Abstract
Genetic analyses indicate that autophagy, an evolutionarily conserved lysosomal degradation pathway, is essential for eukaryotic differentiation and development. However, little is known about whether autophagy contributes to morphogenesis during embryogenesis. To address this question, we examined the role of autophagy in the early development of zebrafish, a model organism for studying vertebrate tissue and organ morphogenesis. Using zebrafish that transgenically express the fluorescent autophagy reporter protein, GFP-LC3, we found that autophagy is active in multiple tissues, including the heart, during the embryonic period. Inhibition of autophagy by morpholino knockdown of essential autophagy genes (including atg5, atg7, and becn1) resulted in defects in morphogenesis, increased numbers of dead cells, abnormal heart structure, and reduced organismal survival. Further analyses of cardiac development in autophagy-deficient zebrafish revealed defects in cardiac looping, abnormal chamber morphology, aberrant valve development, and ectopic expression of critical transcription factors including foxn4, tbx5, and tbx2. Consistent with these results, Atg5-deficient mice displayed abnormal Tbx2 expression and defects in valve development and chamber septation. Thus, autophagy plays an essential, conserved role in cardiac morphogenesis during vertebrate development.
Keywords: autophagy, zebrafish, heart development, tbx2, atg5, atg7, becn1
Introduction
Autophagy is an evolutionarily conserved process by which cytoplasmic constituents including macromolecules (such as proteins, glycogens, lipids, and nucleic acids) and organelles (such as mitochondria, peroxisomes, and endoplasmic reticulum) are degraded by the lysosome.1 The autophagy pathway plays an essential role in enabling eukaryotic organisms to adapt to nutrient deprivation and other forms of environmental stress; it generates the building blocks necessary for macromolecular synthesis, energy production, and cell survival in such settings. By serving as a cellular “disposal” mechanism that removes misfolded or unfolded proteins and damaged organelles, the autophagy pathway also maintains organelle integrity and protein quality control. The nutrient recycling and organelle/protein quality control functions of autophagy likely contribute to its many important roles in physiology and protection against diseases.2,3 Indeed, autophagy genes are important in metazoan survival during starvation; protein, carbohydrate, and lipid metabolism; life-span extension; and protection against cancer, neurodegenerative disorders, infection, and other diseases.
Another key function of autophagy is its role in differentiation and development.4-7 From an evolutionary perspective, autophagy may have arisen as a mechanism to protect unicellular organisms against starvation and other forms of environmental stress (such as overcrowding and extreme temperatures). However, the stimulus to degrade organelles may have led to other evolutionary advantages, including the ability to undergo differentiation and development. Both differentiation and development require cells to undergo major morphological changes and thus, require mechanisms for the degradation and recycling of obsolete cellular components. Therefore, it may not be a coincidence that certain differentiation and developmental events, especially in lower eukaryotic organisms, are triggered or influenced by autophagy-inducing environmental stressors; autophagy may be mechanistically involved in these processes. In support of this concept, autophagy genes are essential for several starvation-induced differentiation and developmental processes, including sporulation in yeast, dauer development in C. elegans, fruiting body formation in Dictyostelium discoideum, and developmental transitions in protozoan parasites (reviewed in refs. 4–7).
The role of autophagy in development also extends to several other (environmental stress-independent) processes that occur in multicellular organisms. For example, abnormalities in larval development, synapse formation, and eye development are observed in autophagy gene mutant Drosophila.8-12 In C. elegans, autophagy genes play a role in apoptotic corpse clearance in embryonic development,13 the L1 larval stage,14 and the adult gonad,15 in degrading germline P granules in somatic cells,16 and in the selective elimination of paternal mitochondria.17,18 In mice, autophagy genes play an essential role in preimplantation development, survival during the neonatal starvation period, and in the differentiation of several specific cell lineages, including adipocytes, erythrocytes, and lymphocytes (T cells and B-1a cells) (reviewed in ref. 7).
One surprising finding is that mice lacking several different essential autophagy genes, including Atg3, Atg5, Atg7, Atg9, and Atg16L1, are reported to lack apparent anatomical abnormalities (reviewed in ref. 6). The most obvious interpretation of this finding is that the autophagy machinery is not required for tissue morphogenesis during embryonic development. However, this finding is puzzling as one would predict that autophagy, a pathway that can both eliminate pre-existing structures and provide materials for building new structures, is an ideal candidate for involvement in developmental processes that require cellular and tissue remodeling. Thus, the question arises as to whether the absence of apparent anatomical abnormalities in several autophagy gene knockout mouse strains truly reflects the lack of a role for autophagy in morphogenesis or whether other factors, such as gene redundancy, activation of compensatory pathways, or difficulties in detection of phenotypic abnormalities may be confounding variables. Of note, in the zebrafish, Hu et al. reported that knockdown of atg5 leads to abnormal morphogenesis of brain regionalization and body plan.19
In contrast to mice lacking Atg3, Atg5, Atg7, Atg9, and Atg6l1, mice lacking 3 other genes involved in autophagy, Becn1, Ambra1, and Rb1cc1/Fip200, die during embryogenesis (reviewed in refs. 5 and 7). becn1−/− embryos die around embryonic d 7.5, with massive cell death and failure to close the proamniotic cavity. Mice lacking Ambra1 are embryonic lethal at d E10–14 and show defective neural tube development and hyperproliferation of neural tissues. (In accord with this result, Benato and coworkers reported that knockdown of the zebrafish Ambra1 orthologs causes defects in body morphogenesis and brain development20). rb1cc1−/− mice are embryonic lethal at d E13.5–16.5, and display defects in heart and liver development. The discrepancy between the embryonic lethal phenotypes of becn1−/−, ambra1−/−, and rb1cc1−/− mice vs. the embryonic viability of the atg3−/−, atg5−/−, atg7−/−, atg9−/−, and atg16l1−/− mice is generally attributed to autophagy-independent functions of the former group of genes. However, other explanations are possible, and additional studies in more tractable model systems are needed to better determine whether the autophagy pathway functions in developmental patterning and tissue morphogenesis during early vertebrate embryonic development.
To address this question, we used morpholino oligonucleotides (MOs) to knock down expression of 3 different autophagy genes, becn1, atg5, and atg7 in developing zebrafish (Danio rerio) embryos. Becn1 (also known as Beclin 1) functions in autophagy as part of the class III phosphatidylinositol 3-kinase complex involved in autophagic vesicle nucleation, and Atg5 and Atg7 are part of the ubiquitin-like protein conjugation systems that function in autophagosome membrane expansion and completion.21 The zebrafish presents unique advantages among vertebrates for the study of developmental patterning and tissue morphogenesis (reviewed in refs. 22 and 23). Unlike the mammalian embryo, the zebrafish does not require a functional cardiovascular system during embryogenesis as oxygen diffusion from the surrounding medium is sufficient for survival.24 Therefore, the organs of mutant zebrafish embryos, especially the heart, can be more readily observed than those in mouse embryos.
We found that MO knockdown of different autophagy genes led to developmental defects, including structural cardiac defects. Autophagy-deficient zebrafish displayed defective cardiac looping, mispatterning of genes normally expressed in the atrioventricular (AV) valve, and upregulation of the genes encoding several heart transcription factors, including Foxn4 and Tbx5, in microarray analyses of cardiac-enriched tissue. Consistent with these findings, autophagy morphants exhibited increased, ectopic expression of tbx2, a downstream target of foxn4 and tbx5. Similarly, cardiac atrioventricular canal (AVC) defects and Tbx2 misexpression were also observed in autophagy-deficient atg5−/− mutant mice. These data demonstrate an unequivocal role for autophagy genes in tissue patterning and morphogenesis during vertebrate development.
Results
Autophagy is active in early embryonic development in zebrafish
To study the role of autophagy in early embryonic development in zebrafish, the temporal pattern of autophagy activation during zebrafish development was monitored. Unlike in mouse embryos, in which autophagy induction was observed at the 1-cell stage stage,25 it has been reported that autophagy is induced after 32 h post-fertilization (hpf) in zebrafish because of delayed expression of proteins essential for autophagy.26 To confirm the temporal expression pattern of autophagy transcripts, we prepared RNA from different stages of embryonic development and used RT-PCR to detect the expression of atg5, becn1, atg7, and ulk1b (Fig. 1A). All 4 transcripts were detected in 1-cell stage-embryos, indicating that they were maternally deposited, as well as at the sphere (3 hpf), germ ring (6 hpf) and 5-somite (12 hpf) stages and at 24 and 48 hpf. atg5 and becn1 transcripts appeared to decrease at the early gastrulation (germ ring) stage, similar to what was reported for atg5,19 ambra1a, and ambra1b.20 He and coworkers previously reported that several transcripts including ulk1b are not expressed before 23 hpf.26 The inconsistency with the previous report may be due to different RT-PCR conditions. He et al. amplified the full-length RNA for autophagy gene expression detection, but here, we amplified shorter 2-exon fragments of each transcript. It is possible that amplifying shorter segments of transcripts results in more sensitive detection than amplification of the full-length transcripts. Hu and coworkers also identified atg5 transcripts at early developmental time points, including 0.2, 5, 10, 18, and 24 hpf.19
Figure 1. Autophagy is present during early zebrafish development and efficiently inhibited by autophagy gene knockdown. (A) Autophagy transcripts are expressed during early development in zebrafish. RT-PCR was performed using RNA isolated from 1-cell, sphere, germ ring, 5-somite, 24 hpf, and 2 dpf stage wild-type embryos using gene-specific primers. nRT, no-RT negative control. (B) Early autophagosomes (left) and autolysosomes (right) were detected in 10-somite stage embryos by electron microscopy. Scale bars, left: 200 nm, right: 500 nm. (C) GFP-Lc3 puncta were visualized by confocal microscopy in 15-somite stage embryos from Tg(cmv::GFP-lc3) fish. Left, z-stack images of somites. Right, GFP-Lc3 puncta in somites merged with bright-field image. Scale bars, left: 30 μm, right: 30 μm. (D) Expression of autophagy proteins after autophagy morpholino (MO) injection. Embryos were injected at the 1-cell stage with control or autophagy-specific MOs, and lysates were prepared for immunoblot analysis at 48 hpf. Top panel, immunoblot with anti-Atg5 antibody. Bottom panel, immunoblot with anti-Becn1 antibody. Actin is shown as a loading control. (E and F) Autophagy gene knockdown impairs autophagy in zebrafish. Primary cells were prepared from Tg(cmv:GFP-lc3) after MO injection and GFP-Lc3 puncta were visualized with confocal microscopy. (E) Representative images of GFP-Lc3 puncta in embryos injected with autophagy gene-specific and control MOs. (F) Quantification of data shown in (E). Representative results from one of 3 separate experiments, expressed as the mean ± SEM of 50 to 100 cells in each group. ***P < 0.001, **P < 0.01, *P < 0.05 for autophagy morphants vs. control morphants; one-tailed t test. (G) Representative immunoblots to detect Lc3 levels in control- and autophagy MO-injected fish. Lysates were prepared from primary cells for immunoblot after MO injection. Actin is shown as a loading control. Similar results were observed in 3 independent experiments. (H) Quantification of the ratio of Lc3-II/Actin for 3 independent experiments, including representative results shown in (G). Bars represent mean ± SEM. **P < 0.01, *P < 0.05 for autophagy morphants vs. control morphants; 2-tailed t test.
To confirm the occurrence of autophagy in early embryonic development of zebrafish, 10-somite stage embryos were monitored for autophagosome structures by electron microscopy. We detected both early autophagosomes and autolysosomes in somite tissue (Fig. 1B), indicating the presence of autophagy at this stage. To further evaluate whether autophagy occurs in vivo, we used the fluorescent autophagy reporter transgenic fish line Tg(cmv:GFP-lc3).26 During autophagy induction, LC3-I (which is diffusely distributed in the cytoplasm) becomes conjugated with phosphatidylethanolamine to form LC3-II, which stably associates with the autophagosomal membrane. GFP-tagged LC3-II accumulates in punctate structures (autophagosomes), thus serving as a useful in vivo marker of autophagy.21 Using confocal microscopy, we noted the accumulation of GFP-Lc3 puncta in zebrafish by the 15-somite stage indicating that similar to other organisms, autophagy is active during early development (Fig. 1C). Some caution is warranted in interpreting this result, owing to the possibility that expression of GFP-Lc3 from the CMV promoter might lead to enhanced puncta formation due to high expression levels. However, taken together with the other data including electron microscopy, our results likely indicate the presence of autophagy in early zebrafish embryos.
Autophagy gene knockdown results in developmental defects
To investigate the role of autophagy in embryo morphogenesis, we used antisense morpholino oligonucleotides (MOs) to inhibit translation of zebrafish mRNAs encoding 3 key components of the autophagy machinery, atg5, atg7, and becn1. We confirmed protein knockdown by immunoblot analysis of whole embryos at 48 hpf (Fig. 1D). Atg5 is normally conjugated to Atg12 via a conjugation reaction that requires the action of the E1-like enzyme Atg7. MO knockdown of Atg5 led to loss of free Atg5 and to the expected loss of Atg12–Atg5 complex formation (Fig. 1D, top panel). Loss of the Atg12–Atg5 complex was similarly observed in atg7 morphants, suggesting efficient knockdown of Atg7 protein expression by atg7 MO injection. Embryos injected with becn1 MOs did not show a complete block in formation of the Atg12–Atg5 complex, an expected result since Becn1 is not required for Atg12–Atg5 complex formation; however, we did observe a decrease in the total amount of the conjugate. Compared with control morphants, becn1 morphants had decreased Becn1 protein expression (Fig. 1D, lower panel). Thus, atg5, atg7, and becn1 MOs successfully inhibit expression of their target autophagy protein in zebrafish embryos.
Next, we evaluated whether autophagy MOs inhibit autophagy in vivo, using a fluorescent autophagy reporter strain of fish, Tg(cmv:GFP-lc3).26 To efficiently quantify the effects of autophagy MOs on zebrafish autophagy, we prepared primary cells from blastula-stage Tg(cmv:GFP-lc3) embryos injected with control or autophagy MOs and counted the number of GFP-Lc3 puncta per cell. Primary cells derived from blastula-stage autophagy morphants displayed reduced numbers of GFP-Lc3 puncta, indicating that knockdown of atg5, becn1, or atg7 inhibited autophagosome accumulation in vivo (Fig. 1E and F). As further evidence that autophagy-directed MOs inhibit autophagy, we performed immunoblots on protein lysates from primary cell cultures of MO-injected embryos, using an antibody specific for Lc3 (Fig. 1G and H). Autophagy MOs efficiently blocked Lc3-II conversion, indicating inhibition of autophagy in vivo.
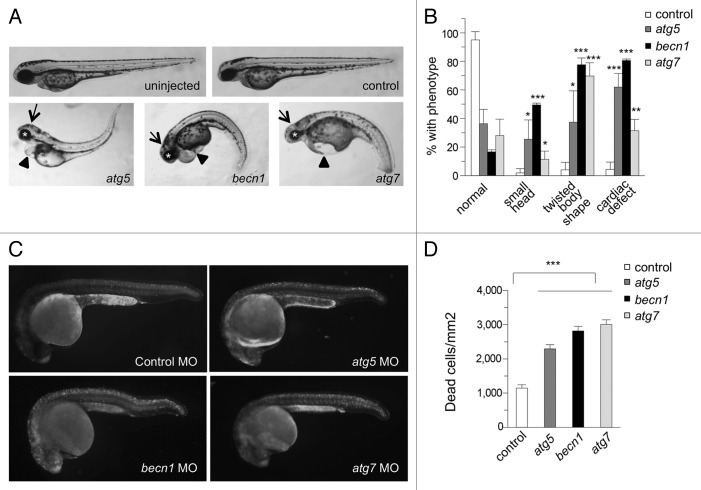
After demonstrating that the autophagy-specific MOs inhibit autophagy in vivo, we assessed the effects of autophagy inhibition on early zebrafish development. Autophagy morphants displayed reproducible abnormal developmental phenotypes including small heads (arrows) and eyes (asterisks), twisted body shapes, and pericardial edema (arrowheads) at 2 d post fertilization (dpf) (Fig. 2A and B). In general, the morphological defects were similar when comparing atg5, becn1, or atg7 knockdown, though there were some differences in penetrance, with becn1 morphants the most severely affected overall, and body morphology defects prominent in atg7 (as well as becn1) morphants. Most strikingly, 62% of atg5 morphants, 80% of becn1 morphants, and 31% of atg7 morphants exhibited cardiac defects such as pericardial edema, defective blood flow through the heart, defective heart looping, enlarged atria, or linearized hearts (Fig. S1). In comparison, more than 95% of fish were normal after control MO injection.
Figure 2. Autophagy gene knockdown in zebrafish results in developmental defects and increased numbers of dead cells. (A and B) Morphology of autophagy morphants at 2 dpf. (A) Representative images of autophagy morphants, which displayed small heads (arrows), abnormal eyes (asterisks), twisted body shapes and cardiac defects (arrowheads) at 2 dpf. (B) Quantification of phenotypes depicted in (A). Results represent the mean ± SEM of 3 independent experiments. ***P < 0.001, **P < 0.01, *P < 0.05 for autophagy morphants vs. control morphants; one-tailed t test. (C and D) Acridine orange (AO) staining of control and autophagy gene morphants at 1 dpf. AO-positive cells were counted in an area spanning from the end of the yolk extension to the end of tail. (D) Quantification of data shown in (C). Results represent the mean ± SEM of more than 40 embryos in each group. ***P < 0.001 for autophagy morphants vs. control morphants; one-tailed t test.
We took several complementary approaches to ensure that our results reflected on-target effects of MOs. First, in addition to the translation-blocking MOs described above, we also injected splice blocking-MOs designed to inhibit splicing of specific exon-intron junctions in the atg5, atg7, or becn1 primary mRNA transcript. Similar abnormal phenotypes were observed in embryos injected with splice blocking-MOs as found in translation-blocking MO-injected autophagy morphants, and the incidence of abnormal phenotypes increased in relationship to the injected MO concentration (Fig. S2A and S2B). Splicing inhibition by MO injection was confirmed by RT-PCR (Fig. S2C).
MO injection can induce abnormal developmental phenotypes due to off-target effects that are attributed to Tp53-mediated cellular toxicity.27 To exclude the possibility that the phenotypes of autophagy morphants were due to these off-target effects, we injected control and autophagy MOs into homozygous tp53 mutant (tp53M214K) fish28 (Fig. S3A and S3B). More than 80% of control morphants were normal at 2 dpf. Except for a slight decrease in the percentage of atg7 knockdown embryos exhibiting small head size, autophagy morphants continued to exhibit developmental defects. 100% of atg5 morphants, more than 80% of becn1 morphants and slightly more than 30% of atg7 morphants showed cardiac defects. Overall, the cardiac defects due to autophagy MOs were not significantly affected by loss of Tp53 function, indicating that the cardiac defects observed in a wild-type background of zebrafish were not caused by Tp53-dependent off-target effects. The finding of similar phenotypes using 2 independent MOs and in wild-type and tp53-deficient backgrounds strongly suggests that the observed phenotypes were due to autophagy gene knockdown.
Autophagy gene knockdown results in increased numbers of dead cells
Autophagy genes are required for the clearance of dead cells during embryonic development in mice,29 chick,30 and nematodes,13 and mice with a null mutation in the autophagy gene Ambra1 have increased cell proliferation and cell death in the central nervous system.31 To determine whether autophagy genes play a similar role in the regulation of cell death or dead cell clearance during zebrafish embryonic development, we stained embryos at 1 dpf with acridine orange (AO), and quantified the number of dead cells per unit area in a defined region of the embryo. Compared with control morphants, autophagy morphants showed a greater than 2-fold increase in numbers of dead cells (Fig. 2C and D). A similar magnitude of increase in numbers of dead cells was seen with injection of autophagy MOs into tp53 mutant (tp53M214K) embryos (Fig. S3C and S3D). These results indicate that autophagy may play an important role in either preventing cell death and/or the removal of dead cells during zebrafish development.
Interestingly, when we followed control or autophagy morphants for long-term survival, we saw striking effects of autophagy gene knockdown. More than 60% of control morphants survived for more than 4 wk. In contrast, survival was poor for autophagy morphants, with approximately 40% of atg5 morphants and less than 10% of becn1 or atg7 morphants surviving more than 10 d (Fig. S4). As the translation-blocking effects of MOs generally wear off within 4–5 d after injection,32 these data suggest that autophagy during the first 4–5 d of development is essential for long-term fish viability.
Autophagy occurs during cardiac development
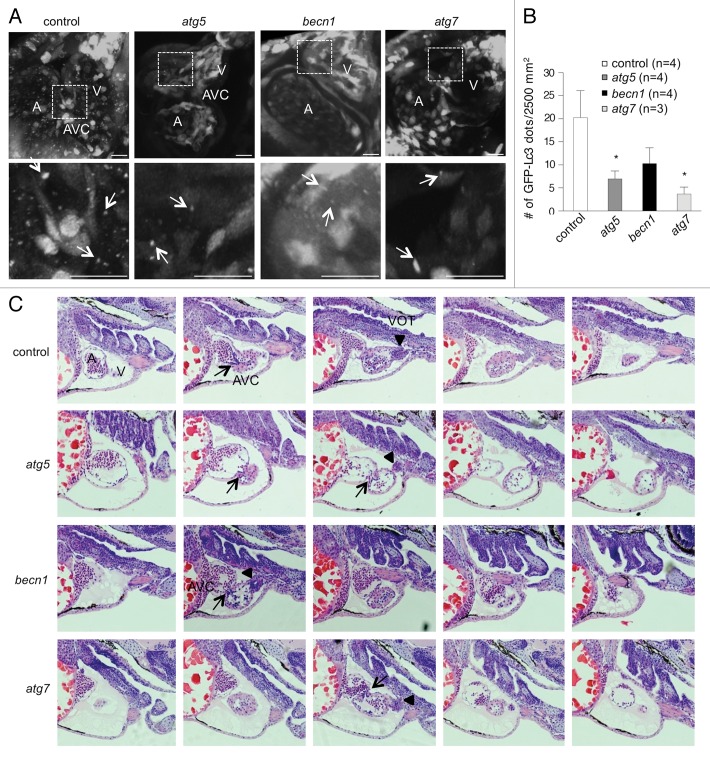
The striking cardiac phenotypes observed in autophagy morphants prompted us to further investigate the role of autophagy during cardiac morphogenesis. To determine whether autophagy occurs during normal cardiac development in zebrafish, embryos from the Tg(cmv:GFP-lc3) autophagy reporter strain were monitored for GFP-Lc3 puncta formation. In control morphant hearts, autophagosomes were found in cardiomyocytes, endocardial cells, the AVC, and the ventricular outflow tract, indicating that autophagy occurs during cardiac development (Fig. 3A and B). At 3 dpf, atg5 and atg7 morphants showed significantly reduced numbers of autophagosomes throughout the entire heart (Fig. 3B). becn1 morphants also showed a trend toward reduced numbers of autophagosomes in the heart that did not reach statistical significance. Imaging of embryos at 2 dpf also showed the presence of autophagosomes in the heart, which appeared to be decreased in the autophagy morphant embryos (Fig. S5).
Figure 3. Autophagy gene knockdown results in abnormal cardiac development. (A and B) Autophagy is active during normal cardiac development and impaired by autophagy MOs. (A) Tg(cmv:GFP-lc3) embryos were injected with either control or autophagy MOs and imaged at 3 dpf using confocal microscopy. Representative images showing GFP-Lc3 puncta (white arrows) in atrium (A), ventricle (V) and AV canal (AVC). The areas delineated by the white dashed lines are shown at higher magnification in the bottom panel. (B) Quantification of data shown in (A). Results represent the mean ± SEM of more than 3 embryos in each group. *P < 0.05 vs. control morphants; one-tailed t test. (C) Hematoxylin and eosin staining of serial sagittal sections of hearts from representative control or autophagy morphants at 3 dpf. The head is positioned to the right. In the control morphant heart, the atrium, AV canal (arrow), ventricle, and ventricular outflow tract (arrow head) (from left to right) are visualized. All autophagy morphant hearts display pericardial edema. The atg5 and becn1 morphant hearts shown have enlarged atria compared with those from the control morphant. The becn1 morphant heart displays an accumulation of blood cells in the atrium. All autophagy morphant hearts shown contain the atrium and ventricle in the same plane indicating incorrect cardiac looping. VOT, ventricular outflow tract.
Inhibition of autophagy induces cardiac defects
To better characterize the cardiac defects in autophagy morphants, histological analysis was performed with control or autophagy MO-injected embryos at 3 dpf. During zebrafish heart development, cardiac progenitor cells migrate from the lateral mesoderm and form a linear heart tube around 24 hpf. The heart tube subsequently tilts slightly to the left and undergoes looping around 2 dpf, representing a structural change from a linearized tube to a 3-dimensional chamber (reviewed in ref. 33). Serial sections of hearts from autophagy morphants showed enlarged atria compared with those of control morphants, and incorrect looping at 3 dpf (Fig. 3C). In control morphants, atria were properly placed caudally and to the left of the ventricles. However, in autophagy morphants, atria were enlarged and cardiac looping was defective, resulting in atria and ventricles appearing in the same histological plane. Additionally, all 3 autophagy morphants exhibited accumulation of blood cells in the atrium compared with the ventricle, suggesting that autophagy inhibition might cause defects in cardiac valve development and circulatory defects.
Cardiac morphogenesis can be influenced by several cardiac-extrinsic developmental factors such as blood circulation and vascular development. Vascular defects can cause secondary cardiac defects by increasing cardiac afterload. To exclude vascular defects as a possible source of the cardiac phenotype of autophagy morphants, we monitored vascular development of morphants using the Tg(fli1:EGFP) line, which expresses GFP in endothelial cells including vasculature, blood cells, and endocardial cells.34 Autophagy morphants did not show any vasculature defects at 3 dpf (Fig. S6), suggesting that the cardiac defects in autophagy morphants are not an indirect consequence of vascular abnormalities.
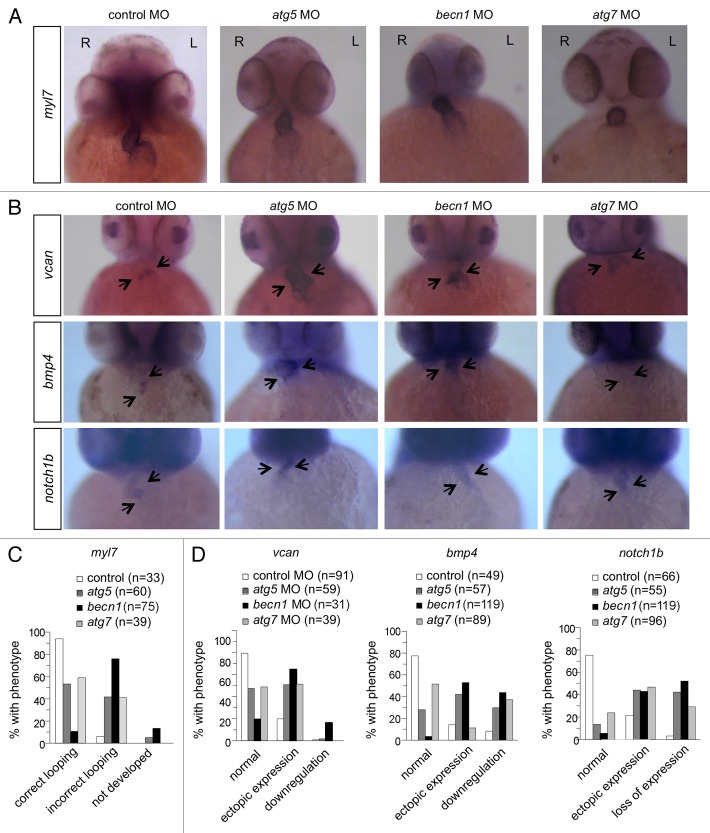
As autophagy morphants showed incorrect looping and potential cardiac valve defects by histological analysis (Fig. 3C), we sought to further confirm these defects using in situ hybridization with cardiac-specific and AV valve-specific markers. To assess cardiac looping, we performed in situ hybridization to detect gene expression of myl7/cmlc2 (myosin, light polypeptide 7, regulatory) at 48 hpf. Control morphants displayed a normal looping pattern, resulting in the ventricle being placed slightly rostral and to the right of the atrium (Fig. 4). In contrast, autophagy morphant hearts showed defects including failure to develop a distinct heart tube in some cases, and failure of looping resulting in co-linear orientation of the 2 chambers in other cases (Fig. 4A and C).
Figure 4. Autophagy gene knockdown results in abnormal cardiac looping and valve development. (A and C) In situ hybridization with a myl7 probe of morphant hearts at 48 hpf. (A) Representative images of a control morphant showing correct looping with the atrium placed left and caudal, and the ventricle right and rostral, and of autophagy morphants, showing linearized hearts without looping. (C) Quantification of data shown in (A). More than 30 embryos were analyzed in each group. (B and D) In situ hybridization of morphants with probes that detect cardiac valve markers at 2 dpf. (B) Representative images of vcan, bmp4, and notch1b expression in the hearts of control and autophagy mutants. (D) Quantification of data shown in (B). More than 30 embryos in each group were analyzed. R, right; L, left.
To further analyze cardiac valve development in autophagy morphants, we performed in situ hybridization to detect expression of 3 genes, vcan, bmp4, and notch1b, which have been shown to be important in cardiac valve development (Fig. 4B and D). Atrioventricular valve development is a highly regulated process, involving the regulation of multiple signaling networks in cardiomyocytes and endocardial cells, and resulting in endothelial cells undergoing epithelial-mesenchymal transition to populate the endocardial cushions, which give rise to the valves.35-37 In control MO-injected embryos, the expression of vcan, bmp4, and notch1b is confined to the region of the AV valve. In contrast, these genes were misexpressed after knockdown of autophagy genes. vcan was ectopically expressed throughout the ventricle in atg5 and atg7 morphants, and its expression was increased in the AV valve and in parts of the atrium and ventricle in becn1 morphants. The cardiomyocyte marker bmp4 and the endocardial marker notch1b were ectopically expressed, most strikingly in the ventricle, in about 50% of autophagy morphants. In some autophagy morphants, the expression of bmp4 and notch1b was markedly decreased or absent (Fig. 4B and D). Thus, autophagy gene knockdown leads to defective cardiac looping, defective valve formation and ectopic expression of valve markers, suggesting that inhibition of autophagy disrupts gene regulatory networks critical for proper cardiac morphogenesis.
Autophagy knockdown alters the gene expression pattern of the developing heart
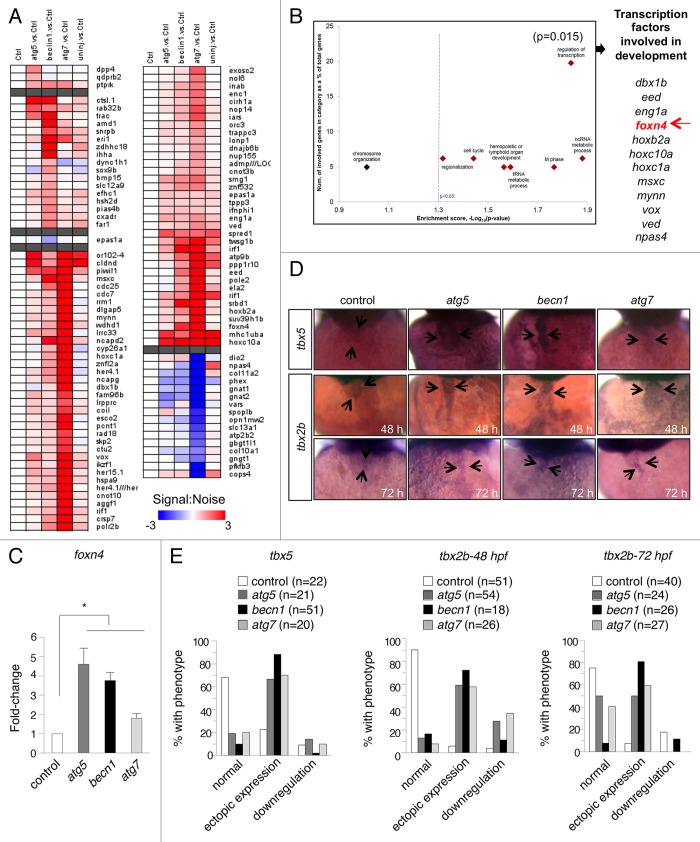
To further examine the effects of autophagy inhibition on cardiac gene expression, we purified RNA from the hearts of control and autophagy morphants at 3 dpf and profiled gene expression by microarray analyses. We identified 109 genes (Fig. 5A) that were differentially expressed (P < 0.05 and log2fold-change > 2×) between control MO- and autophagy MO-injected hearts, and also informative and predictive of an altered autophagy state, as evaluated by achieving an area under the receiver-operating characteristic curve (AUROC) of > 0.75, which indicates favorable true-positive rates over false-positives. Transcripts that were differentially regulated in autophagy morphants included a large number of genes that are important for cardiac development and function, including slc12a9, nup155, lrpprc, snrpb, crsp7/MED26, and spred1. The differentially expressed genes were enriched for fundamental biological processes including regionalization, cell cycle, hematopoiesis, tRNA metabolism, and noncoding RNA metabolic processes. Most strikingly, we observed among differentially expressed genes in morphants an overrepresentation (hypergeometric P = 0.015) of transcriptional factors with roles in development, including foxn4, dbx1b, eed, eng1a, hoxb2a, hoxc10a, hoxc1a, msxc, mynn, vox, ved, and npas4 (Fig. 5B). The magnitude of these effects was greatest for atg7 morphants, but similar changes were also observed in atg5 and becn1 morphants. We confirmed the differential expression of dbx1b, eed, hoxb2a, her15.1, mynn, msxc, and znfl2a by quantitative RT-PCR of RNA from purified hearts (Fig. S7). These data demonstrate that developmental cardiac gene transcription programs are aberrant in autophagy morphant zebrafish.
Figure 5. Autophagy gene knockdown results in upregulation of transcription factors that are involved in development. (A) Heat map of altered transcripts from the hearts of autophagy morphants at 3 dpf. 109 genes were identified which are informative and predictive of various altered autophagy states. (B) Enrichment analysis of biological processes overrepresented in genes that are differentially expressed and informative of the altered autophagy state. Of interest is the set of transcription factors that are involved in development, including foxn4 (arrow). (C) Real-time PCR measurement of foxn4 mRNA levels in purified hearts of autophagy morphants at 3 dpf. Results represent the mean ± SEM of triplicate samples. *P < 0.05 vs. control morphants; one-tailed t test. (D and E) In situ hybridization to detect tbx5 and tbx2b expression in morphant hearts. (D) Representative images of control and autophagy morphants, with the autophagy morphants showing stronger expression of tbx5 and tbx2b throughout the heart (arrows). (E) Quantification of data shown in (D). More than 18 embryos in each group were analyzed.
We performed additional studies to validate the abnormal expression of a key gene important for cardiac development, foxn4, previously shown to be critical for proper AVC formation in zebrafish.38 To validate the differential expression of foxn4 in autophagy morphant fish, we performed quantitative RT-PCR on purified heart mRNA (Fig. 5C). In agreement with the microarray results, foxn4 expression was upregulated in all 3 autophagy morphants (Fig. 5C). tbx5 normally cooperates with foxn4 to regulate expression of tbx2b during formation of the AVC. Misexpression of tbx2b causes defective AV canal development and impaired cardiac function.38 In situ hybridization showed that autophagy gene knockdown caused increased expression and mislocalization of tbx5 in the developing heart (Fig. 5D and E). Consistent with this result, autophagy morphants exhibited expanded expression of tbx2b throughout the atrium and ventricle as well as in the AVC (Fig. 5D and E). Taken together, these data indicate a critical role for autophagy in regulating cardiac development and proper patterning of the AV canal.
Atg5 knockout mice exhibit cardiac defects
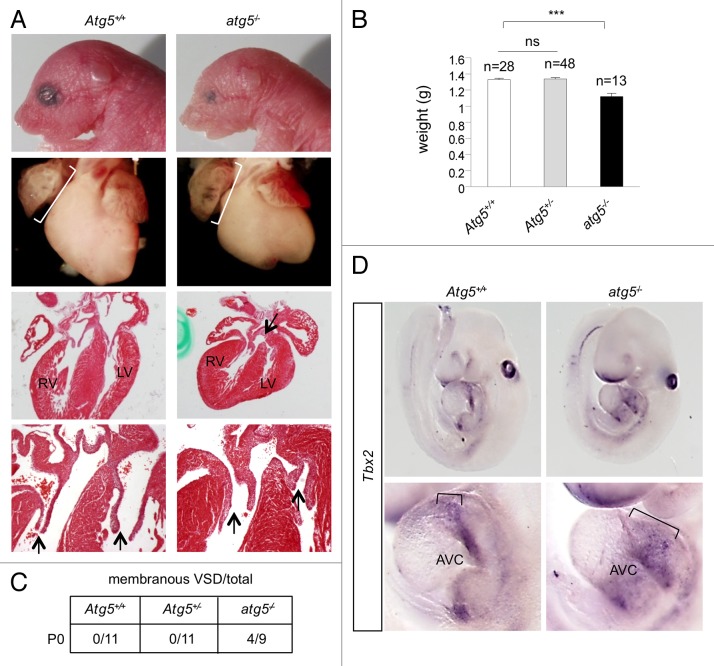
Our data above indicate that autophagy knockdown induces developmental cardiac defects in zebrafish. We next asked if this role of autophagy is conserved in mammals, especially in light of the striking conservation of the autophagy pathway and its functions across diverse species. To investigate whether autophagy plays a role in mammalian cardiac development, we first imaged the hearts of E9.5 embryos from GFP-LC3 transgenic mice39 and observed GFP puncta, indicating the presence of autophagy during cardiac development (Fig. S8). Next, we examined postnatal d 0 (P0) Atg5-deficient mice. atg5−/− mice have previously been shown to die within several hours of birth, due to defective nutrient and energy homeostasis.40 atg5−/− mice derived from crosses of Atg5+/− heterozygotes were significantly smaller than Atg5+/+ or Atg5+/− mice at P0 (Fig. 6A and B). atg5−/− pups frequently had small heads and morphologically abnormal eyes, reminiscent of the defects seen in autophagy morphant zebrafish. Upon macroscopic analysis of P0 hearts, we noted enlarged right atria (relative to the size of total heart mass) in all Atg5−/− mice examined (Fig. 6A, middle). Upon histological analysis, 4 out of 9 atg5−/− mice were found to have AV canal defects including abnormal valve morphology and a membranous ventricular septal defect (VSD) at P0 (Fig. 6A and C). Six of the 9 atg5−/− mice exhibited abnormal, thickened valves (Fig. 6A). Atg5+/+ mice or Atg5+/− mice did not show the same cardiac developmental abnormalities (n = 11 in each group), although 1 out of 11 in each of the Atg5+/+ and Atg5+/− groups exhibited a small muscular VSD.
Figure 6.atg5−/− mice exhibit defective cardiac development. (A) Representative images of atg5−/− and Atg5+/+ mice (at d P0). Top row, photomicrographs of heads and eyes; second row from top, photomicrographs of hearts; third and fourth rows, hematoxylin and eosin sections of hearts. Brackets in third row denote enlarged right atrium; arrow in third row denotes ventricular septal defect. Arrows in the fourth row denote AV valve thickening. (B) Body weights of atg5−/−, Atg5+/− and Atg5+/+ mice. Results represent mean ± SEM ***P < 0.005 for indicated comparison; one-tailed t test. (C) Summary of prevalence of membranous ventricular septal defect in mice of the indicated genotype at d P0. (D) In situ hybridization to detect Tbx2 expression in atg5−/− and Atg5+/+ mice (at d E9.5). Top panels, photomicrographs of Tbx2 expression in the whole embryo; lower panels, photomicrographs of Tbx2 expression in hearts. Brackets in lower panel denote expanded Tbx2 expression in atrioventricular cushion (AVC). Results shown are representative of results from at least 4 embryos per genotype.
To determine whether impairment of autophagy in the murine heart leads to similar gene expression abnormalities as those we observed in the zebrafish autophagy morphants, we isolated E9.5 embryos from crosses of Atg5+/− heterozygotes and performed in situ hybridization for Tbx2 (Fig. 6D). At this stage, Tbx2 expression is normally restricted to the myocardium of the AVC, the inner curvature, and the outflow tract.41,42 Consistent with the results in zebrafish, all atg5−/− mice displayed abnormal Tbx2 expression; one mutant exhibited stronger expression, and three others displayed expanded, ectopic Tbx2 expression. In contrast, 4/4 Atg5+/− and 4/5 Atg5+/+ littermates displayed normal Tbx2 expression. These results indicate that the requirement of autophagy for developmental cardiac patterning is conserved from fish to mammals.
Discussion
Here we demonstrated that autophagy plays an essential role in cardiac morphogenesis during vertebrate development. Autophagy is induced during the period of rapid early embryonic growth and organogenesis, and knockdown of autophagy genes causes severe developmental defects. The autophagy pathway is active in the developing heart, and inhibition of autophagy disrupts the gene expression program responsible for cardiac morphogenesis, leading to misregulation of key cardiac-specific transcription factors and ectopic expression of cardiac patterning genes. The result is aberrant development of the cardiac chambers and the atrioventricular canal, as well as increased embryonic lethality. While previous studies have indicated that specific autophagy factors are required for development of the Drosophila eye,12,43 the zebrafish brain,19,20 and for mouse development (reviewed in ref. 7), our results provide the first evidence that autophagy contributes to the cardiac morphogenic program during vertebrate development. We further found that mice deficient in Atg5 exhibit defective development of the AV canal and membranous VSDs. These findings are consistent with defects of endocardial cushion development. Consistent with a role for autophagy in early developmental morphogenesis of the heart, we showed that autophagy-deficient atg5−/− mice display aberrant expression of the cardiac patterning gene Tbx2. These observations strongly resemble the cardiac phenotype of zebrafish deficient in autophagy, indicating that the essential role of ATG genes in developmental morphogenic patterning is conserved throughout vertebrate evolution.
In mouse embryos, autophagy is activated during the oocyte-to-embryo transition that occurs after fertilization in preimplantation embryos.25 The viable phenotype of many mouse autophagy gene mutants has been attributed to maternally supplied autophagy factors.7 In zebrafish, the yolk serves as a source of nutrients during early development. Using the biochemical conversion of Lc3-I to Lc3-II as a marker of active autophagy, He and coworkers reported that autophagy was detectable beginning at 32 hpf.26 We detected autophagy in somite-stage (approximately 15 hpf) embryos, as evidenced by the presence of GFP-Lc3 puncta, which mark autophagosomes in the CMV:GFP-Lc3 transgenic reporter line. The earlier appearance of autophagy in our experiments may have to do with the increased sensitivity of confocal microscopy to detect the occurrence of autophagy in individual cells, as opposed to the biochemical assay used by He et al. In fact, we detected autophagosome formation in primary cultures of cells prepared from shield-stage embryos (6 hpf). While it is possible that autophagy in this setting is a cellular response to in vitro culture and the absence of the yolk, our results indicated that embryonic cells are competent for autophagy from an early time in development. Taken together with our detection of autophagy gene transcripts, both maternally supplied and at several stages in the first 24 h of development, as well as our demonstration of robust autophagy occurring in intact, somite-stage embryos, our results indicated that autophagy is part of the early developmental program in zebrafish embryos, during the period of active tissue morphogenesis and organogenesis.
It is perhaps somewhat surprising that extensive autophagy occurs during early fish development, given that in amniotes, the yolk serves as a ready supply of nutrients. In this setting, autophagy may serve to support the high metabolic demands of tissues undergoing large-scale morphogenesis and cell movement, which could fulfill energetic needs beyond those supported by relatively slow diffusion of nutrients from the yolk. Alternatively, autophagy in developing zebrafish tissues may reflect an intrinsic role of the pathway in cellular differentiation, similar to what has been described in yeast sporulation, Dictyostelium fruiting body formation, erythrocyte maturation, and the differentiation of trypanosomes.7
Whether it serves to support the metabolic demands of tissue undergoing morphogenesis, a cellular differentiation program, or both processes, autophagy is necessary for normal development in zebrafish, as evidenced by the pleiotropic effects of autophagy gene knockdown. Knockdown of atg5, becn1, or atg7 all led to similar defects in the embryos, including microcephaly, microphthalmia, and abnormal body shape. Autophagy gene knockdown in tp53-deficient strains produced nearly identical results, further ruling out off-target effects.
Many autophagy genes are essential for functioning of the pathway, as shown by the phenotypes of loss-of-function mutations of individual autophagy genes in yeast, nematodes, and mammalian cells.7,44-46 Reflecting the nonredundant functions of autophagy genes, knockdown either of atg5, becn1, or atg7 alone were sufficient to inhibit autophagy and induce developmental defects. We did observe subtle differences in the knockdown phenotypes of different morphants; for example atg7 morphants exhibited a lower percentage of embryos with defective cardiac looping. These differences among the atg5, becn1, and atg7 morphants could be due to different efficiencies of knockdown, though based on immunoblot detection of protein levels, all MOs appeared to be equally effective in decreasing autophagy gene expression. Alternatively, the amount and half-life of maternally expressed autophagy genes may differ for the three genes we studied. It is also possible that individual autophagy genes are expressed at different levels and/or required to different degrees in specific tissues or organs. In mice, different autophagy mutants exhibit a range of developmental defects, from early embryonic lethality to full viability (reviewed in ref. 7). Interestingly, despite the overall similar phenotypes of atg5, becn1, or atg7 knockdown, the autophagy MOs had distinct effects on the long-term survival of zebrafish. Even beyond 5 d, after which time MOs generally no longer provide effective knockdown,47 the morphants continued to have impaired survival. Poor survival did not directly correlate with the severity of the embryonic phenotype, as atg7 morphants exhibited the most rapid fall in viability despite having a somewhat milder apparent developmental phenotype. Taken together, despite variation in the penetrance of abnormal phenotypes in different autophagy morphants, these results indicate a role for autophagy in embryonic morphogenesis and in physiological processes that are essential for long-term viability.
In the zebrafish, cardiac development begins about 12 hpf, with the specification of bilateral stripes of cardiac-progenitor cells in the lateral mesoderm.48,49 The progenitors migrate medially and fuse to form the linear heart tube, which then undergoes looping and folding movements, resulting in the formation of 2 distinct chambers. During this process, from approximately 37 hpf to 72 hpf, a set of endocardial cells in the atrioventricular canal (AVC) region undergo morphological changes and delamination which, over the course of the next several days, results in formation of the AV valve leaflets.50,51 Molecular markers of this change include the restriction of bmp4 and vcan to the AVC myocardium, and notch1b to the AVC endocardium.35
Having noted the striking cardiac phenotype resulting from autophagy knockdown, we examined the genes that were differentially expressed in the knockdown embryos, and found enrichment for a number of fundamental cellular pathways, including cell cycle, metabolism, and M-phase. Most significantly, we saw altered expression of a large number of genes encoding transcriptional regulators. In this group of genes, foxn4 was of particular interest as it regulates development of the AVC, cooperating with tbx5 to regulate expression of tbx2b.38 Rather than the normally restricted expression in the AVC, both tbx5 and tbx2b were ectopically expressed throughout the heart tube after autophagy knockdown. Thus, proper functioning of the autophagy pathway is required for the normal morphological and gene expression changes driving AVC formation.
We identified autophagy in multiple regions of developing hearts. The sensitivity of AVC development to autophagy inhibition may reflect a need for autophagy to contribute to the energy metabolism in cells undergoing dramatic morphological changes and movements. Alternatively, autophagy may be required to regulate one or more signaling pathways necessary to restrict expression of the valve development program to a specific set of cells. In the absence of autophagy, misexpression of key AVC transcription factors leads to continued ectopic expression of downstream targets such as notch1b, bmp4a, and vcan outside the AVC, and results in impaired cardiac development.
To understand whether these findings are specific to zebrafish or represent a more generally conserved role for autophagy in cardiac development, we carefully examined the phenotype of Atg5-deficient mice. Atg5 knockout mice are born at normal ratios, but die soon after birth from suckling defects and an impaired starvation response.40 The cardiac phenotype of these mice has not been studied in detail. Compared with wild-type or Atg5-heterozygous siblings, atg5−/− mice weighed less and had smaller heads and smaller eyes, strongly reminiscent of the developmental phenotypes of autophagy-deficient zebrafish. Most significantly, at P0 nearly half of the atg5−/− mice exhibited severe AVC developmental defects, including abnormal valve development and membranous VSD. At earlier stages of development, atg5−/− mice show abnormal expression of the transcription factor, Tbx2. Therefore, impaired autophagy leads to defects in the transcriptional patterning and morphogenesis of the AVC, and this effect is conserved across vertebrates.
There are several implications of this work. First, we have revealed an unanticipated role for autophagy in cardiac morphogenesis during development. Autophagy is present in developing tissues, and knockdown of autophagy genes causes profound defects in the development of tissues such as eye, brain, and heart, which must undergo major morphological changes during development. Second, we show that the effect of deficient autophagy on developmental patterning of the AVC is conserved from fish to mammals. These results indicate that autophagy genes should be examined as candidates for the many unexplained cases of human congenital cardiac defects. Furthermore, as autophagy has previously been invoked as having both adaptive and maladaptive effects in hearts under hemodynamic stress,52-54 it will be of interest to study whether autophagic control of developmental gene expression, as we have described here, may also play a role in response to cardiac stress and injury.
Materials and Methods
Zebrafish strains and lines
Embryos from wild-type AB strain were used for MO injection. Fish were raised and maintained under standard conditions.55 Autophagy reporter fish Tg(cmv:GFP-lc3) (gift of Dr Daniel J Klionsky, University of Michigan) were previously described.26 Tg(fli1:GFP)34 were a gift of Dr Nathan Lawson (University of Massachusetts) and tp53M214K mutants28 were a gift of Dr Tom Look, Dana-Farber Cancer Institute.
Mouse strains
Autophagy reporter GFP-LC3 mice39 and Atg5 knockout mice40 have been previously described. atg5−/− mice were generated by obtaining offspring of Atg5+/− crosses. All animal protocols were approved by the UT Southwestern Medical Center Institutional Animal Care and Use Committee.
Zebrafish embryonic cell cultures
Approximately 50 autophagy MO- or control MO-injected embryos were used for primary cell culture. Injected embryos at the shield stage were sterilized with 70% ethanol, and dissociated in trypsin/EDTA solution (GIBCO, 15400). Primary cells were plated on a Lab-tek chamber slide (Nunc, 177437) in LDF (Leibovitz 50%, DMEM 35%, F-12 15%) medium containing antibiotics and incubated at 25 °C.
Zebrafish RNA preparation and RT-PCR
RNA from 1-cell, sphere, germ ring, 5-somite, 24 hpf and 2 dpf stages of zebrafish embryos was purified by Trizol (Invitrogen, 15596026) according to the manufacturer’s instructions. cDNA was synthesized using a RT2 HT first strand kit (Qiagen, 330411). To detect different autophagy transcripts, PCR reactions were performed. The primer sequences used are listed in the Supplementary Methods.
Morpholino injections
Translation-blocking and splice-blocking MOs directed against atg5, becn1, and atg7 MOs were obtained from Gene-Tools. Control injections were done with standard control MO from Gene-Tools. Embryos were injected at the 1-cell stage with 0.3–1 mM of MOs. The MO sequences used are listed in the Supplementary Methods.
Western blot analyses
Embryos at the 2 dpf stage were anesthetized with tricaine and incubated for 30 min on ice in lysis buffer containing 150 mM NaCl, 10mM Tris pH7.4, 0.2% Triton X-100, 0.3% NP-40, 0.2 mM Na3VO4 and protease inhibitors (Roche, 11697498001). After centrifugation at 13,000 × g, supernatant fractions were used for immunoblotting. Primary cells were prepared after autophagy or control MO injection and used for immunoblotting. Antibodies to Atg5 (Novus, NB110-53818), Becn1 (Beclin 1) (Santa Cruz Biotechnology, sc-11427), LC3 (Novus, NB100-2220) and Actb/β-actin antibody (Millipore, MAB1501R) were used for immunoblotting.
Acridine orange staining
1-dpf embryos were washed with PBS and incubated in PBS containing 2 μg/ml acridine orange (Fisher Biotech, BP116-10) for 30 min.56 After staining, embryos were washed 5 times with E3 buffer (5 mM NaCl, 0.17 mM KCl, 0.33 mM CaCl2, 0.33 mM MgOS4). Mounted embryos in 3% methylcellulose (Sigma, M0387) were visualized using a Leica MZ16 FA fluorescence stereo dissecting microscope (Leica).
Microscopy imaging
Images of whole-mount zebrafish embryos were taken with a Nikon Coolpix 4500 mounted on a Leica MZ125 stereo dissecting microscope. Images of GFP-Lc3 in primary cells or zebrafish embryos were acquired on a Zeiss LSM 510 META confocal microscope. The embryos were treated with 0.003% 1-phenyl-2-thiourea to reduce pigment at 24 hpf. For confocal imaging, embryos were maintained in 0.002% tricaine and mounted in 3% low-melt agarose. Confocal z-stack images were used and analyzed by IMARIS.
Transmission electron microscopy
Ten somite stage-embryos were fixed with 2.5% glutaraldehyde in 0.1 M cacodylate buffer pH 7.4 for 3 h. Sections were imaged using a JEOL 1200 EX electron microscope at 120 KV equipped with a Sis Morada 11 megapixel mount CCD camera.
Survival assay
More than 100 embryos were injected with autophagy or control MOs. The number of surviving injected fish was counted daily for a month.
Histological analysis
Injected zebrafish embryos were fixed at 3 dpf in 4% paraformaldehyde (PFA) for 48 h and sagittal sections were prepared. Postnatal pups were sacrificed and the heart was dissected, fixed in 4% PFA, and dehydrated. Transverse sections were stained with hematoxylin and eosin for tissue morphology.
In situ hybridization
Whole mount in situ hybridizations were performed using standard protocols with zebrafish embryos at 2 dpf to detect myl7, vcan, bmp4, notch1b, and tbx2b and at 3 dpf to detect tbx2b and tbx5. Primers used for riboprobe synthesis are listed in the Supplementary Methods. T7 primer (TAATACGACT CACTATAGGG AGA) was added to reverse primers to facilitate antisense riboprobe synthesis. Riboprobe against vcan was kindly provided by Dr Didier Stainier (UCSF). Mouse embryos at E9.5 were used for in situ hybridization to detect Tbx2 using a previously described Riboprobe.57
Microarray analyses
Zebrafish hearts were purified from control MO- or autophagy MO-injected embryos as described.58 RNA from each group of injected embryos was purified at 3 dpf by Trizol, digested with DNase, and further purified using RNeasy columns (Qiagen, 74104). RNA analyses were performed at the microarray core facility at the Harvard and Partners Healthcare Center for Genetics and Genomics. The quantity, purity, and integrity of RNA were evaluated by UV spectrophotometry and RNA-nano Bioanalyzer (Agilent). Sample processing and hybridization on Zebrafish Genome GeneChip microarrays (Affymetrix) were performed according to the manufacturer’s instructions. Probeset-level normalization was achieved using the MAS5.0 algorithm.59 Data analysis and linear modeling were performed in the R programming language, utilizing functions from Linear Models for Microarray Data.60 For each probe, a moderated t-statistic (with standard errors moderated across genes) was computed using a Bayesian model. For each transcript, signal-to-noise ratios (SNR) were computed for each respective morphant compared with the control group. The expression SNR profiles of transcripts identified as differentially expressed (P < 0.05 and fold-change > 2×) were further subjected to feature (attribute) selection61,62 using Information Gain (Kullback-Leibler divergence or relative entropy) to identify transcripts that are most informative of the respective altered autophagy state. The accuracy of class prediction using a Java implementation of a naïve Bayes classifier62,63 incorporating these features was evaluated using 10-fold cross-validation. Enrichment analysis of biological processes was performed by computing the hypergeometric test as previously described.64 Biological process annotations and assignments were obtained from Gene Ontology (GO). Microarray data have been deposited at www.ncbi.nlm.nih.gov/geo, accession number GSE51541
Real-time PCR analyses
Total RNA prepared from heart was used for quantitative RT-PCR. cDNA was synthesized using RT2 HT first strand kit (Qiagen, 330411). Quantitative PCR reaction for foxn4 was performed with QuantiFast SYBR Green PCR kit (Qiagen, 204054) using an Applied Biosystems 7500 Real-time thermocycler. All signals were analyzed and normalized to actb/β-actin mRNA. Primers for foxn4 were obtained from Qiagen and primers for actb have been described previously.65
Supplementary Material
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank Ashley Ratley for zebrafish care. This work was supported by grant 5R01CA135731 and a grant from the Amon G Carter Foundation to JFA; and R01CA109618 and CPRIT RP120718-P1 to BL. We thank Daniel Klionsky, Noboru Mizushima, and Tom Look for providing critical reagents and Haley Harrington for providing assistance with manuscript preparation.
Glossary
Abbreviations:
- AV
atrioventricular
- AVC
atrioventricular canal
- dpf
days post-fertilization
- hpf
hours post-fertilization
- MO
morpholino oligonucleotide
References
- 1.Mizushima N. Autophagy in protein and organelle turnover. Cold Spring Harb Symp Quant Biol. 2011;76:397–402. doi: 10.1101/sqb.2011.76.011023. [DOI] [PubMed] [Google Scholar]
- 2.Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–75. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Levine B, Klionsky DJ. Development by self-digestion: molecular mechanisms and biological functions of autophagy. Dev Cell. 2004;6:463–77. doi: 10.1016/S1534-5807(04)00099-1. [DOI] [PubMed] [Google Scholar]
- 5.Cecconi F, Levine B. The role of autophagy in mammalian development: cell makeover rather than cell death. Dev Cell. 2008;15:344–57. doi: 10.1016/j.devcel.2008.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mizushima N, Komatsu M. Autophagy: renovation of cells and tissues. Cell. 2011;147:728–41. doi: 10.1016/j.cell.2011.10.026. [DOI] [PubMed] [Google Scholar]
- 7.Mizushima N, Levine B. Autophagy in mammalian development and differentiation. Nat Cell Biol. 2010;12:823–30. doi: 10.1038/ncb0910-823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Juhász G, Csikós G, Sinka R, Erdélyi M, Sass M. The Drosophila homolog of Aut1 is essential for autophagy and development. FEBS Lett. 2003;543:154–8. doi: 10.1016/S0014-5793(03)00431-9. [DOI] [PubMed] [Google Scholar]
- 9.Berry DL, Baehrecke EH. Growth arrest and autophagy are required for salivary gland cell degradation in Drosophila. Cell. 2007;131:1137–48. doi: 10.1016/j.cell.2007.10.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Denton D, Shravage B, Simin R, Mills K, Berry DL, Baehrecke EH, Kumar S. Autophagy, not apoptosis, is essential for midgut cell death in Drosophila. Curr Biol. 2009;19:1741–6. doi: 10.1016/j.cub.2009.08.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shen W, Ganetzky B. Autophagy promotes synapse development in Drosophila. J Cell Biol. 2009;187:71–9. doi: 10.1083/jcb.200907109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen SF, Kang ML, Chen YC, Tang HW, Huang CW, Li WH, Lin CP, Wang CY, Wang PY, Chen GC, et al. Autophagy-related gene 7 is downstream of heat shock protein 27 in the regulation of eye morphology, polyglutamine toxicity, and lifespan in Drosophila. J Biomed Sci. 2012;19:52. doi: 10.1186/1423-0127-19-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang S, Jia K, Wang Y, Zhou Z, Levine B. Autophagy genes function in apoptotic cell corpse clearance during C. elegans embryonic development. Autophagy. 2013;9:138–49. doi: 10.4161/auto.22352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li W, Zou W, Yang Y, Chai Y, Chen B, Cheng S, Tian D, Wang X, Vale RD, Ou G. Autophagy genes function sequentially to promote apoptotic cell corpse degradation in the engulfing cell. J Cell Biol. 2012;197:27–35. doi: 10.1083/jcb.201111053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ruck A, Attonito J, Garces KT, Núnez L, Palmisano NJ, Rubel Z, Bai Z, Nguyen KC, Sun L, Grant BD, et al. The Atg6/Vps30/Beclin 1 ortholog BEC-1 mediates endocytic retrograde transport in addition to autophagy in C. elegans. Autophagy. 2011;7:386–400. doi: 10.4161/auto.7.4.14391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang Y, Yan L, Zhou Z, Yang P, Tian E, Zhang K, Zhao Y, Li Z, Song B, Han J, et al. SEPA-1 mediates the specific recognition and degradation of P granule components by autophagy in C. elegans. Cell. 2009;136:308–21. doi: 10.1016/j.cell.2008.12.022. [DOI] [PubMed] [Google Scholar]
- 17.Al Rawi S, Louvet-Vallée S, Djeddi A, Sachse M, Culetto E, Hajjar C, Boyd L, Legouis R, Galy V. Postfertilization autophagy of sperm organelles prevents paternal mitochondrial DNA transmission. Science. 2011;334:1144–7. doi: 10.1126/science.1211878. [DOI] [PubMed] [Google Scholar]
- 18.Sato M, Sato K. Degradation of paternal mitochondria by fertilization-triggered autophagy in C. elegans embryos. Science. 2011;334:1141–4. doi: 10.1126/science.1210333. [DOI] [PubMed] [Google Scholar]
- 19.Hu Z, Zhang J, Zhang Q. Expression pattern and functions of autophagy-related gene atg5 in zebrafish organogenesis. Autophagy. 2011;7:1514–27. doi: 10.4161/auto.7.12.18040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Benato F, Skobo T, Gioacchini G, Moro I, Ciccosanti F, Piacentini M, Fimia GM, Carnevali O, Dalla Valle L. Ambra1 knockdown in zebrafish leads to incomplete development due to severe defects in organogenesis. Autophagy. 2013;9:476–95. doi: 10.4161/auto.23278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mizushima N, Yoshimori T, Levine B. Methods in mammalian autophagy research. Cell. 2010;140:313–26. doi: 10.1016/j.cell.2010.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dodd A, Curtis PM, Williams LC, Love DR. Zebrafish: bridging the gap between development and disease. Hum Mol Genet. 2000;9:2443–9. doi: 10.1093/hmg/9.16.2443. [DOI] [PubMed] [Google Scholar]
- 23.Liu J, Stainier DY. Zebrafish in the study of early cardiac development. Circ Res. 2012;110:870–4. doi: 10.1161/CIRCRESAHA.111.246504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pelster B, Burggren WW. Disruption of hemoglobin oxygen transport does not impact oxygen-dependent physiological processes in developing embryos of zebra fish (Danio rerio) Circ Res. 1996;79:358–62. doi: 10.1161/01.RES.79.2.358. [DOI] [PubMed] [Google Scholar]
- 25.Tsukamoto S, Kuma A, Murakami M, Kishi C, Yamamoto A, Mizushima N. Autophagy is essential for preimplantation development of mouse embryos. Science. 2008;321:117–20. doi: 10.1126/science.1154822. [DOI] [PubMed] [Google Scholar]
- 26.He C, Bartholomew CR, Zhou W, Klionsky DJ. Assaying autophagic activity in transgenic GFP-Lc3 and GFP-Gabarap zebrafish embryos. Autophagy. 2009;5:520–6. doi: 10.4161/auto.5.4.7768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Robu ME, Larson JD, Nasevicius A, Beiraghi S, Brenner C, Farber SA, Ekker SC. p53 activation by knockdown technologies. PLoS Genet. 2007;3:e78. doi: 10.1371/journal.pgen.0030078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Berghmans S, Murphey RD, Wienholds E, Neuberg D, Kutok JL, Fletcher CD, Morris JP, Liu TX, Schulte-Merker S, Kanki JP, et al. tp53 mutant zebrafish develop malignant peripheral nerve sheath tumors. Proc Natl Acad Sci U S A. 2005;102:407–12. doi: 10.1073/pnas.0406252102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Qu X, Zou Z, Sun Q, Luby-Phelps K, Cheng P, Hogan RN, Gilpin C, Levine B. Autophagy gene-dependent clearance of apoptotic cells during embryonic development. Cell. 2007;128:931–46. doi: 10.1016/j.cell.2006.12.044. [DOI] [PubMed] [Google Scholar]
- 30.Mellén MA, de la Rosa EJ, Boya P. The autophagic machinery is necessary for removal of cell corpses from the developing retinal neuroepithelium. Cell Death Differ. 2008;15:1279–90. doi: 10.1038/cdd.2008.40. [DOI] [PubMed] [Google Scholar]
- 31.Fimia GM, Stoykova A, Romagnoli A, Giunta L, Di Bartolomeo S, Nardacci R, Corazzari M, Fuoco C, Ucar A, Schwartz P, et al. Ambra1 regulates autophagy and development of the nervous system. Nature. 2007;447:1121–5. doi: 10.1038/nature05925. [DOI] [PubMed] [Google Scholar]
- 32.Bill BR, Petzold AM, Clark KJ, Schimmenti LA, Ekker SC. A primer for morpholino use in zebrafish. Zebrafish. 2009;6:69–77. doi: 10.1089/zeb.2008.0555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stainier DY. Zebrafish genetics and vertebrate heart formation. Nat Rev Genet. 2001;2:39–48. doi: 10.1038/35047564. [DOI] [PubMed] [Google Scholar]
- 34.Lawson ND, Weinstein BM. In vivo imaging of embryonic vascular development using transgenic zebrafish. Dev Biol. 2002;248:307–18. doi: 10.1006/dbio.2002.0711. [DOI] [PubMed] [Google Scholar]
- 35.Beis D, Bartman T, Jin SW, Scott IC, D’Amico LA, Ober EA, Verkade H, Frantsve J, Field HA, Wehman A, et al. Genetic and cellular analyses of zebrafish atrioventricular cushion and valve development. Development. 2005;132:4193–204. doi: 10.1242/dev.01970. [DOI] [PubMed] [Google Scholar]
- 36.Chang CP, Neilson JR, Bayle JH, Gestwicki JE, Kuo A, Stankunas K, Graef IA, Crabtree GR. A field of myocardial-endocardial NFAT signaling underlies heart valve morphogenesis. Cell. 2004;118:649–63. doi: 10.1016/j.cell.2004.08.010. [DOI] [PubMed] [Google Scholar]
- 37.Hurlstone AF, Haramis AP, Wienholds E, Begthel H, Korving J, Van Eeden F, Cuppen E, Zivkovic D, Plasterk RH, Clevers H. The Wnt/beta-catenin pathway regulates cardiac valve formation. Nature. 2003;425:633–7. doi: 10.1038/nature02028. [DOI] [PubMed] [Google Scholar]
- 38.Chi NC, Shaw RM, De Val S, Kang G, Jan LY, Black BL, Stainier DY. Foxn4 directly regulates tbx2b expression and atrioventricular canal formation. Genes Dev. 2008;22:734–9. doi: 10.1101/gad.1629408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mizushima N, Yamamoto A, Matsui M, Yoshimori T, Ohsumi Y. In vivo analysis of autophagy in response to nutrient starvation using transgenic mice expressing a fluorescent autophagosome marker. Mol Biol Cell. 2004;15:1101–11. doi: 10.1091/mbc.E03-09-0704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kuma A, Hatano M, Matsui M, Yamamoto A, Nakaya H, Yoshimori T, Ohsumi Y, Tokuhisa T, Mizushima N. The role of autophagy during the early neonatal starvation period. Nature. 2004;432:1032–6. doi: 10.1038/nature03029. [DOI] [PubMed] [Google Scholar]
- 41.Christoffels VM, Hoogaars WM, Tessari A, Clout DE, Moorman AF, Campione M. T-box transcription factor Tbx2 represses differentiation and formation of the cardiac chambers. Dev Dyn. 2004;229:763–70. doi: 10.1002/dvdy.10487. [DOI] [PubMed] [Google Scholar]
- 42.Harrelson Z, Kelly RG, Goldin SN, Gibson-Brown JJ, Bollag RJ, Silver LM, Papaioannou VE. Tbx2 is essential for patterning the atrioventricular canal and for morphogenesis of the outflow tract during heart development. Development. 2004;131:5041–52. doi: 10.1242/dev.01378. [DOI] [PubMed] [Google Scholar]
- 43.Zirin J, Perrimon N. Drosophila as a model system to study autophagy. Semin Immunopathol. 2010;32:363–72. doi: 10.1007/s00281-010-0223-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Meléndez A, Tallóczy Z, Seaman M, Eskelinen EL, Hall DH, Levine B. Autophagy genes are essential for dauer development and life-span extension in C. elegans. Science. 2003;301:1387–91. doi: 10.1126/science.1087782. [DOI] [PubMed] [Google Scholar]
- 45.Shintani T, Mizushima N, Ogawa Y, Matsuura A, Noda T, Ohsumi Y. Apg10p, a novel protein-conjugating enzyme essential for autophagy in yeast. EMBO J. 1999;18:5234–41. doi: 10.1093/emboj/18.19.5234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kametaka S, Okano T, Ohsumi M, Ohsumi Y. Apg14p and Apg6/Vps30p form a protein complex essential for autophagy in the yeast, Saccharomyces cerevisiae. J Biol Chem. 1998;273:22284–91. doi: 10.1074/jbc.273.35.22284. [DOI] [PubMed] [Google Scholar]
- 47.Nasevicius A, Ekker SC. The zebrafish as a novel system for functional genomics and therapeutic development applications. Curr Opin Mol Ther. 2001;3:224–8. [PubMed] [Google Scholar]
- 48.Serbedzija GN, Chen JN, Fishman MC. Regulation in the heart field of zebrafish. Development. 1998;125:1095–101. doi: 10.1242/dev.125.6.1095. [DOI] [PubMed] [Google Scholar]
- 49.Lee RK, Stainier DY, Weinstein BM, Fishman MC. Cardiovascular development in the zebrafish. II. Endocardial progenitors are sequestered within the heart field. Development. 1994;120:3361–6. doi: 10.1242/dev.120.12.3361. [DOI] [PubMed] [Google Scholar]
- 50.Stainier DY, Beis D, Jungblut B, Bartman T. Endocardial cushion formation in zebrafish. Cold Spring Harb Symp Quant Biol. 2002;67:49–56. doi: 10.1101/sqb.2002.67.49. [DOI] [PubMed] [Google Scholar]
- 51.Scherz PJ, Huisken J, Sahai-Hernandez P, Stainier DY. High-speed imaging of developing heart valves reveals interplay of morphogenesis and function. Development. 2008;135:1179–87. doi: 10.1242/dev.010694. [DOI] [PubMed] [Google Scholar]
- 52.Nakai A, Yamaguchi O, Takeda T, Higuchi Y, Hikoso S, Taniike M, Omiya S, Mizote I, Matsumura Y, Asahi M, et al. The role of autophagy in cardiomyocytes in the basal state and in response to hemodynamic stress. Nat Med. 2007;13:619–24. doi: 10.1038/nm1574. [DOI] [PubMed] [Google Scholar]
- 53.Zhu H, Tannous P, Johnstone JL, Kong Y, Shelton JM, Richardson JA, Le V, Levine B, Rothermel BA, Hill JA. Cardiac autophagy is a maladaptive response to hemodynamic stress. J Clin Invest. 2007;117:1782–93. doi: 10.1172/JCI27523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang ZV, Ferdous A, Hill JA. Cardiomyocyte autophagy: metabolic profit and loss. Heart Fail Rev. 2013;18:585–94. doi: 10.1007/s10741-012-9350-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Westerfield M. The zebrafish book. A guide for the laboratory use of zebrafish (Danio rerio). 4th ed, Eugene, Oregon: Univ of Oregon Press, 2000 [Google Scholar]
- 56.Verduzco D, Amatruda JF. Analysis of cell proliferation, senescence, and cell death in zebrafish embryos. Methods Cell Biol. 2011;101:19–38. doi: 10.1016/B978-0-12-387036-0.00002-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bollag RJ, Siegfried Z, Cebra-Thomas JA, Garvey N, Davison EM, Silver LM. An ancient family of embryonically expressed mouse genes sharing a conserved protein motif with the T locus. Nat Genet. 1994;7:383–9. doi: 10.1038/ng0794-383. [DOI] [PubMed] [Google Scholar]
- 58.Burns CG, MacRae CA. Purification of hearts from zebrafish embryos. Biotechniques. 2006;40:278–81. doi: 10.2144/000112135. [DOI] [PubMed] [Google Scholar]
- 59.Hubbell E, Liu WM, Mei R. Robust estimators for expression analysis. Bioinformatics. 2002;18:1585–92. doi: 10.1093/bioinformatics/18.12.1585. [DOI] [PubMed] [Google Scholar]
- 60.Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3:e3. doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- 61.Xing EP, Jordan MJ, Karp RM. Feature selection in high-dimensional genomic data”,in Eighteenth International Conference on Machine Learning. In: Brodley CE, Danyluk AP, eds. Eighteenth International Conference on Machine Learning Morgan Kaufmann, 2001:601-8 [Google Scholar]
- 62.Witten IH, Frank E, Hall MA. Data mining: practical machine learning tools and techniques. Williamstown, MA, USA: Morgan Kaufmann, 2011 [Google Scholar]
- 63.Langley P, Iba W, Thompson K. An Analysis of Bayesian Classifiers. In: Swartout WR, ed. Tenth National Conference on Artificial Intelligence: MIT Press, 1992:223-8 [Google Scholar]
- 64.Hitomi J, Christofferson DE, Ng A, Yao J, Degterev A, Xavier RJ, Yuan J. Identification of a molecular signaling network that regulates a cellular necrotic cell death pathway. Cell. 2008;135:1311–23. doi: 10.1016/j.cell.2008.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Neumann JC, Chandler GL, Damoulis VA, Fustino NJ, Lillard K, Looijenga L, Margraf L, Rakheja D, Amatruda JF. Mutation in the type IB bone morphogenetic protein receptor Alk6b impairs germ-cell differentiation and causes germ-cell tumors in zebrafish. Proc Natl Acad Sci U S A. 2011;108:13153–8. doi: 10.1073/pnas.1102311108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.