CA: Elsevier [Google Scholar]
- Milne R. G., Ramasso E., Lenzi R., Masenga V., Sarindu N., Clark M. F. (1995). Pre- and post-embedding immunogold labelling and electron microscopy in plant host tissue of three antigenically unrelated MLOs: primula yellows, tomato big bud and bermudagrass white leaf. Eur J Plant Pathol 101, 57–67 [Google Scholar]
- Rong R., Rao S., Scott S. W., Carner G. R., Tainter F. H. (2002). Complete sequence of the genome of two dsRNA viruses from Discula destructiva. Virus Res 90, 217–224 [DOI] [PubMed] [Google Scholar]
- Ronquist F., Huelsenbeck J. P. (2003). MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19, 1572–1574 [DOI] [PubMed] [Google Scholar]
- Sabanadzovic S., Valverde R. A., Brown J. K., Martin R. R., Tzanetakis I. E. (2009). Southern tomato virus: the link between the families Totiviridae and Partitiviridae. Virus Res 140, 130–137 [DOI] [PubMed] [Google Scholar]
- Sambrook J., Fritsch E. F., Maniatis T. (1989). Molecular Cloning: a Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory [Google Scholar]
- Sanger F., Nicken S., Coulson A. R. (1977). DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A 74, 5463–5467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soding J. (2005). Protein homology detection by HMM–HMM comparison. Bioinformatics 21, 951–960 [DOI] [PubMed] [Google Scholar]
- Stamatakis A., Hoover P., Rougemont J. (2008). A rapid bootstrap algorithm for the RAxML Web servers. Syst Biol 57, 758–771 [DOI] [PubMed] [Google Scholar]
- Turina M., Desvoyes B., Scholthof K. B. G. (2000). A gene cluster encoded by Panicum mosaic virus is associated with virus movement. Virology 266, 120–128 [DOI] [PubMed] [Google Scholar]
- Turina M., Prodi A., Van Alfen N. K. (2003). Role of the Mf1–1 pheromone precursor gene of the filamentous ascomycete Cryphonectria parasitica. Fungal Genet Biol 40, 242–251 [DOI] [PubMed] [Google Scholar]
- Turina M., Ciuffo M., Lenzi R., Rostagno L., Mela L., Derin E., Palmano S. (2006). Characterization of four viral species belonging to the family Potyviridae isolated from Ranunculus asiaticus. Phytopathology 96, 560–566 [DOI] [PubMed] [Google Scholar]
- Vaira A. M., Milne R. G., Accotto G. P., Luisoni E., Masenga V., Lisa V. (1997). Partial characterization of a new virus from ranunculus with a divided RNA genome and circular supercoiled thread-like particles. Arch Virol 142, 2131–2146 [DOI] [PubMed] [Google Scholar]
- Wheeler D. L., Barrett T., Benson D. A., Bryant S. H., Canese K., Chetvernin V., Church D. M., Dicuccio M., Edgar R., et al. (2008). Database resources of the National Center for Biotechnology Information. Nucleic Acids Res 36, D13–D21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whelan S., Goldman N. (2001). A general empirical model of protein evolution derived from multiple protein families using a maximum-likelihood approach. Mol Biol Evol 18, 691–699 [DOI] [PubMed] [Google Scholar]
- White P. S., Morales F., Roossinck M. J. (1995). Interspecific reassortment of genomic segments in the evolution of Cucumoviruses. Virology 207, 334–337 [DOI] [PubMed] [Google Scholar]
- Yutin N., Makarova K. S., Mekhedov S. L., Wolf Y. I., Koonin E. V. (2008). The deep archaeal roots of eukaryotes. Mol Biol Evol 25, 1619–1630 [DOI] [PMC free article] [PubMed] [Google Scholar]