Abstract
Modifications of histones, the chief protein components of the chromatin, have emerged as critical regulators of life and death. While the “apoptotic histone code” came to light a few years ago, accumulating evidence indicates that autophagy, a cell survival pathway, is also heavily regulated by histone-modifying proteins. In this review we describe the emerging “autophagic histone code” and the role of histone modifications in the cellular life vs. death decision.
Keywords: histone posttranslational modifications, histone code, HAT, HDAC, HDM, HMT, KAT8/hMOF/MYST1, EHMT2/G9a, SUV420H2
Introduction
Wrap and pack: Histones are chief protein components of chromatin
Chromatin is the polymer formed by DNA together with related histone proteins. It is organized in repeats of nucleosomes that contain an octamer of histones formed by the 4 core histones—H2A, H2B, H3, and H4, and 2 laps of, on average, around 147 DNA base pairs that wrap around the histone octamer.1,2 (Fig. 1). The nucleosomal structure resembles “beads on a string,” where the “beads” represent the individual nucleosomes and the “string” represents the linker DNA. Linker histones such as histone H1, and other nonhistone proteins can bind to the nucleosomal arrays to compact the chromatin further, thereby contributing to the formation of higher-order chromatin structures.
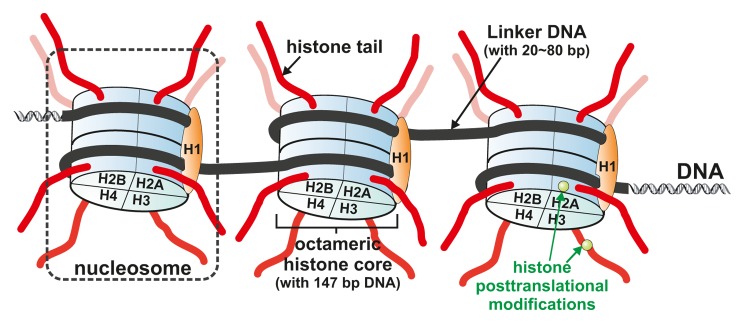
Figure 1. Chromatin structure. Chromatin is made of repeating units of nucleosomes, which consist of ~147 base pairs of DNA wrapped around a histone octamer consisting of 2 copies each of the core histones H2A, H2B, H3, and H4. Linker histone H1 is positioned on top of the nucleosome core particles stabilizing higher order chromatin structure. The histones are subject to a wide variety of posttranslational modifications, primarily on their N-terminal tails, but also in their globular core region.
Decorating histones and their tails: Histone posttranslational modifications
Like other proteins, histones are subject to posttranslational modifications (PTMs), including phosphorylation, ubiquitination, methylation, and acetylation. These modifications and the corresponding demodifications are all executed and governed by specific enzyme complexes.3 Important groups of enzymes in this histone regulatory network are the histone acetyltransferases (HATs or KATs for lysine specificity), the histone deacetylases (HDACs), histone methyltransferases (HMTs) and histone or lysine demethylases (KDMs). The most well- recognized action of a HAT is the addition of an acetyl group to lysines on the N terminus of histones, by which they mediate chromatin decondensation. HDACs substract the acetyl groups and induce a tighter binding of histones to DNA.4,5 Histone methylation is more complex, as methylation comes in 3 steps, mono-, di- and trimethylation, and the effect of lysine methylation of a histone depends strictly on which lysine is methylated and to what extent. Histone PTMs are especially prevalent within the histone N-terminal tails, which protrude from the nucleosomal surface, but are also on the histone globular core region.6,7 These histone PTMs have the potential to affect the overall structure of chromatin with clear functional consequences.5,8 Histone modifications contribute to the regulation of transcription, transcriptional elongation, higher order chromatin structure, and other fundamental events. An early demonstration showed that PTMs can alter the electrostatic charge of the histone thus altering the DNA binding ability, and thereby regulating the packaging of chromatin and the access of DNA-binding proteins such as transcription factors. Although still valid, recent studies have demonstrated many other models for the effects resulting from chromatin modification. A major role for the PTMs is to function as signal platforms, or “beacons,” to recruit specific modules to local chromatin, in a context- and modification-dependent manner. Such modules may initiate or repress transcription directly, but many new studies also suggest that such modules can influence the stepwise recruitment of secondary modules and thereby transcription rate, protection of DNA, and sensitivity to signals in a more fine-tuned manner.9
What would histone posttranslational modifications do during autophagy?
Macroautophagy, hereafter referred to as autophagy, is a conserved, typically catabolic, process that leads to the lysosomal degradation of cytoplasmic components, abnormal protein aggregates, and damaged organelles.10,11 Autophagy involves dynamic membrane-rearrangements controlled by a set of autophagy-related (ATG) proteins.12 The kinase mechanistic target of rapamycin (MTOR) is a central regulator for autophagy induction, where activated MTOR suppresses autophagy, and negative regulation of MTOR promotes it. Paradoxically, while autophagy is considered to be a protective process, it can be involved in cell death as well;13 however, it is yet unclear what regulates this life or death decision.14 Autophagy is implicated in a number of physiological processes important for human health and disease.10 Therefore, understanding the pathways regulating the autophagic life and death decision and its cellular long-term effects might help to improve autophagy-based clinical treatments.15,16 A role for histone PTMs in autophagy was not obvious at first; indeed until recently, nuclear regulation of autophagy was not expected as reports indicated that cells without a nucleus can still accumulate autophagic vesicles as a response to various inducers of autophagy.17 However, in recent years, compelling evidence revealed that de facto the nucleus is a major regulator of autophagy. Indeed, the autophagic process appears to encompass transcriptional and epigenetic programs, including histone PTMs. Here we will review histone PTMs that have been directly or indirectly linked to autophagy, and their functional consequence on this biological process.
Autophagy-Related Histone Modifications
First attempts, first observations, and a first hint
First attempts to link autophagy to histone modifications came from the use of HDAC inhibitors, which promote histone hyperacetylation and thereby affect gene expression patterns. In 2004, Shao and coworkers presented data indicating that the HDAC inhibitors butyrate and suberoylanilide hydroxamic acid (SAHA) can induce autophagic cell death in a range of human cancer cell lines.18 Of note, these investigations were performed before the discovery that the acetylation level of histones can regulate autophagy, which led to a focus on the interpretation of the effects of HDAC inhibitors toward nonhistone targets.19-21 However, a first report of histone PTMs occurring upon autophagy induction, came from work by Madeo, Kroemer, and coworkers. In aging yeast, they determined that the cytoprotective induction of autophagy by spermidine relies on the inhibition of HAT activity, in particular Iki3 and Sas3, which causes global hypoacetylation of histone H3, and therefore probably reflects the repression of gene expression. In contrast, some genes, for example ATG genes, seem to be protected from this global hypoacetylation, and therefore maintain accessible promoters, allowing their selective transcription. The authors propose that this might be an important mechanism for saving resources during starvation.22 Nevertheless, to our knowledge, to identify the first hint on the potential regulation of autophagy by histone PTMs, one has to look back to the beginning of this millennium. Indeed in 2001, Yamaki and colleagues demonstrated, that yeast lacking the histone chaperone ASF1A (anti-silencing function 1A histone chaperone), undergo cell death exhibiting a plethora of autophagic bodies.23 In yeast, acetylation of histone H3 lysine 56 (H3K56ac) is catalyzed by the HAT Rtt109, which requires Asf1 for function.24
H3K56ac
Histone H3 lysine 56 is located at a rather unique position in the nucleosome; this mark localizes at the entry and exit points of a nucleosome. Noteworthy the histone-DNA interactions at these sites are weakened by H3K56 acetylation.25 Furthermore, acetylation of H3K56 has a crucial role for the integration of histone variants and for packaging of DNA into chromatin following DNA replication and repair. H3K56ac also contributes to the control of gene expression.25-28 Remarkably the use of a histone H3-H4 yeast mutant library identified specifically the H3K56ac residue as being reduced by the TOR inhibitor rapamycin. It was proposed that TOR signaling regulates H3K56ac through repression of the H3K56 HDACs Hst3 and Hst4.28 In humans, both EP300/KAT3B/p300 and KAT2A/GCN5 are responsible for the acetylation of H3K56.29 In fact, knockdown of EP300 can stimulate autophagy, whereas overexpression of EP300 inhibits starvation-induced autophagy.30 However, the actual effect of the EP300 histone acetyltransferase activity on autophagy remains to be determined, since EP300 regulates the acetylation of various known components of the autophagy machinery, including ATG5, ATG7, MAP1LC3, and ATG12.30 Furthermore, it was proposed that KAT2A can generate acetylated substrates for the autophagic pathway and/or directly promote autophagy.31 The mechanism regulating deacetylation of H3K56 is controversial; indeed 5 different HDACs—HDAC1, HDAC2 SIRT1, SIRT2, and SIRT3—have been described to deacetylate this residue.8,29,32,33 Remarkably all of these HDACs are regulators of autophagy, reversing acteylation of nonhistone targets such as ATGs, and certainly also due to their action on intrinsic natural histone targets.19,34,35 In summary, acetylation of H3K56, catalyzed by EP300 or KAT2A (Rtt109 in yeast) and displaced by various HDACs including SIRT1 to SIRT3 and HDAC1/2 (Hst3 and Hst4 in yeast), appears to be positively regulated by the MTOR-signaling pathway and repressed upon autophagy induction (Fig. 2).
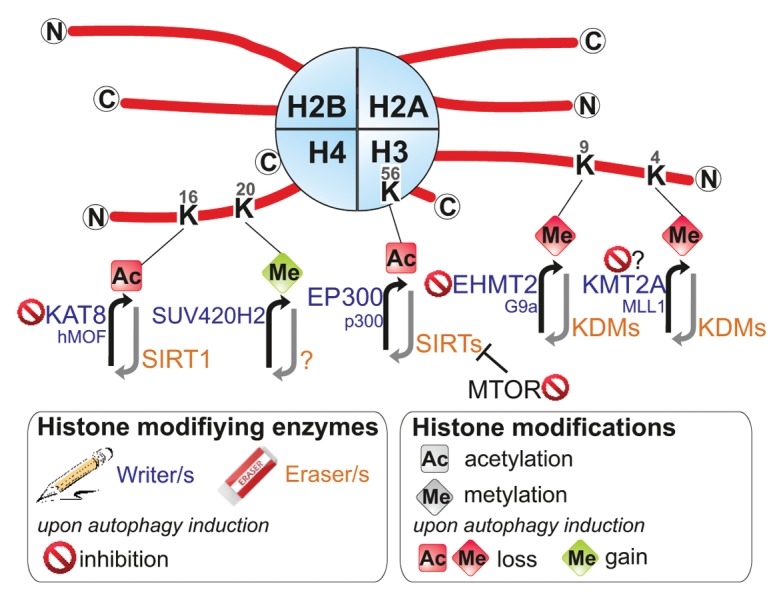
Figure 2. Autophagic histone modifications. Posttranslational modifications on histone tails that occur in autophagic cells are represented. These covalent modifications are added or removed by histone modifying enzymes often termed as “writers” and “erasers.” The modifications shown are discussed in the main text. ac, acetylated; me, methylated.
H4K16ac
Unlike most histone modifications, acetylation of histone H4 lysine 16 (H4K16ac) plays a central role for the regulation of higher-order chromatin structures beyond the level of nucleosomes. In particular, H4K16ac influences the functional interactions between histones and the chromatin fiber, providing a potential regulatory mechanism for chromatin folding and playing an important role in transcription.36 In multiple cell types, the induction of autophagy, by various stimuli, has been coupled to a reduction of H4K16ac. In addition, genome-wide H4K16 deacetylation is associated predominantly with the downregulation of autophagy-related genes, such as ATG9A, GABARAPL2, MAP1LC3, ULK1, ULK3, and VMP1.37 Remarkably, whereas many HATs show either little substrate specificity or preference for other residues, KAT8/hMOF/MYST1 is necessary for the majority of H4K16 acetylation. KAT8’s antagonizer is the HDAC SIRT1, which targets a plethora of nonhistone proteins, but has H4K16 as its primary histone target.38 It appears that KAT8 downregulation is part of the autophagy program, which in turn contributes to reduction of H4K16ac.37 The observed dramatic changes in the levels of H4K16 acetylation, through the downregulation of KAT8, and associated transcriptional gene regulation suggested that there might be a functional role for this epigenetic change during autophagy. Investigations of the autophagic flux revealed that inhibition of the autophagic H4K16 deacetylation does not inhibit autophagy, but on the contrary increases autophagic flux. The observed increase in autophagic flux is in accordance with the genome-wide investigations, which provide compelling evidence that H4K16 deacetylation is directly linked to a downregulation of autophagy-related genes.37 In summary, KAT8 is a substrate for autophagy, and its degradation and the consequent reduction of H4K16 acetylation, and the H4K16 deacetylation-dependent regulation of autophagy-related genes, in turn, regulates the autophagic flux.
H3K4me3
Di- and trimethylation of histone H3 at lysine 4 (H3K4me2/me3) is involved in transcriptional competence and activation. The highest H3K4me3 levels occur near the transcriptional start sites of highly expressed genes.39 H3K4 is methylated by the SET1 and mixed lineage leukemia family of HMTs40 and removed by KDM1A/LSD1 (lysine (K)-specific demethylase 1A) and KDM5/JARID1 [lysine (K)-specific demethylase 5] family of histone lysine demethylases (KDMs).41 Genome-wide investigations showed that the H4K16ac and H3K4me3 histone marks reside within single nucleosomal units in human cells.42,43 The coexistence of H4K16ac with H3K4me3 marks is consistent with the identification of multiple molecular interactions between the enzymes that are responsible for installing these marks, KMT2A/MLL1 [lysine (K)-specific methyltransferase 2A] and KAT8.42,44,45 In agreement with this molecular connection, autophagy also leads to a reduction in H3K4me3 levels in a wide variety of cell lines from yeast to humans. The disappearance of H3K4me3 alongside H4K16ac is likely to be involved in a general transcriptional inhibition to save energy during prolonged starvation. Interestingly, the WNT/β-catenin signaling pathway has recently been reported as a negative regulator of autophagy by inhibiting the transcription of SQSTM1/p62. During autophagy WNT is released from the SQSTM1 promoter and degraded, increasing SQSTM1 gene expression and autophagy levels.46 Furthermore, WNT interacts with, and induces, mixed lineage leukemia that promotes H3K4me3 levels at promoters.47,48 By blocking WNT in propagating tumor cells a strong reduction in global H3K4me3 levels is observed.48 Hence, the degradation of WNT during autophagy is potentially involved in the downregulation of global H3K4me3 levels.
H4K20me3
H4K20 methylation is associated with transcriptional repression.49 Hence, H4K20me3 is present in constitutive heterochromatin regions,49,50 especially in chromosomal regions that contain silenced genes.51 The methylation of H4K20 is performed by a series of HMTs, including SETD8 and SUV420 (SUV420H1 and SUV420H2). SETD8 is responsible for the majority of the monomethylation on H4K20. H4K20me1 then acts as a substrate for the SUV4-20 enzymes, which are responsible for the methylation reactions to H4K20me2/3. PHF8 (PHD finger protein 8) has been identified as a H4K20 demethylase. Interestingly, serum starvation, a prominent inducer of autophagy, has a marked impact on the global level of H4K20me3. Thus, Kourmouli et al. described how a decrease in serum in the culture medium from 10% to 0.1% leads to a significant, serum concentration dependent, increase in H4K20me3. Furthermore, the amount of brightly stained cells increases from 10% up to 90% under serum starvation.50 Supporting the role of H4K20me3 as a functional component of autophagy regulation is the fact that the previously described autophagy regulatory H4K16ac mark is heavily connected to H4K20me3. Recently it was suggested that H4K20me is deposited through a preceding deacetylation of H4K16ac and this process determines the levels of H4K20me3 throughout the genome.52 The H4K16ac-H4K20me3 connection is evident particularly in the control of RNA polymerase II (RNAP II/Pol II) pausing. This regulatory mechanism is an important regulatory step in transcription where RNA synthesis begins but “pauses” 20–50 nucleotides downstream of the transcriptional start site.53 It is expected that thousands of human genes, about 20–30% of the genome, are regulated by RNAP II pausing. In this respect, H4K20me3 acts by counteracting H4K16ac, enhancing RNAP II pausing. Therefore, during autophagy an increase in H4K20me3 together with a decrease in H4K16ac can act in conjunction to inhibit the expression of a subset of genes under control of RNAP II pausing.
H3K9me2
The most recent newcomer to the field of autophagy-regulated histone modifications is the dimethylation of lysine 9 on histone H3 (H3K9me2). Recently, 2 papers described the removal of H3K9me2 from the promoters of several autophagy-related genes and how this modification can affect the outcome of autophagy.54 H3K9 can be mono-, di- or trimethylated by a whole set of different histone methyltransferases (HMTs) such as SUV39H1 [suppressor of variegation 3-9 homolog 1 (Drosophila)] or EHMT2/G9a (euchromatic histone-lysine N-methyltransferase 2), or acetylated by HATs such as KAT2A and KAT2B/PCAF.55 The decrease of H3K9 acetylation levels is required for an increase of H3K9me2. H3K9 can be demethylated by several members of the family of histone lysine demethylases (KDMs). Methylation of this residue is mainly associated with transcriptional repression. EHMT2 associates with the promoters of core autophagy genes including LC3B and WIPI1, and also with TP53INP2/DOR, acting as a transcriptional repressor. However, when autophagy is induced, EHMT2 leaves the promoters and allows demethylation as well as acetylation of H3K9 thereby increasing the expression levels of autophagy-related proteins.54 Furthermore, the specific EHMT2 inhibitor BRD4770 increases the level of BNIP3 (BCL2/adenovirus E1B 19kDa interacting protein 3) and even LC3 at higher doses. Also, when TP53-mutant PANC-1 cancer cells are cotreated with the autophagy inducer gossypol, autophagy and even autophagy-related cell death are enhanced.56 In summary, the EHMT2-mediated methylation level of H3K9 acts as a repressor of autophagy under normal conditions. However, after autophagy induction EHMT2 is readily removed from promoters of autophagy-related genes leading to disappearance of H3K9me2, increase in H3K9ac and thus transcription of genes. When autophagy is induced alongside a persistent reduction in EHMT2, autophagy seems to become overstimulated resulting in cell death with autophagic features.
Decision makers? Survival vs. death histone codes
Allis and Turner proposed the “histone code” hypothesis that encapsulates the role of histone modifications for chromatin structure and the corresponding regulation of nuclear functions.57,58 According to this hypothesis, the combination of specific covalent posttranslational histone modifications influences the chromatin structure and leads to an impact on the transcriptional outputs.57 This hypothesis was further developed with the concept that various combinations of histone posttranslational modifications are in control of specific chromatin-related functions and processes.59 The recently described connections between autophagy and histone modifications make it appear likely that they are more than “useless” bystanders of the autophagic process. Specific histone modifications have also been linked to apoptotic chromatin changes, providing evidence for the existence of an apoptotic histone code.5 Similar to this “death code,” a set of changes in histone modifications occur during autophagy, creating an autophagy histone code, in other words a “survival code,” which determines the destiny of the cell (Fig. 2).
Autophagy is usually seen as a cellular survival process to cope with a limited supply of nutrients, damaged organelles, or stress conditions. However, its overstimulation can also lead to autophagic cell death.13,14 Histone modifications are involved in early stages, namely the removal of H3K9me2, and late stages, namely H4K16 deacetylation, of autophagy ensuring a level of autophagy required for survival. Noteworthy, the same modifications that are involved in autophagy also show a remarkable inverse correlation with the apoptotic histone marks indicating that an interference with the autophagic histone code can push the cell toward cell death (Fig. 3). Moreover, the same histone modifications known to date to take part in the “survival histone code” or the “death histone code” are also involved in carcinogenesis, thus representing histone onco-modifications. This is rather unsurprising as deregulations of autophagy and apoptosis are both hallmarks of cancer. While the inhibition of the apoptotic program is generally beneficial for cancer cells the situation is more complicated with autophagy. In general, during early cancer development a high level of autophagy can result in cancer cell removal, whereas established tumors rely on an increased autophagy for resistance to stress, including a limited nutrient supply.12
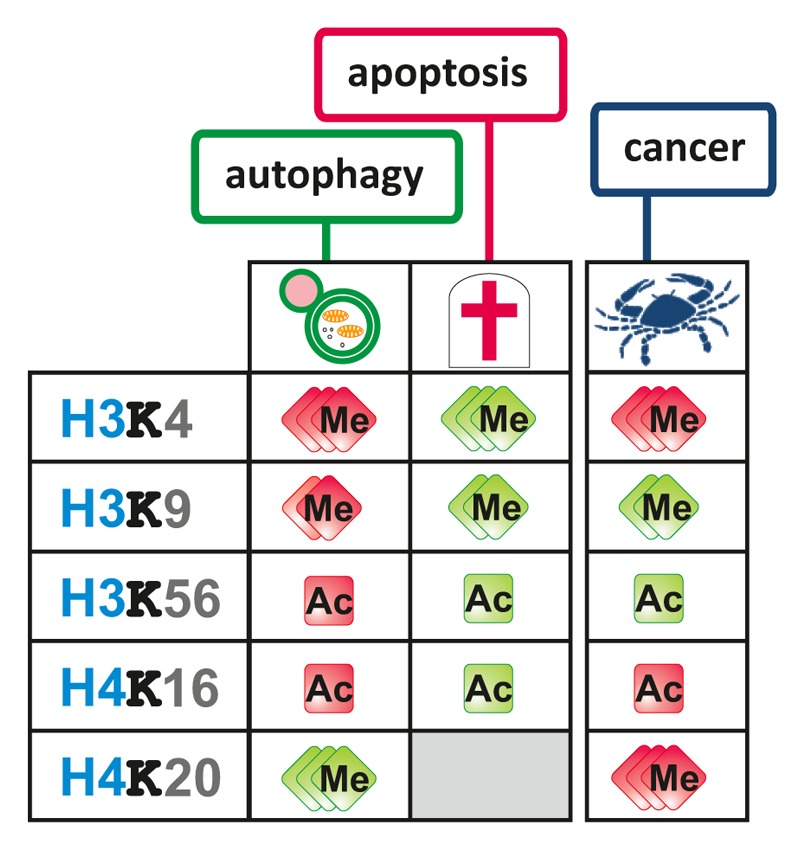
Figure 3. Autophagic vs. apoptotic histone codes. Overview of autophagy-related histone modifications that are also linked to apoptosis and/or carcinogenesis. Autophagy-related histone modifications display an intriguing inverse relationship to apoptosis-related histone modifications ac, acetylated; H, histone; K, lysine; me, methylated.
Others and we have shown that antagonizing the survival marks during autophagy can indeed induce cell death.37,56 Both removal of the HMT EHMT2 by the specific chemical inhibitor BRD4770, decreasing the level of the repressive H3K9me2 histone modification, or increase of the active H4K16ac histone modification (through overexpression of KAT8, or antagonizing SIRT1 using valproic acid, Ex527 or siRNA) during autophagy, result in cancer cell death. Inhibition of EHMT2 or induction of H4K16ac both lead to a more open chromatin structure eliminating the tight control of autophagy-related gene expression potentially resulting in an autophagic overstimulation and finally cell death.
Perspectives
Dysregulation of the histone codes
Overall it appears that histone modifications finally claim their place as central regulators of autophagy. Within the last few months, the nucleus returned to the spotlight of autophagy research28,37,54,60 and the HATs and HDACs that were long known to be involved in the cytosolic control of autophagy seem to regulate their name-giving targets, the histones, upon autophagy induction as well. There is striking but unsurprising correlation in regulatory histone modifications during autophagy, apoptosis, and cancer development. Deregulation of both apoptosis and autophagy and also histone modifications are tightly linked to cancer. Thus, distortion of the action of a few histone-modifying enzymes can have enormous impact on disease progression, whereas their restoration or modification could be beneficial for a plethora of human conditions including cancer. There is no doubt that the histone survival code only represents the tip of the iceberg of regulatory autophagic histone modifications. While the characterization of the “death histone code” is still in its infancy, the “survival histone code” is just born. Moreover, how these histone PTMs contribute to autophagy (transcription, silencing, pausing, autophagy flux control, etc.) remains to be elucidated in detail.
Disclosure of Potential Conflicts of Interest
The authors declare no conflict of interest.
Acknowledgments
We apologize to authors whose primary references could not be cited owing to space limitations. This work was supported by the Swedish Childhood Cancer Foundation, the Swedish Cancer Society, the Swedish Research Council, the Cancer Society in Stockholm, and the Karolinska Institutet Foundations.
Glossary
Abbreviations:
- ac
acetylated
- ATG
autophagy-related
- EHMT2
euchromatic histone-lysine N-methyltransferase 2
- H2A
histone 2A
- H2B
histone 2B
- H3
histone 3
- H4
histone 4
- HAT
histone acetyltransferase
- HDAC
histone deacetylase
- HMT
histone methyltransferase
- K
lysine
- KAT
K (lysine) acetyltransferase
- KDM
histone lysine demethylase
- (MAP1)LC3
microtubule-associated protein 1 light chain 3
- MTOR
mechanistic target of rapamycin
- me
methylated
- RNAP II/Pol II
RNA polymerase II
- PTM
posttranslational modification
- S
serine
- SAHA
suberoylanilide hydroxamic acid
- SIRT
sirtuin
- Suv
suppressor of variegation
- WNT
wingless-type MMTV integration site family
References
- 1.Luger K, Mäder AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–60. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 2.Kornberg RD, Lorch Y. Twenty-five years of the nucleosome, fundamental particle of the eukaryote chromosome. Cell. 1999;98:285–94. doi: 10.1016/S0092-8674(00)81958-3. [DOI] [PubMed] [Google Scholar]
- 3.Tan M, Luo H, Lee S, Jin F, Yang JS, Montellier E, Buchou T, Cheng Z, Rousseaux S, Rajagopal N, et al. Identification of 67 histone marks and histone lysine crotonylation as a new type of histone modification. Cell. 2011;146:1016–28. doi: 10.1016/j.cell.2011.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Allis CD, Berger SL, Cote J, Dent S, Jenuwien T, Kouzarides T, Pillus L, Reinberg D, Shi Y, Shiekhattar R, et al. New nomenclature for chromatin-modifying enzymes. Cell. 2007;131:633–6. doi: 10.1016/j.cell.2007.10.039. [DOI] [PubMed] [Google Scholar]
- 5.Füllgrabe J, Hajji N, Joseph B. Cracking the death code: apoptosis-related histone modifications. Cell Death Differ. 2010;17:1238–43. doi: 10.1038/cdd.2010.58. [DOI] [PubMed] [Google Scholar]
- 6.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 7.Tropberger P, Schneider R. Scratching the (lateral) surface of chromatin regulation by histone modifications. Nat Struct Mol Biol. 2013;20:657–61. doi: 10.1038/nsmb.2581. [DOI] [PubMed] [Google Scholar]
- 8.Füllgrabe J, Kavanagh E, Joseph B. Histone onco-modifications. Oncogene. 2011;30:3391–403. doi: 10.1038/onc.2011.121. [DOI] [PubMed] [Google Scholar]
- 9.Yun M, Wu J, Workman JL, Li B. Readers of histone modifications. Cell Res. 2011;21:564–78. doi: 10.1038/cr.2011.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–75. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang Z, Klionsky DJ. Eaten alive: a history of macroautophagy. Nat Cell Biol. 2010;12:814–22. doi: 10.1038/ncb0910-814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Levine B, Yuan J. Autophagy in cell death: an innocent convict? J Clin Invest. 2005;115:2679–88. doi: 10.1172/JCI26390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen Y, Klionsky DJ. The regulation of autophagy - unanswered questions. J Cell Sci. 2011;124:161–70. doi: 10.1242/jcs.064576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Michaud M, Martins I, Sukkurwala AQ, Adjemian S, Ma Y, Pellegatti P, Shen S, Kepp O, Scoazec M, Mignot G, et al. Autophagy-dependent anticancer immune responses induced by chemotherapeutic agents in mice. Science. 2011;334:1573–7. doi: 10.1126/science.1208347. [DOI] [PubMed] [Google Scholar]
- 16.Jiang H, White EJ, Conrad C, Gomez-Manzano C, Fueyo J. Autophagy pathways in glioblastoma. Methods Enzymol. 2009;453:273–86. doi: 10.1016/S0076-6879(08)04013-5. [DOI] [PubMed] [Google Scholar]
- 17.Morselli E, Mariño G, Bennetzen MV, Eisenberg T, Megalou E, Schroeder S, Cabrera S, Bénit P, Rustin P, Criollo A, et al. Spermidine and resveratrol induce autophagy by distinct pathways converging on the acetylproteome. J Cell Biol. 2011;192:615–29. doi: 10.1083/jcb.201008167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shao Y, Gao Z, Marks PA, Jiang X. Apoptotic and autophagic cell death induced by histone deacetylase inhibitors. Proc Natl Acad Sci U S A. 2004;101:18030–5. doi: 10.1073/pnas.0408345102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bánréti A, Sass M, Graba Y. The emerging role of acetylation in the regulation of autophagy. Autophagy. 2013;9:819–29. doi: 10.4161/auto.23908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lin SY, Li TY, Liu Q, Zhang C, Li X, Chen Y, Zhang SM, Lian G, Liu Q, Ruan K, et al. GSK3-TIP60-ULK1 signaling pathway links growth factor deprivation to autophagy. Science. 2012;336:477–81. doi: 10.1126/science.1217032. [DOI] [PubMed] [Google Scholar]
- 21.Yi C, Ma M, Ran L, Zheng J, Tong J, Zhu J, Ma C, Sun Y, Zhang S, Feng W, et al. Function and molecular mechanism of acetylation in autophagy regulation. Science. 2012;336:474–7. doi: 10.1126/science.1216990. [DOI] [PubMed] [Google Scholar]
- 22.Eisenberg T, Knauer H, Schauer A, Büttner S, Ruckenstuhl C, Carmona-Gutierrez D, Ring J, Schroeder S, Magnes C, Antonacci L, et al. Induction of autophagy by spermidine promotes longevity. Nat Cell Biol. 2009;11:1305–14. doi: 10.1038/ncb1975. [DOI] [PubMed] [Google Scholar]
- 23.Yamaki M, Umehara T, Chimura T, Horikoshi M. Cell death with predominant apoptotic features in Saccharomyces cerevisiae mediated by deletion of the histone chaperone ASF1/CIA1. Genes Cells. 2001;6:1043–54. doi: 10.1046/j.1365-2443.2001.00487.x. [DOI] [PubMed] [Google Scholar]
- 24.Han J, Zhou H, Horazdovsky B, Zhang K, Xu RM, Zhang Z. Rtt109 acetylates histone H3 lysine 56 and functions in DNA replication. Science. 2007;315:653–5. doi: 10.1126/science.1133234. [DOI] [PubMed] [Google Scholar]
- 25.Masumoto H, Hawke D, Kobayashi R, Verreault A. A role for cell-cycle-regulated histone H3 lysine 56 acetylation in the DNA damage response. Nature. 2005;436:294–8. doi: 10.1038/nature03714. [DOI] [PubMed] [Google Scholar]
- 26.Watanabe S, Radman-Livaja M, Rando OJ, Peterson CL. A histone acetylation switch regulates H2A.Z deposition by the SWR-C remodeling enzyme. Science. 2013;340:195–9. doi: 10.1126/science.1229758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xu F, Zhang K, Grunstein M. Acetylation in histone H3 globular domain regulates gene expression in yeast. Cell. 2005;121:375–85. doi: 10.1016/j.cell.2005.03.011. [DOI] [PubMed] [Google Scholar]
- 28.Chen H, Fan M, Pfeffer LM, Laribee RN. The histone H3 lysine 56 acetylation pathway is regulated by target of rapamycin (TOR) signaling and functions directly in ribosomal RNA biogenesis. Nucleic Acids Res. 2012;40:6534–46. doi: 10.1093/nar/gks345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Das C, Lucia MS, Hansen KC, Tyler JK. CBP/p300-mediated acetylation of histone H3 on lysine 56. Nature. 2009;459:113–7. doi: 10.1038/nature07861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee IH, Finkel T. Regulation of autophagy by the p300 acetyltransferase. J Biol Chem. 2009;284:6322–8. doi: 10.1074/jbc.M807135200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Robert T, Vanoli F, Chiolo I, Shubassi G, Bernstein KA, Rothstein R, Botrugno OA, Parazzoli D, Oldani A, Minucci S, et al. HDACs link the DNA damage response, processing of double-strand breaks and autophagy. Nature. 2011;471:74–9. doi: 10.1038/nature09803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vempati RK, Jayani RS, Notani D, Sengupta A, Galande S, Haldar D. p300-mediated acetylation of histone H3 lysine 56 functions in DNA damage response in mammals. J Biol Chem. 2010;285:28553–64. doi: 10.1074/jbc.M110.149393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yuan J, Pu M, Zhang Z, Lou Z. Histone H3-K56 acetylation is important for genomic stability in mammals. Cell Cycle. 2009;8:1747–53. doi: 10.4161/cc.8.11.8620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xie HJ, Noh JH, Kim JK, Jung KH, Eun JW, Bae HJ, Kim MG, Chang YG, Lee JY, Park H, et al. HDAC1 inactivation induces mitotic defect and caspase-independent autophagic cell death in liver cancer. PLoS One. 2012;7:e34265. doi: 10.1371/journal.pone.0034265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ng F, Tang BL. Sirtuins’ modulation of autophagy. J Cell Physiol. 2013;228:2262–70. doi: 10.1002/jcp.24399. [DOI] [PubMed] [Google Scholar]
- 36.Shogren-Knaak M, Ishii H, Sun JM, Pazin MJ, Davie JR, Peterson CL. Histone H4-K16 acetylation controls chromatin structure and protein interactions. Science. 2006;311:844–7. doi: 10.1126/science.1124000. [DOI] [PubMed] [Google Scholar]
- 37.Füllgrabe J, Lynch-Day MA, Heldring N, Li W, Struijk RB, Ma Q, Hermanson O, Rosenfeld MG, Klionsky DJ, Joseph B. The histone H4 lysine 16 acetyltransferase hMOF regulates the outcome of autophagy. Nature. 2013;500:468–71. doi: 10.1038/nature12313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vaquero A, Sternglanz R, Reinberg D. NAD+-dependent deacetylation of H4 lysine 16 by class III HDACs. Oncogene. 2007;26:5505–20. doi: 10.1038/sj.onc.1210617. [DOI] [PubMed] [Google Scholar]
- 39.Shi Y, Lan F, Matson C, Mulligan P, Whetstine JR, Cole PA, Casero RA, Shi Y. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell. 2004;119:941–53. doi: 10.1016/j.cell.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 40.Ruthenburg AJ, Allis CD, Wysocka J. Methylation of lysine 4 on histone H3: intricacy of writing and reading a single epigenetic mark. Mol Cell. 2007;25:15–30. doi: 10.1016/j.molcel.2006.12.014. [DOI] [PubMed] [Google Scholar]
- 41.Klose RJ, Zhang Y. Regulation of histone methylation by demethylimination and demethylation. Nat Rev Mol Cell Biol. 2007;8:307–18. doi: 10.1038/nrm2143. [DOI] [PubMed] [Google Scholar]
- 42.Ruthenburg AJ, Li H, Milne TA, Dewell S, McGinty RK, Yuen M, Ueberheide B, Dou Y, Muir TW, Patel DJ, et al. Recognition of a mononucleosomal histone modification pattern by BPTF via multivalent interactions. Cell. 2011;145:692–706. doi: 10.1016/j.cell.2011.03.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang Z, Zang C, Cui K, Schones DE, Barski A, Peng W, Zhao K. Genome-wide mapping of HATs and HDACs reveals distinct functions in active and inactive genes. Cell. 2009;138:1019–31. doi: 10.1016/j.cell.2009.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dou Y, Milne TA, Tackett AJ, Smith ER, Fukuda A, Wysocka J, Allis CD, Chait BT, Hess JL, Roeder RG. Physical association and coordinate function of the H3 K4 methyltransferase MLL1 and the H4 K16 acetyltransferase MOF. Cell. 2005;121:873–85. doi: 10.1016/j.cell.2005.04.031. [DOI] [PubMed] [Google Scholar]
- 45.Katoh H, Qin ZS, Liu R, Wang L, Li W, Li X, Wu L, Du Z, Lyons R, Liu CG, et al. FOXP3 orchestrates H4K16 acetylation and H3K4 trimethylation for activation of multiple genes by recruiting MOF and causing displacement of PLU-1. Mol Cell. 2011;44:770–84. doi: 10.1016/j.molcel.2011.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Petherick KJ, Williams AC, Lane JD, Ordóñez-Morán P, Huelsken J, Collard TJ, Smartt HJ, Batson J, Malik K, Paraskeva C, et al. Autolysosomal β-catenin degradation regulates Wnt-autophagy-p62 crosstalk. EMBO J. 2013;32:1903–16. doi: 10.1038/emboj.2013.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sierra J, Yoshida T, Joazeiro CA, Jones KA. The APC tumor suppressor counteracts beta-catenin activation and H3K4 methylation at Wnt target genes. Genes Dev. 2006;20:586–600. doi: 10.1101/gad.1385806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wend P, Fang L, Zhu Q, Schipper JH, Loddenkemper C, Kosel F, Brinkmann V, Eckert K, Hindersin S, Holland JD, et al. Wnt/β-catenin signalling induces MLL to create epigenetic changes in salivary gland tumours. EMBO J. 2013;32:1977–89. doi: 10.1038/emboj.2013.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schotta G, Lachner M, Sarma K, Ebert A, Sengupta R, Reuter G, Reinberg D, Jenuwein T. A silencing pathway to induce H3-K9 and H4-K20 trimethylation at constitutive heterochromatin. Genes Dev. 2004;18:1251–62. doi: 10.1101/gad.300704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kourmouli N, Jeppesen P, Mahadevhaiah S, Burgoyne P, Wu R, Gilbert DM, Bongiorni S, Prantera G, Fanti L, Pimpinelli S, et al. Heterochromatin and tri-methylated lysine 20 of histone H4 in animals. J Cell Sci. 2004;117:2491–501. doi: 10.1242/jcs.01238. [DOI] [PubMed] [Google Scholar]
- 51.Henckel A, Nakabayashi K, Sanz LA, Feil R, Hata K, Arnaud P. Histone methylation is mechanistically linked to DNA methylation at imprinting control regions in mammals. Hum Mol Genet. 2009;18:3375–83. doi: 10.1093/hmg/ddp277. [DOI] [PubMed] [Google Scholar]
- 52.Serrano L, Martínez-Redondo P, Marazuela-Duque A, Vazquez BN, Dooley SJ, Voigt P, Beck DB, Kane-Goldsmith N, Tong Q, Rabanal RM, et al. The tumor suppressor SirT2 regulates cell cycle progression and genome stability by modulating the mitotic deposition of H4K20 methylation. Genes Dev. 2013;27:639–53. doi: 10.1101/gad.211342.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kapoor-Vazirani P, Kagey JD, Vertino PM. SUV420H2-mediated H4K20 trimethylation enforces RNA polymerase II promoter-proximal pausing by blocking hMOF-dependent H4K16 acetylation. Mol Cell Biol. 2011;31:1594–609. doi: 10.1128/MCB.00524-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Artal-Martinez de Narvajas A, Gomez TS, Zhang JS, Mann AO, Taoda Y, Gorman JA, Herreros-Villanueva M, Gress TM, Ellenrieder V, Bujanda L, et al. Epigenetic regulation of autophagy by the methyltransferase G9a. Mol Cell Biol. 2013;33:3983–93. doi: 10.1128/MCB.00813-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jin Q, Yu LR, Wang L, Zhang Z, Kasper LH, Lee JE, Wang C, Brindle PK, Dent SY, Ge K. Distinct roles of GCN5/PCAF-mediated H3K9ac and CBP/p300-mediated H3K18/27ac in nuclear receptor transactivation. EMBO J. 2011;30:249–62. doi: 10.1038/emboj.2010.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yuan Y, Tang AJ, Castoreno AB, Kuo SY, Wang Q, Kuballa P, Xavier R, Shamji AF, Schreiber SL, Wagner BK. Gossypol and an HMT G9a inhibitor act in synergy to induce cell death in pancreatic cancer cells. Cell Death Dis. 2013;4:e690. doi: 10.1038/cddis.2013.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–5. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 58.Turner BM. Histone acetylation and an epigenetic code. Bioessays. 2000;22:836–45. doi: 10.1002/1521-1878(200009)22:9<836::AID-BIES9>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 59.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–80. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 60.Jenuwein T, Allis CD. The return of the nucleus: transcriptional and epigentic control of autophagy. Nature Rev Mol Cell Biol. 2014;15:65–74. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]


