Abstract
Amyotrophic lateral sclerosis (ALS) is a devastating neurodegenerative disorder caused by selective motor neuron degeneration. Abnormal protein aggregation and impaired protein degradation pathways may contribute to the disease pathogenesis. Although it has been reported that autophagy is altered in patients and animal model of ALS, little is known about the role of autophagy in motor neuron degeneration in this disease. Our previous study shows that rapamycin, an MTOR-dependent autophagic activator, accelerates disease progression in the SOD1G93A mouse model of ALS. In the present report, we have assessed the role of the MTOR-independent autophagic pathway in ALS by determining the effect of the MTOR-independent autophagic inducer trehalose on disease onset and progression, and on motor neuron degeneration in SOD1G93A mice. We have found that trehalose significantly delays disease onset prolongs life span, and reduces motor neuron loss in the spinal cord of SOD1G93A mice. Most importantly, we have documented that trehalose decreases SOD1 and SQSTM1/p62 aggregation, reduces ubiquitinated protein accumulation, and improves autophagic flux in the motor neurons of SOD1G93A mice. Moreover, we have demonstrated that trehalose can reduce skeletal muscle denervation, protect mitochondria, and inhibit the proapoptotic pathway in SOD1G93A mice. Collectively, our study indicated that the MTOR-independent autophagic inducer trehalose is neuroprotective in the ALS model and autophagosome-lysosome fusion is a possible therapeutic target for the treatment of ALS.
Keywords: amyotrophic lateral sclerosis, Cu/Zn superoxide dismutase 1, autophagy, trehalose, autophagosome-lysosome fusion
Introduction
Amyotrophic lateral sclerosis is an adult-onset fatal neurodegenerative disorder, characterized by the progressive loss of upper and lower motor neurons, as well as by the formation of misfolded protein inclusions in the motor neurons and other neurons.1-3 The majority of ALS cases are sporadic but around 10% are familial.1 Mutations in SOD1 (superoxide dismutase 1, soluble) account for approximately 20% of familial ALS.2 Although the causes of most cases of ALS are not fully understood, it is believed that a toxic gain of function resulting from abnormal SOD1 protein accumulation is probably one of the mechanisms leading to this disease.3 Therefore, strategies to limit the toxic protein aggregation by accelerating the degradation of SOD1 protein may have therapeutic potential.
In eukaryotic cells, there are 2 major systems for degradation of cytoplasmic proteins: the ubiquitin-proteasome system and the autophagy-lysosome pathway.4 It is reported that mutant SOD1 can be degraded by autophagy and/or proteasome pathways in both neuronal and non-neuronal cells.5 Previous studies have demonstrated the accumulation of autophagic vacuoles in the spinal cord of SOD1G93A mice and ALS patients by using different modulators to alter the autophagy level, indicating the possible role of autophagy in ALS progression and pathogenesis.6-8 Furthermore, activation of autophagy may be protective in certain conditions in some neurodegenerative diseases by enhancing the removal of toxic protein aggregates.9 On the other hand, emerging evidence supports the point of view that dysfunction of autophagy contributes to neurodegeneration.10,11 Several reports indicate that impaired clearance of autophagosomes may occur in Alzheimer, Parkinson, and Huntington diseases.10,12 To date, it is not known whether the accumulated autophagic vacuoles in ALS are the result of autophagy induction or autophagic flux impairment.13 Moreover, we also found that SQSTM1/p62 is accumulated in the spinal cord of SOD1G93A mice accompanied by an increased number of autophagic vacuoles.14 Hence, it is worthwhile to investigate if directly manipulating the autophagic flux affects ALS onset and progression.15
Our previous study has found that treatment of SOD1G93A mice with rapamycin, a classic mechanistic target of rapamycin (MTOR)-dependent autophagic activator, actually exacerbated motor neuron loss and exaggerated disease progression, although rapamycin induced further accumulation of autophagic vacuoles which unexpectedly failed to reduce the level of mutant SOD1 aggregation in the ALS mice.14 A later report also shows that rapamycin had no beneficial effect in disease onset and survival of the SOD1 H46R/H48Q mouse model of ALS.16 Furthermore, rapamycin-induced autophagy aggravates neuromuscular pathology in a valosin-containing protein mutant mouse model of ALS.17 In addition, lithium, a compound used as a mood stabilizer, has the effect of promoting autophagy through the MTOR-independent pathway.18 However, there are contradicting reports that lithium may improve or accelerate disease progression in clinical patients and Sod1 mutant-mouse models of ALS.19-22 Thus, these results led us to determine the effects of MTOR-independent autophagic inducers in ALS.
Trehalose is a nonreducing disaccharide present in a wide variety of organisms, including bacteria, yeast, insects, fungi, and plants, but not produced in mammalian cells.23 It has been reported that trehalose increased neuron survival by removing MAPT/tau inclusions in mutant P301S MAPT transgenic mice.24 Furthermore, trehalose was found to accelerate the clearance of mutant HTT (huntingtin) and SNCA/α-synuclein in vitro.25 Here, we administrate trehalose in adult SOD1G93A mice to study the effects and possible mechanisms of this MTOR-independent autophagic inducer on motor neuron survival, and explore the role of autophagic flux in the pathogenesis of ALS.
Results
Trehalose delayed disease onset and prolonged the life span in SOD1G93A mice
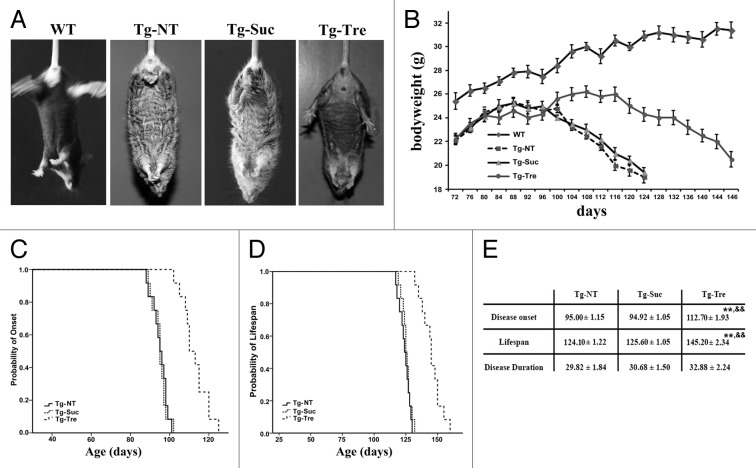
SOD1G93A mice usually develop a progressive paresis involving primarily the hind limbs with atrophy of the skeletal musculature at around 90 d of age.26 Compared with wild-type (WT) mice, SOD1G93A mice showed a typical hind limbs-clasping phenotype at 120 d (Fig. 1A). Tg-Tre mice can hold its hind limbs outward, while Tg-Suc mice did not change the post (Fig. 1A). Moreover, we tested the body weight of the mice during disease progression. As shown in Figure 1B, there was an obvious body weight loss in the 3 groups of SOD1G93A mice following disease progression. Tg-Tre mice exhibited a significantly reduction in weight loss compared with the age-matched Tg-Suc or Tg-NT mice.
Figure 1. Effects of trehalose on the phenotype, disease onset, and life span in SOD1G93A mice. (A) The different hind limb-clasping phenotype in 120 d old SOD1G93A mice; (B) The body weight curves in the 4 mouse groups; The results of Kaplan-Meier survival analysis (SPSS 17.0) showed the probability of disease onset (C) and the probability of survival (D) in the Tg-NT, Tg-Suc, Tg-Tre mice; (E) The data of disease duration, onset, and life span in NT, Suc or Tre mice. There were 6 mice in the WT group, and 12 mice in each group of Tg-NT, Tg-Suc, and Tg-Tre. P values were analyzed by one-way ANOVA. Data are presented as mean ± SEM **P < 0.01 as compared with Tg-NT group; &&P < 0.01 as compared with Tg-Suc group.
To explore whether trehalose treatment can influence disease onset and progression, we measured the motor function by conducting rotarod test in the SOD1G93A mice. We found that trehalose treatment significantly delayed disease onset in Tg-Tre mice compared with Tg-Suc or Tg-NT mice (112.70 ± 1.93 vs. 94.92 ± 1.05 or 95.0 ± 1.15 d, respectively, both P < 0.01). No significant delay in disease onset was found between Tg-Suc and Tg-NT mice (94.92 ± 1.05 vs. 95.0 ± 1.15 d, P > 0.05) (Fig. 1C). Moreover, Tg-Tre mice showed between a 20 d to 21 d extension in life span compared with Tg-Suc or Tg-NT mice, respectively (145.20 ± 2.34 vs. 125.60 ± 1.05 or 124.10 ± 1.22 d, respectively, both P < 0.01). There was no significant difference between Tg-Suc and Tg-NT in life span (125.60 ± 1.05 vs. 124.10 ± 1.22 d, P > 0.05) (Fig. 1D). We also compared disease duration, which is defined as the time period starting from disease onset to the death of the mice, showing that trehalose treatment did not affect disease duration compared with sucrose or absence of treatment (Fig. 1E).
Trehalose protected motor neuron survival in SOD1G93A mice
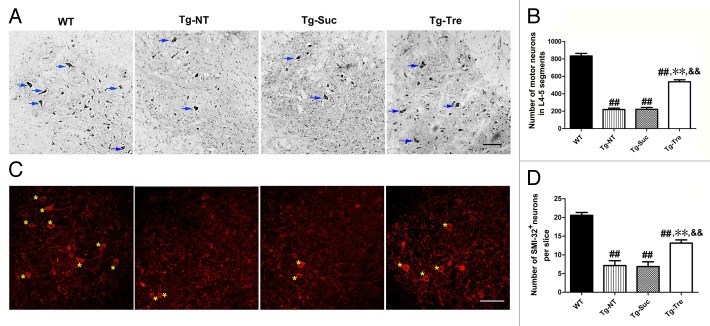
To determine the effect of trehalose on motor neuron survival, we performed Nissl staining to examine motor neuron survival in the L4-5 spinal cord of SOD1G93A mice. We found there was significantly lower motor neuron survival in the 120 d Tg-NT mice compared with the WT mice (219.80 ± 16.03 vs. 834.50 ± 29.25, P < 0.001) (Fig. 2B). The number of motor neurons surviving in Tg-Tre mice was more than twice that of the Tg-Suc or Tg-NT mice (538.5 ± 22.41 vs. 222.3 ± 20.52 or 219.80 ± 16.03, respectively, both P < 0.01). There was no difference in motor neuron survival between Tg-Suc and Tg-NT mice (222.30 ± 20.52 vs. 219.80 ± 16.03, P > 0.05) (Fig. 2A and B).
Figure 2. Effects of trehalose on motor neuron survival in SOD1G93A mice. (A) Representative photomicrographs of motor neurons in the anterior horn of spinal cords of 4 mouse groups by Nissl staining; (C) Representative photomicrographs of motor neurons in the anterior horn of spinal cord of 4 mouse groups by SMI-32 immunostaining. There were 3 mice in each group; asterisks mark the motor neurons. Scale bar: 100 μm; (B) The number of motor neurons in L4-5 segments by Nissl staining; (D) The mean number of SMI-32 positive motor neurons in both sides of one slice of an L4-5 segment. Data were analyzed using one-way ANOVA followed by Tukey post hoc test. All values are presented as mean ± SEM ##P < 0.001 as compared with WT mice; **P < 0.01 as compared with Tg-NT mice; &&P < 0.01 as compared with Tg-Suc mice.
Furthermore, we performed immunostaining with SMI-32, which was considered as a neuron-specific biomarker in the nervous system, to assess the morphology and survival of motor neurons per slice in the spinal cord of SOD1G93A mice. The result of SMI-32 immunostaining also showed trehalose treatment could protect the motor neurons in the SOD1G93A mice (Fig. 2C). Statistic analysis showed that there was an obvious increase in the number of SMI-32-positive neurons per slice in Tg-Tre mice compared with Tg-Suc or Tg-NT mice (13.14 ± 0.86 vs. 6.86 ± 1.21 or 7.14 ± 1.30, respectively, both P < 0.01) (Fig. 2D). These results showed that trehalose treatment could protect motor neurons in the SOD1G93A mice.
Trehalose reduced oxidative stress and attenuated skeletal muscle denervation
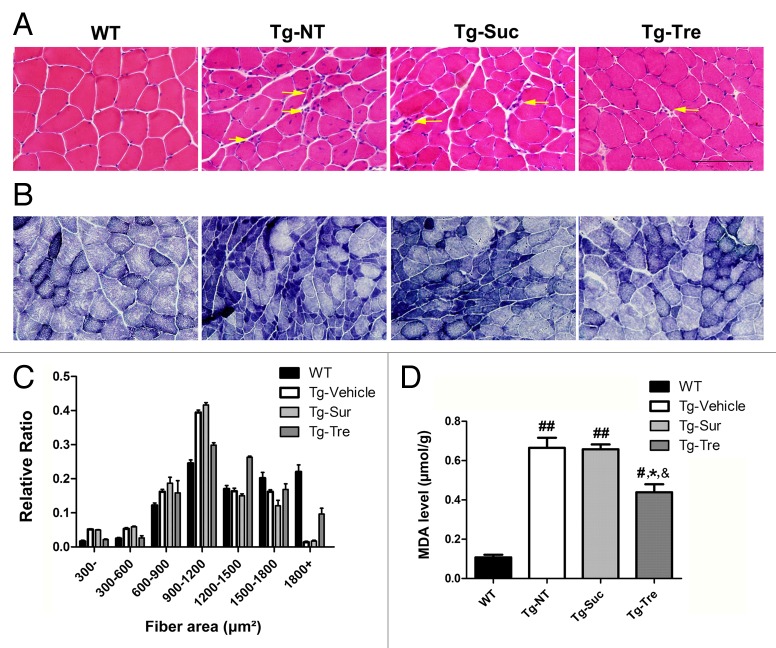
Muscle staining in cryosections revealed obvious structure and functional changes in Tg-NT and Tg-Suc mice as compared with WT mice (Fig. 3). Morphological analysis of muscles with hematoxylin-eosin (HE) staining showed typical features of muscle degeneration that included atrophic fibers, hematoxylin inclusions, and central nuclei in Tg-NT and Tg-Suc mice (Fig. 3A, marked with arrows). Statistical analysis showed that about 63% of the muscular fiber area was < 1200 μm2, 35% of the fiber area was ≥ 1200 μm2 and only 2% of the fiber area was ≥ 1800 μm2 in Tg-NT and Tg-Suc mice, as compared with 59% of muscular fiber area of ≥ 1200 μm2 and more than 25% of the fiber area of ≥ 1800 μm2 in WT mice (Fig. 3C). Trehalose treatment attenuated muscle degeneration and increased the fiber area of ≥ 1800 μm2 to 10% in SOD1G93A mice (Fig. 3A and C). Nicotinamide adenine dinucleotide hydrogen (NADH) staining showed grouped type I or type II myofibers and also the increased (dark blue) oxidative metabolism in Tg-NT and Tg-Suc mice (Fig. 3B). Trehalose treatment reduced the dark blue-stained oxidative muscle fibers and attenuated the grouped myofibers in the SOD1G93A mice (Fig. 3B). To further detect oxidative stress in the muscle of SOD1G93A mice, we tested malondialdehyde (MDA) levels in the 4 mouse groups. In Tg-NT mice, the level of MDA was significantly higher in the gastrocnemius muscle compared with WT mice (0.66 ± 0.05 vs. 0.11 ± 0.02 μmol/g, P < 0.01) (Fig. 3D). Tg-Tre mice showed lower MDA levels in skeletal muscle as compared with Tg-Suc or Tg-NT mice (0.44 ± 0.04 vs. 0.65 ± 0.02 or 0.66 ± 0.05 μmol/g, both P < 0.05) (Fig. 3D).
Figure 3. Effects of trehalose on skeletal muscle pathology in SOD1G93A mice. HE (A) and NADH staining (B) of gastrocnemius muscle sections in 4 mouse groups. Arrows marked the significant grouped atrophic fibers and hematoxylin inclusions. Scale bar: 100 μm; (C) The fiber area of gastrocnemius muscle in 4 mouse groups; (D) Levels of MDA in gastrocnemius muscle of 4 mouse groups. There were 3 mice in each group. Data were analyzed using one-way ANOVA followed by Tukey post hoc test. All values are presented as mean ± SEM ##P < 0.01 and #P < 0.05 as compared with WT mice; *P < 0.05 as compared with Tg-NT mice; &P < 0.05 as compared with Tg-Suc mice.
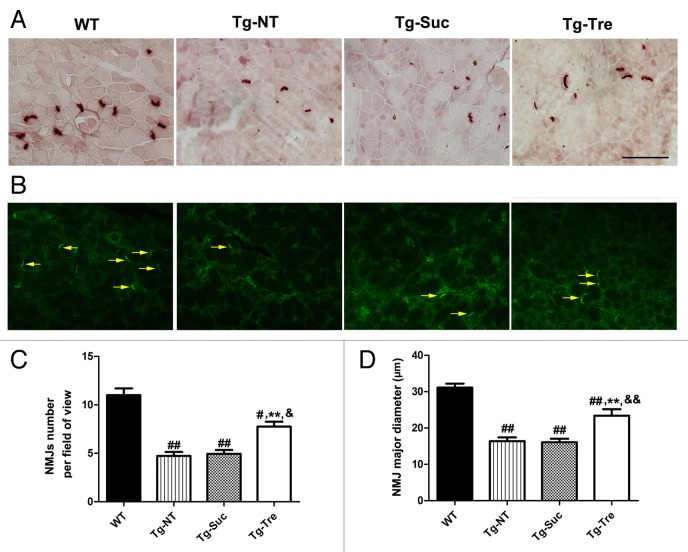
To examine muscle function and degeneration, we used nonspecific esterase (NSE) staining and ACHE (acetylcholinesterase) immunostaining to detect the number and morphology of neuromuscular junctions (NMJs) in the 4 mouse groups. Results of NSE and ACHE staining showed that NMJs in Tg-NT, Tg-Suc and Tg-Tre mice was strikingly reduced compared with WT mice (Fig. 4A and B). Quantitative analysis indicated that the number of NMJs in Tg-NT mice was significantly decreased compared with WT mice (4.73 ± 0.40 vs. 11.70 ± 0.67, P < 0.01) (Fig. 4C). Furthermore, the diameter of NMJs in Tg-NT mice was also significantly reduced compared with WT mice (16.38 ± 1.03 vs. 31.10 ± 1.10 μm, P < 0.01) (Fig. 4D). Trehalose treatment increased the number of NMJs in the gastrocnemius muscle of the ALS mice (4.94 ± 0.40) to 7.47 ± 0.52 per field (Fig. 4C). Furthermore, the diameter of NMJs in Tg-Tre mice was significantly larger than that of the Tg-Suc or Tg-NT mice (23.40 ± 1.78 vs. 16.07 ± 0.98 or 16.38 ± 1.03 μm, both P < 0.01) (Fig. 4D). Taken together, our results suggest that trehalose treatment can significantly reduce oxidative stress and protect skeletal muscles from denervation in SOD1G93A mice.
Figure 4. Effects of trehalose on the NMJs of gastrocnemius muscle in SOD1G93A mice. NSE staining (A) and ACHE immunostaining (B) of gastrocnemius muscle sections in 4 mouse groups. Arrows marked the ACHE-positive NMJs in the muscle slides. Scale bar: 100 μm; (C) Quantitative analysis of the number of NMJs per field in 4 mouse groups; (D) Statistical analysis of the mean major diameter of NMJs in different mouse groups. There were 3 mice in each group. Data were analyzed using one-way ANOVA followed by Tukey post hoc test. Values are presented as mean ± SEM ##P < 0.01 and #P < 0.05 as compared with WT mice; **P < 0.01 as compared with Tg-NT mice; &&P < 0.01 and &P < 0.05 as compared with Tg-Suc mice.
Trehalose regulated the MTOR-independent autophagy pathway
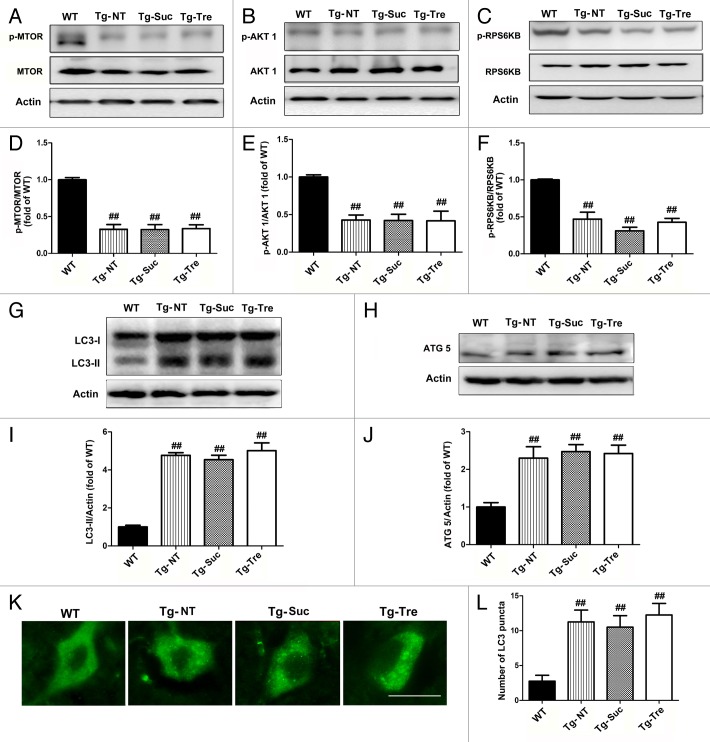
It is known that trehalose is an MTOR-independent autophagy enhancer.27 To investigate the effects of trehalose on autophagy in the SOD1G93A mice, we measured the protein levels of LC3-II, autophagy-related 5 (ATG5), and MTOR pathway-related proteins, such as p-MTOR, p-RPS6KB (phospho-Thr389-specific 70-kDa ribosomal protein S6 kinase, 70 kDa, polypeptide 1) and p-AKT1. Quantitative analysis of western blots showed that p-MTOR, p-RPS6KB and p-AKT1 protein levels were decreased to 32.67%, 47.0%, and 42.67% in SOD1G93A mice as compared with age-matched WT mice (Fig. 5D–F). As previously reported,25 trehalose treatment did not affect the levels of MTOR pathway proteins in SOD1G93A mice. No significant difference was found with the protein levels of p-MTOR/MTOR, p-RPS6KB/RPS6KB, and p-AKT1/AKT1 between Tg-Tre mice and Tg-NT or Tg-Suc mice (Fig. 5A–C). Our results further indicate that trehalose does not change the MTOR pathway activity in SOD1G93A mice.
Figure 5. Effects of trehalose on the MTOR-independent pathway and autophagy-related proteins. Western blot analysis of protein levels of p-MTOR (A), p-AKT1 (B), and p-RPS6KB (C) in the 4 mouse groups (WT, Tg-NT, Tg-Suc, Tg-Tre). Quantitative analysis of the ratio of p-MTOR/MTOR (D), p-AKT1/AKT1 (E) and p-RPS6KB/RPS6KB (F) in the 4 mouse groups. There were 3 mice in each group; ##P < 0.01 when compared with WT mice. Western blot analysis of protein levels of LC3-II (G) and ATG5 (H) in 4 mouse groups. Quantitative analysis of LC3-II/LC3-I (I) and ATG5 (J). There were 3 mice in each group; ##P < 0.01 as compared with WT mice. (K) Immunostaining of LC3 in the motor neurons of 4 mouse groups. Scale bar: 20 μm; (L) Quantitative analysis of LC3 puncta numbers in the 4 different mouse groups. There were 3 mice in each group. Data were analyzed using one-way ANOVA followed by Tukey post hoc test. All values are presented as mean ± S.E.M, ##P < 0.01 as compared with WT mice.
Compared with the control (sucrose) treatment, trehalose administration did not change the ATG12–ATG5 conjugate levels in SOD1G93A mice, which is consistent with the previous report that trehalose has no significant effect on the ATG-related protein levels in vitro25 (Fig. 5H). However, it was notable in our study that the level of LC3-II was slightly but not significantly increased in Tg-Tre mice as compared with Tg-NT or Tg-Suc mice (Fig. 5G). Moreover, the result of LC3 immunofluorescent staining also revealed a slight increase of LC3 positive puncta in the motor neurons of Tg-Tre mice as compared with Tg-NT or Tg-Suc mice (12.25 ± 3.14 vs. 10.50 ± 3.31 or 11.25 ± 3.40 per cell, respectively, both P > 0.05) (Fig. 5K and L).
Trehalose reduced protein aggregation in motor neurons
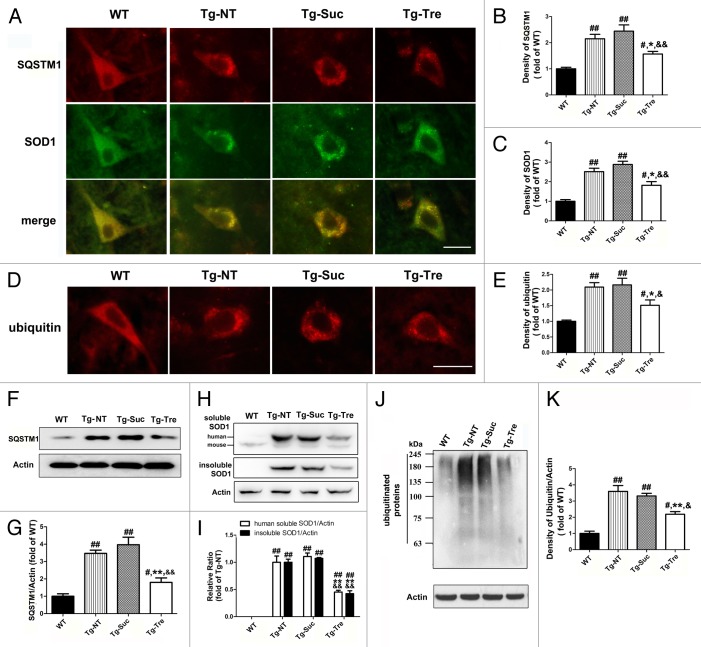
Immunostaining revealed the colocalization of SQSTM1 and SOD1 aggregation in the motor neurons of Tg-NT mice, while no costaining was detected in the WT controls (Fig. 6A). However, there was a reduction in SQSTM1 protein aggregation in Tg-Tre mouse motor neurons as compared with Tg-NT or Tg-Suc mice (Fig. 6A). The density of SQSTM1 immunostaining in motor neurons of Tg-Tre mice was reduced to 63.85% of that observed in Tg-Suc mice (Fig. 6B). Quantitative analysis of western blots further demonstrated a significant reduction in SQSTM1 levels in Tg-Tre mice compared with Tg-Suc (-54.62%, P < 0.01) or Tg-NT mice (-48.26%, P < 0.01) (Fig. 6F and G). Furthermore, we observed a significant decrease in SOD1 aggregates in Tg-Tre mice compared with Tg-NT or Tg-Suc mice (Fig. 6A). The density of SOD1 immunostaining in the motor neurons of Tg-Tre mice was reduced to 64.93% of the density observed in Tg-Suc mice (Fig. 6C). Western blot analysis revealed a 59.13% and 54.56% decrease of insoluble SOD1 protein in the spinal cord of Tg-Tre mice as compared with Tg-NT and Tg-Suc mice, respectively (both P < 0.01) (Fig. 6H and I). Moreover, The level of human soluble SOD1 showed a significant reduction in Tg-Tre mice as compared with Tg-NT and Tg-Suc mice (−49.35% and −54.80%, respectively, both P < 0.01) (Fig. 6H and I).
Figure 6. Effects of trehalose on SQSTM1 level and protein aggregation in SOD1G93A mice. (A) Double labeling of SQSTM1 and SOD1 in the motor neurons of WT and SOD1G93A mice. (D) Immunostaining of ubiquitin in motor neurons. Scale bar: 20 μm. Quantitative analysis of SQSTM1 density (B), SOD1 density (C) and ubiquitin density (E) in motor neurons. Western blot analysis of SQSTM1 level (F), soluble and insoluble SOD1 protein level (H) and high molecular weight bands of ubiquitinated proteins (J) in the 4 different mouse groups. Quantitative analysis of SQSTM1 protein levels (G), human soluble SOD1 and insoluble SOD1 protein (I) and ubiquitinated proteins levels (K) in the spinal cord of SOD1G93A mice. There were 3 mice in each group. Data were analyzed using one-way ANOVA followed by Tukey post hoc test. Values are presented as mean ± SEM ##P < 0.01 and #P < 0.05 as compared with WT mice; **P < 0.01 and *P < 0.05 as compared with Tg-NT mice; &&P < 0.01 and &P < 0.05 as compared with Tg-Suc mice.
In addition, compared with the sucrose treatment or the absence of treatment, trehalose administration resulted in a significant decrease of ubiquitinated positive protein aggregation in the motor neuron of SOD1G93A mice (−39.27%, P < 0.01 and −34.13%, P < 0.05) (Fig. 6D). The density of ubiquitin fluorescent immunostaining in the motor neurons of Tg-Tre mice was reduced to 69.90% of that seen in Tg-Suc mice (Fig. 6E).Western blotting showed a significant reduction of ubiquitinated protein levels of high molecular weight in the spinal cord of Tg-Tre mice (Fig. 6J and K). These results indicate that trehalose can induce the degradation of aggregated proteins in the motor neurons of SOD1G93A mice. To test whether trehalose treatment affects the expression of the Sod1 gene in the spinal cord of SOD1G93A mice, we performed real-time PCR in Tg-Tre mice to compare with the Tg-NT, Tg-Suc, and WT mice. Our results showed that trehalose treatment did not change the mRNA level of the Sod1 gene in the spinal cord of SOD1G93A mice (Fig. S1). Taking these results together, we believe that trehalose most likely affects soluble and insoluble SOD1 protein degradation without altering Sod1 gene expression in the ALS mouse model.
Trehalose protected mitochondria and ameliorated autophagic flux defects
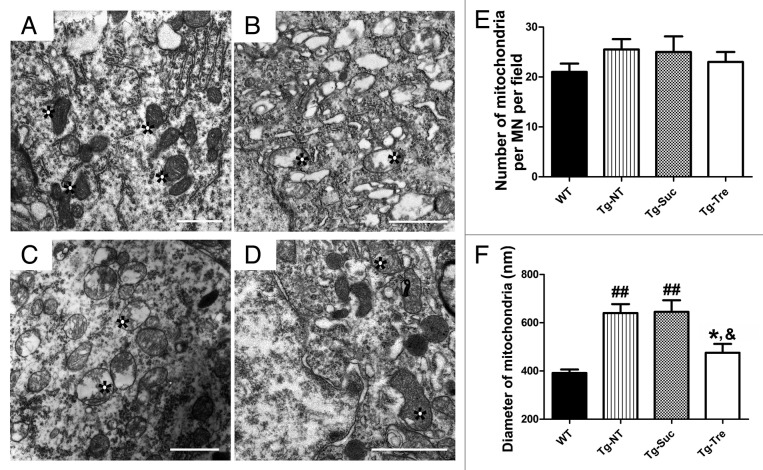
Transmission EM showed that the mitochondria in the motor neurons of Tg mice appeared to have broken crista, and degenerating swollen mitochondria were often detected (Fig. 7B and C, marked with asterisks). Trehalose treatment seemed to rescue mitochondrial degeneration in the motor neurons of SOD1G93A mice (Fig. 7D, marked with asterisks). Quantitative analysis showed that the average diameter of mitochondria was increased in the motor neurons of Tg-NT mice compared with WT mice (640.40 ± 37.32 vs. 391.50 ± 14.73 nm, P < 0.01) (Fig. 7F). In Tg-Tre mice, the average diameter of mitochondria was significantly reduced to 475.60 nm (compared with Tg-Suc or Tg-NT, both P < 0.05). We also counted the number of mitochondria per motor neuron in these mice. However, we found that Tg-NT, Tg-Suc and Tg-Tre mice did not differ significantly from WT mice in the number of mitochondria (Fig. 7E).
Figure 7. EM analysis of mitochondria in the motor neurons of SOD1G93A mice. Representative EM photomicrographs of mitochondrial structure in WT (A), Tg-NT (B), Tg-Suc (C) and Tg-Tre mice (D). Quantitative analysis of the number (E) and the mean diameter of mitochondria (F) in the motor neuron of 4 mouse groups. There were 3 mice in each group; asterisks marked mitochondria. Data were analyzed using one-way ANOVA followed by Tukey post hoc test. Values are presented as mean ± SEM ##P < 0.01 as compared with WT mice; *P < 0.05 as compared with Tg-NT mice; &P < 0.05 as compared with Tg-Suc mice. All scale bars: 1 μm.
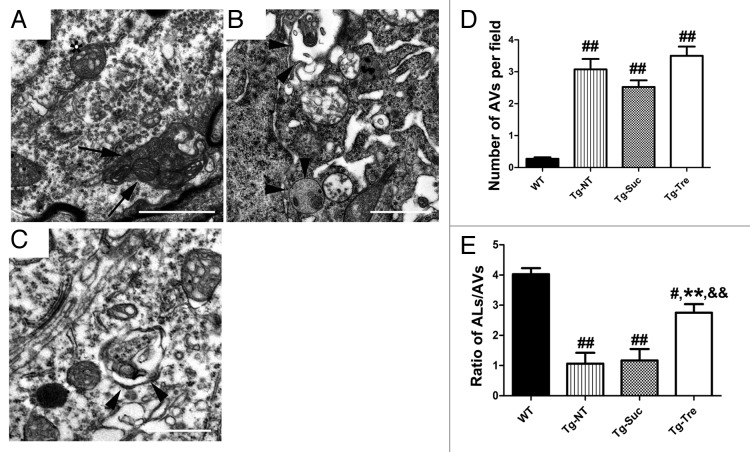
To further determine the ultra structure alterations in SOD1G93A mice after trehalose treatment, we used EM to quantitatively examine the number of autophagosomes and autolysosomes and other changes in the motor neurons of SOD1G93A mice in the different treatment groups. The classic autophagosome could be found in the cell body and axon of motor neurons in the Tg-Tre mice (Fig. 8A, marked with arrow). These double-membrane structures contain mitochondria and parts of cytoplasm. More importantly, there were obvious autolysosome aggregations in the motor neuron soma and axon of the Tg-Tre mice (Fig. 8B and C, marked with arrowheads). We then analyzed the number of autophagosomes and autolysosomes in the motor neurons of all 4 mouse groups. Our results demonstrated a significantly reduced autolysosome/autophagosome ratio in Tg-NT or Tg-Suc mice as compared with WT mice (Fig. 8E). Trehalose treatment significantly improved the autolysosome/autophagosome ratio in the motor neurons of SOD1G93A mice (Fig. 8E). Furthermore, the number of autolysosomes in the motor neurons of Tg-Tre mice was slightly but not significantly increased compared with Tg-Suc or Tg-NT mice (3.50 ± 0.29 vs. 2.52 ± 0.21 or 3.07 ± 0.32 per field, both P > 0.05) (Fig. 8D). Collectively, EM findings further indicate that the autophagic flux defect is ameliorated by trehalose treatment in the SOD1G93A mice.
Figure 8. EM analysis of autophagosomes and autolysosomes in the motor neurons of SOD1G93A mice. Double-membrane autophagosome (A) and single-membrane autolysosomes (B and C) in motor neurons of trehalose-treated mice. Arrow marks the typical autophagosome; arrowheads mark the autolysosomes. (D) Quantitative analysis of the number of autophagosomes per field of 4 mouse groups. (E) Quantitative analysis of autophagosome/autolysosome ratios per motor neuron of the 4 different mouse groups. There were 3 mice in each group. Data were analyzed using one-way ANOVA followed by Tukey post hoc test. Values are presented as mean ± SEM ##P < 0.001 and #P < 0.01 as compared with WT mice; **P < 0.01 as compared with Tg-NT mice; &&P < 0.01 as compared with Tg-Suc mice. Scale bars: 1 μm.
Trehalose protected motor neurons against proapoptotic processes
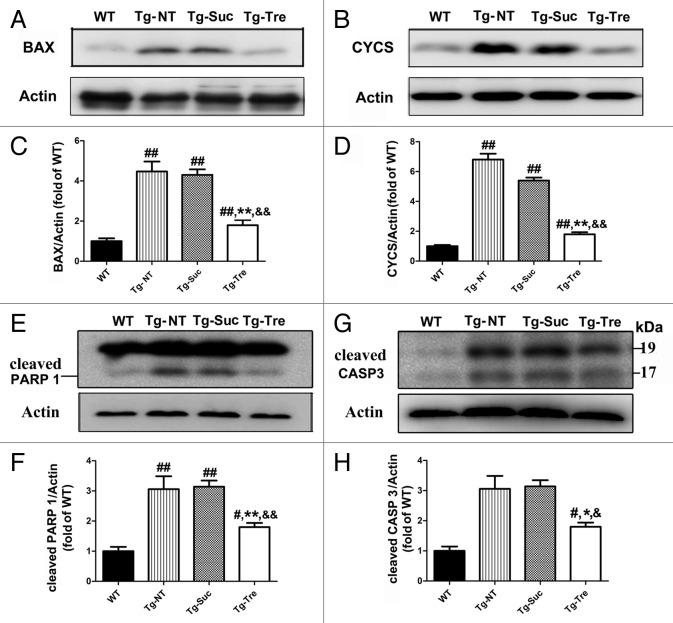
To further determine the effects of trehalose on the mitochondrial function and apoptotic process, we examined the protein levels of BAX, CYCS (cytochrome c, somatic), cleaved CASP3/caspase-3, and cleaved PARP1 in the spinal cord of SOD1G93A mice (Fig. 9A, B, E, and G). The quantitative analysis of western blot results showed that trehalose treatment can decrease the levels of BAX and CYCS to 58.13% and 66.62%, respectively, as compared with the control (Fig. 9C and D). Furthermore, trehalose treatment reduced cleaved PARP1 level to 46.42% compared with the control (Fig. 9F). Moreover, trehalose treatment can significantly inhibit CASP3 activation and reduce the level of cleaved CASP3 compared with the controls (Fig. 9H). No significant difference was found between Tg-NT and Tg-Suc mice on these related proteins. These data indicate that trehalose treatment can protect motor neurons survival through inhibition of the proapoptotic pathway in the SOD1G93A mice.
Figure 9. Effects of trehalose on proapoptotic pathway in the SOD1G93A mice. Western blot analysis of BAX (A), CYCS (B), PARP1 (E), and cleaved CASP3 (G) in the 4 different groups of mice. Quantitative analysis BAX (C), CYCS (D), PARP1 (F) in the spinal cord of the 4 different groups of mice. There were 3 mice in each group. Data were analyzed using one-way ANOVA followed by Tukey post hoc test. Values are presented as mean ± SEM ##P < 0.01 as compared with WT mice; #P < 0.05 as compared with WT mice; **P < 0.01 as compared with Tg-NT mice; *P < 0.05 as compared with Tg-NT mice; &&P < 0.01 when compared with Tg-Suc mice; &P < 0.05 when compared with Tg-Suc mice.
Discussion
Growing evidence supports the view that abnormal protein aggregation is involved in the pathogenesis of ALS.9,11,28,29 However, the contribution of autophagy in the pathology of ALS has not yet been fully elucidated. Herein, we report that trehalose can delay disease onset and prolong the lifespan in the SOD1G93A mouse model of ALS. Pathologically, trehalose protects the motor neurons from degeneration as showing obviously increased motor neurons survival in the anterior horn of spinal cord in the ALS mice. Interestingly, we also found that trehalose significantly improves muscle atrophy by restoring muscle functions and attenuating oxidative stress, which might be a possible mechanism of the neuroprotective effects of trehalose on motor neurons in the ALS model since several recent studies have demonstrated that skeletal muscle might be a primary site of disease to initiate non-autonomous motor neuron degeneration in ALS.30,31
The neuroprotective mechanisms of trehalose in neurodegenerative diseases are largely unknown. It is speculated that the neuroprotective effect of trehalose might be associated with the promotion of aggregated protein degradation.24,25,32 For example, trehalose can enhance the clearance of mutant PABPN1 [poly(A) binding protein, nuclear 1], which causes oculopharyngeal muscular dystrophy.32 In addition, trehalose can induce the degradation of mutant HTT, SNCA, and MAPT inclusions by activating autophagy.24,25 Our findings provide evidence that trehalose is able to reduce SOD1 aggregation and ubiquitinated proteins in the motor neurons of SOD1G93A mice. Particularly, we showed that trehalose can significantly decrease the insoluble SOD1 and high molecular weight ubiquitinated proteins in the spinal cord tissues of ALS mice.
It is well known that the ubiquitin-proteasome and autophagy-lysosome are the 2 major systems participating in the protein degradation in eukaryotic cells.4 Different from the ubiquitin-proteasome system, which serves as an important route for short-lived proteins degradation, autophagy is responsible for the degradation of long-lived proteins or protein complexes.33 Although a recent study reports that impairment of the ubiquitin-proteasome system, but not autophagy failure is involved in motor neuron degeneration in ALS,34 the accumulated evidence suggests that autophagic dysfunction plays a critical pathogenic process leading to ALS.9,10,29 Our previous study and others have reported that treatment with the MTOR-dependent autophagic inducer rapamycin aggravates motor neuron degeneration and accelerates disease progression in different mice models of ALS.14,17 Here, we have demonstrated that the MTOR-independent autophagic activator trehalose is able to protect motor neurons, delay disease onset and prolong life span in the ALS mice. The opposite results of rapamycin and trehalose from our studies in the same mouse model of ALS are likely due to the different effects of those autophagic activators on the status of autophagic flux in the ALS mice.29 The changes of SQSTM1 level and EM analysis from our own study in SOD1G93A mice provide strong evidence of autophagic flux defects in the ALS model.
The SQSTM1 protein is a linkage molecule between LC3-decorated autophagosomes and ubiquitin-conjugated protein aggregates, which is finally degraded in autolysosomes.35 Recently this molecule has been used to monitor autophagic flux changes.36 It is noteworthy that trehalose can reduce SQSTM1 aggregation in the motor neurons of spinal cord of ALS mice, suggesting that trehalose can protect motor neurons probably through the amelioration of the autophagic flux defect. Transmission EM is a gold-standard method in qualitative and quantitative analysis of autophagic structures.37 Using EM, we have shown that trehalose does not change the number of autophagosomes but increases the ratio of autolysosome/autophagosome in the motor neurons of ALS mice. Based on these results, we speculate that trehalose can ameliorate the autophagic flux defect by inducing autophagosome-lysosome fusion. However, we have found that the trehalose treatment slightly but not significantly increases the levels of autophagy-related proteins such as LC3-II and ATG5, which is different from the report of Castillo et al., in which they observe remarkable increased levels of LC3-II in the spinal cord of SOD1G86R mice after trehalose treatment.38 These differences may result from the different routes of drug delivery (oral + intraperitoneal injection was used in Castillo’s study38) and different mutant SOD1 mouse lines (SODG86R mice were used by Castillo and coworkers38). Previous studies report distinct pathogenic mechanisms in mouse models with different SOD1 mutant lines.16,39 In addition to the trehalose-mediated general autophagy activation effect, our study focused on trehalose-induce autophagosome-lysosome fusion, rescuing defective autophagic flux and autophagosome degradation in the motor neurons of SOD1G93A mice.
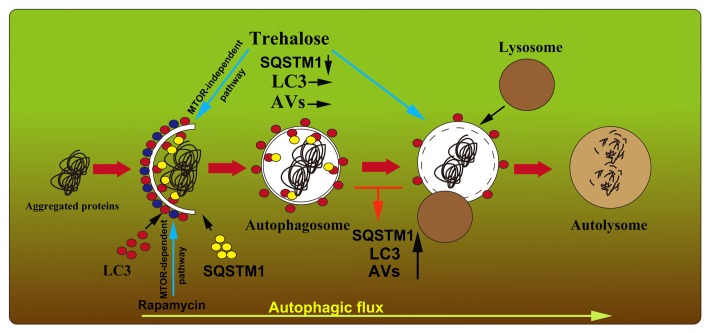
It has been reported that the autophagic flux defect in ALS can be caused by ALS-related mutant genes such as DCTN1, CHMP2B (charged multivesicular body protein 2B), and UBQLN2 mutations.29,40-42 It is thought that microtubule-based vesicular trafficking is essential for the delivery of autophagosome to lysosomes and subsequent fusion, and impaired dynein-mediated trafficking may be associated with the autophagosome-lysosome fusion defect.43,44 Indeed, impairment of the axonal transport is found in mutant SOD1 probably through the inhibition of dynein/dynactin function,45 which may contribute to the autophagic flux defect in the motor neurons of ALS. Other molecules that are required for the fusion of autophagosomes with lysosomes, such as RAB7, the class C Vps/HOPS tethering complex and SNAREs are likely involved in the pathogenetic process of the autophagic flux defect.46-48 However, direct evidence for the relevance of mutant SOD1 and these components is missing. More studies are needed to explore the possible mechanisms of autophagic flux defects in ALS. In this regard, we propose a model of autophagic flux defect and trehalose target in ALS as illustrated in Figure 10. This model can be used to explain the augmented effects of rapamycin in the ALS model,14,16 as well as the beneficial effects of trehalose that not only regulates autophagy24,25,27 but also induces autolysosome formation and degradation, leading to motor neuron protection as shown in the current study.
Figure 10. Proposed model of the autophagic flux dysfunction and the possible roles of trehalose in the motor neurons of SOD1G93A mice. Impaired autophagosome-lysosome fusion might contribute to the degeneration of motor neurons, which leads to the abnormal high SQSTM1 level, LC3 puncta aggregation, and the increase in autophagosomes in the motor neurons of SOD1G93A mice. Rapamycin activates autophagy in an MTOR-dependent pathway and it does not affect the status of autophagic flux in the ALS mice. However, trehalose induces autophagosome formation in an MTOR-independent pathway and rescues the impaired fusion step, which results in SQSTM1 and aggregated autophagosome degradation in the motor neurons of SOD1G93A mice. AVs, autophagic vacuoles.
It is worth noting that a similar autophagosome impairment and the autophagosome-lysosome fusion defect are also found in patients with Alzheimer disease and other neurodegenerative diseases.10,12,49 These similarities suggest a possibility that these neurodegenerative diseases might share a common pathogenetic mechanism resulting from the autophagosome-lysosome fusion defect. Thus, an impairment of the fusion step in the autophagy process, rather than autophagy activation, might be a key point contributing to the pathogenesis and progression of these neurodegenerative diseases.10
Mitochondrial dysfunction is believed to contribute to the pathogenesis of ALS, which causes motor neuron death by increasing generation of reactive oxygen species and initiating the apoptotic pathway.50 As reported previously, consistent with the Sarkar et al. report25 that trehalose leads to protection against proapoptotic damages via the mitochondrial cell death pathway, our findings provide further explanation for the neuroprotective effects of trehalose in the motor neuron survival of ALS mice. Moreover, trehalose is a multifunctional molecule, including its effects as a chemical chaperone, which may play a role to suppress protein misfolding and reduce endoplasmic reticulum stress and stress-related injury.51 Increased ER-stress has been reported in the motor neurons of SOD1 transgenic mice model and clinical ALS patients.52,53 However, Castillo et al. have not found the anti-endoplasmic reticulum stress effect of trehalose in SOD1G86R mice.38
In conclusion, our study provides new evidence to support the autophagic flux defect in the motor neurons of ALS and that the MTOR-independent autophagic inducer trehalose is able to attenuate the autophagic flux defect and improve disease course in the ALS model. The results from the present study in combination with our previous one indicate that the defect in autophagosome-lysosome fusion is a critical pathological event for ALS and the molecules that can repair the fusion defect might be potential therapeutic targets for developing a novel therapy to treat this devastating disease.
Materials and Methods
Transgenic mice and treatment
Transgenic SOD1G93A mice were originally obtained from Jackson Laboratories (002726), which express about 20 copies of mutant human SOD1 with a Gly93Ala substitution (B6SJL-Tg-SOD1G93A-1Gur). The genotypes of the transgenic mice were identified by PCR according to our previously reported reference.14 Male SOD1G93A mice were randomly divided into 3 groups (Tg-NT, Tg-Suc, Tg-Tre) with 21 mice in each group and these mice were housed under the conditions of constant temperature and controlled lighting (light/dark period 12/12 h). We chose 2% trehalose treatment for SOD1G93A mice according to previous reports.24,25 For Tg-Tre mice, 2% trehalose-containing water was given to SOD1G93A mice through ad libitum consumption, starting at 64 d of age and continuing until the day the mice died. We calculated that the average water consumption of each mouse was 5 to 6 ml per day. For Tg-Suc mice, 2% sucrose-containing water was given to mice as a vehicle control. For Tg-NT mice, no treatment was given to SOD1G93A mice. In addition, 15 of the age-matched WT littermates were used as controls. All the transgenic mice were weighed every 4 d starting from 64 d of age until death and the monitoring of WT mice was ended at 146 d of age. Animals care and procedures were performed in accordance with the Laboratory Animal Care Guidelines approved by the Shanghai Institutes for Biological Sciences of the Chinese Academy of Sciences.
Behavioral study
Test of motor function (Rotarod test)
Rotarod performance was measured in SOD1G93A mice starting at 80 d of age. Motor function was assessed as described previously.54 Mice were trained for 1 wk prior to treatment in order for them to adapt to the apparatus. After training, we got their basal motor performance in Rotarod. Next, the mice were tested every 2 d, and the date of disease onset was recorded when the mice could not stay on the rod for 5 min.
Assessment of life span
SOD1G93A mice developed complete paralysis roughly at around 126 d of age.26 For life-span analysis, the date of “death” (or terminal impairment) was defined as the day when the mice could not right itself within 30 s after being placed on its back.55 All the mice were tested every day and the date representing the end-point was recorded.
Immunofluorescent staining
Mice were anesthetized with chloral hydrate and sacrificed by transcardiac perfusion with ice-cold 100 mM phosphate-buffered saline (PBS, pH 7.4) and 4% paraformaldehyde. The spinal cords were removed and postfixed in 4% paraformaldehyde overnight at 4 °C, transferred to 15% and 30% sucrose in PBS, and then frozen at −80 °C. Serial frozen sections of the spinal cord (10 μm) were cut and mounted on gelatin-coated slides. The slides were incubated in 3% bovine serum in PBS-0.3% Triton X-100 for 1 h, followed by overnight incubation at 4 °C with the primary antibodies for LC3B (1:200; Cell Signal, 3868), SOD1 (1:200; abcam, ab16831), SQSTM1 (1:100; BD Transduction Laboratories, 610832), ubiquitin (1:800, Milipore, MAB1510), SMI-32 (1:1,000; Abcam, ab28025) and ACHE (1:200, Santa Cruz, sc-11409). After washing with PBS, the slides were incubated with fluorescent dye Cy2-conjugated goat anti-rabbit (Jackson Immunoresearch, 111-225-045) or Cy3-conjugated goat anti-mouse (Jackson Immunoresearch, 115-165-062), and were then visualized at 600 × magnifications under a fluorescent microscope (Nikon 80i).
Motor neuron survival analysis
The fixed L4-5 spinal cords were cut with a Leica cryostat (Leica) to 10-μm thickness and mounted on gelatin-coated slides. Sections were immunofluorescent stained with 1% cresyl violet (Sigma, C5042) and photographed with a microscope (Olympus, IX81). Anterior horns at both sides on every fourth section from a total of 200 slices per animal were examined by a technician who was blinded to the experimental design. The number of motor neurons was counted according to the following rules: 1) neurons located in the anterior horn ventral to the line tangential to the ventral tip of the central canal; 2) neurons with a maximum diameter of 20 μm or larger and 3) neurons with a distinct nucleolus.56,57 For SMI-32 positive MNs analysis, L4-5 spinal cord was cut to a thickness of 10 μm. Sections were stained for immunofluorescence with SMI-32 antibody according to the method of Immunofluorescence staining and captured by microscope (Olympus, IX81). Twenty slices were collected per animal at an interval of 10 slices. Motor neurons in anterior horns at both sides per slice were counted in a blinded manner.
Extraction of soluble and insoluble proteins
Spinal cords were removed and lysed by sonication in ice-cold extraction buffer (10 mM Tris–HCl, pH 8.0, 1 mM EDTA, 100 mM NaCl, 0.5% NP-40, supplemented with protease inhibitor cocktail (Sigma, P8340), PMSF (1 mM; Sigma, P7626) and 50 mM iodoacetamide (Sigma, I6125). The lysates were then centrifuged at 130,000 g for 10 min at 4 °C using an ultracentrifuge (Beckman-Coulter Optimal L-100 XP). The supernatant fraction was kept as the soluble sample. The pellet fraction was resonicated in extraction buffer and was centrifuged again at 130,000 g for 10 min at 4 °C. Then the remaining pellet fraction was resolved in resuspension buffer (10 mM Tris–HCl, pH 8.0, 1 mM EDTA, 100 mM NaCl, 0.5% NP-40, 0.5% deoxycholic acid (Sigma, D6750) and 2% SDS) and sonicated until the pellet fraction was resuspended in the solution, and this preparation was kept as the insoluble sample. Soluble and insoluble samples were then analyzed by immunoblotting.
Immunoblotting analysis
Mice were anesthetized with chloral hydrate and sacrificed by trans-cardiac perfusion with ice-cold 100 mM PBS. After removing the spinal cords, total protein was extracted from the tissues with RIPA lysis buffer containing 50 mM TRIS-HCl, pH 7.4, 150 mM NaCl, 1% Nonidet P-40 (Sigma, D6750), 1 mM EDTA, 1 mM PMSF, and a protease inhibitor cocktail (Sigma, P8340). The protein concentrations were assayed from the resulting supernatant fractions by the BCA method (Thermo Scientific, 23225). Forty μg of proteins were loaded and separated on 8%, 10%, or 12% SDS-PAGE gels. Proteins were transferred onto 0.45 μm or 0.22 μm polyvinylidene fluoride membranes (Millipore, HVPPEAC12 or GVPPEAC12), blocked in 5% nonfat milk or 5% bovine serum albumin (Sigma, A1933) for 1 h. Membranes were then incubated in the presence of the respective primary antibodies: p-RPS6KB (1:500, Cell Signaling Technology, 9208), RPS6KB (1:1,000, Cell Signaling Technology, 2708), p-MTOR (1:1,000, Cell Signaling Technology, 2971), MTOR (1:1,000, Cell Signaling Technology, 2983), BAX (1:1,000, Cell Signaling Technology, 2772), CYCS (1:1,000, Epitomics, 3895-1), p-AKT1 (1:1,000, Epitomics, 2214-1), AKT1 (1:2,000, Epitomics, 1085-1), LC3B (1:500, Sigma, L7543), poly (ADP-ribose) polymerase, PARP1 (1:1,000, Cell Signaling Technology, 9542), cleaved CASP3 (1:500, Cell Signaling Technology, 9664), ATG5 (1:500, MBL, M153-3), SQSTM1 (1:500, MBL, PM045), SOD1 (1:2,000, abcam, ab16831), ubiquitin (1:500, Milipore, MAB1510). A mouse, anti ACTB/β-actin antibody (1:6,000; Sigma) was used to demonstrate equal protein loading. Peroxidase-conjugated secondary antibodies were used and then protein bands were visualized using a chemiluminescent horseradish peroxidase substrate (ECL, Pierce, 34075) and were quantified with an image analyzer software (Bio-Rad, Image lab 4.1).
Electron microscopy analysis
The tissues of spinal cord (L4-5) were fixed in 2.5% glutaraldehyde in 100 mM PBS and cut into 50-μm thick sections by a vibratome. The sections were postfixed with 1% OsO4, dehydrated and embedded in Durcupan (ACM; Fluka) on a microscope slide and coverslipped. Those sections were further cut by a Reichert ultramicrotome (Leica) into 70-nm thick sections. The ultra thin sections were then stained with uranyl acetate and lead citrate and evaluated with a CM-120 EM (Philips).
Quantitative analysis of autophagosomes, autolysosomes, and mitochondria
The SOD1G93A mice in the 3 groups (Tg-NT, Tg-Suc, Tg-Tre) and age-matched WT littermates were examined by EM. Twenty-five randomly selected EM images per animal were captured at a final magnification of 10,000 × g, and the number of autophagosomes and autolysosomes in each captured field was counted by visual inspection using previously established criteria for identification.12 In brief, autophagosomes were defined as the double-membrane structures, whose inside density was similar to that of the surrounding cytosol. Autolysosomes have only one limiting membrane, and contain cytoplasmic material and /or organelles at various stages of degradation. For mitochondria analysis, at least 20 randomly selected motor neurons per animal were captured at a final magnification of 5,000 × , then the number and the major diameter of mitochondria in each motor neuron was calculated by the technician who was blinded to the experimental design.
Pathological analysis of skeletal muscles
Fresh gastrocnemius muscle (5 × 5 × 10 mm3) was dissected out from the right leg and immersed immediately in liquid nitrogen-cooled isopentance. Cross-sections were cut through the middle of the muscle at 10 μm and stained by HE for morphology analysis. In addition, mitochondrial activity was assessed by NADH reaction. The number and pathological alteration of NMJs were detected by NSE. The procedure was conducted as in a previous report.58,59 Twenty randomly selected NSE images per animal were captured at a final magnification of 200 × , and the number and diameter of NSE-positive NMJs in each captured field was calculated by a researcher who was blinded to the study design. Analysis was performed using a one-way ANOVA test followed by the Turkey post hoc test.
Measurement of MDA levels
Fresh gastrocnemius muscles were dissected, weighted, and frozen in liquid nitrogen. The whole muscles were homogenized in ice-cold TRIS-HCl (20 mM) buffer. The homogenate was centrifuged at 1300 g at 4 °C for 10 min. The supernatant was collected and frozen at −80 °C until the day of detection. The MDA level was measured by fluorescence assay of thiobarbituric acid reactive substance formation following the indications of a commercial kit (Jiancheng, Nanjing, A003-2), detected at 532 nm and quantified with an image analyzer (BioTek, synergy H4). The measured MDA levels were normalized with respect to homogenization volume and muscle weight.
Statistics
Disease onset and life-span analysis were performed by Kaplan-Meier analysis (SPSS 17.0). Other data were analyzed using one-way ANOVA followed by Tukey post hoc test (Prism5, GraphPad software, Inc.). All values were presented as mean ± SEM. Significant differences were defined as P < 0.05.
Supplementary Material
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
This study was funded by research grants from the National Nature Science Foundation (No. 81000541, No. 81200977), National Basic Research Program (No. 2011CB510003), PhD Innovation Fund of Shanghai Jiaotong University school of Medicine (BXJ201218), and Specialized Research Fund for the Doctoral Program of Higher Education of China (20120073110077). We give thanks to the EM Laboratory of Shanghai Medical College of Fudan University for their technical assistance.
Glossary
Abbreviations:
- ACHE
acetylcholinesterase
- ALS
amyotrophic lateral sclerosis
- ATG5
autophagy-related 5
- CHMP2B
charged multivesicular body protein 2B
- EM
electron microscopy
- HE
Hematoxylin-Eosin
- MAP1LC3 (LC3)
microtubule-associated protein 1 light chain 3
- MDA
malondialdehyde
- MTOR
mechanistic target of rapamycin
- NADH
nicotinamide adenine dinucleotide hydrogen
- NMJs
neuromuscular junctions
- NSE
nonspecific esterase
- PARP1
poly (ADP-ribose) polymerase
- p-RPS6KB
phospho-Thr389-specific 70-kDa ribosomal protein S6 kinase, 70 kDa, polypeptide 1
- SOD1
superoxide dismutase 1, soluble
- WT
wild type
References
- 1.Andersen PM. Amyotrophic lateral sclerosis associated with mutations in the CuZn superoxide dismutase gene. Curr Neurol Neurosci Rep. 2006;6:37–46. doi: 10.1007/s11910-996-0008-9. [DOI] [PubMed] [Google Scholar]
- 2.Kirby J, Halligan E, Baptista MJ, Allen S, Heath PR, Holden H, Barber SC, Loynes CA, Wood-Allum CA, Lunec J, et al. Mutant SOD1 alters the motor neuronal transcriptome: implications for familial ALS. Brain. 2005;128:1686–706. doi: 10.1093/brain/awh503. [DOI] [PubMed] [Google Scholar]
- 3.Pasinelli P, Brown RH. Molecular biology of amyotrophic lateral sclerosis: insights from genetics. Nat Rev Neurosci. 2006;7:710–23. doi: 10.1038/nrn1971. [DOI] [PubMed] [Google Scholar]
- 4.Goldberg AL. Protein degradation and protection against misfolded or damaged proteins. Nature. 2003;426:895–9. doi: 10.1038/nature02263. [DOI] [PubMed] [Google Scholar]
- 5.Kabuta T, Suzuki Y, Wada K. Degradation of amyotrophic lateral sclerosis-linked mutant Cu,Zn-superoxide dismutase proteins by macroautophagy and the proteasome. J Biol Chem. 2006;281:30524–33. doi: 10.1074/jbc.M603337200. [DOI] [PubMed] [Google Scholar]
- 6.Li L, Zhang X, Le W. Altered macroautophagy in the spinal cord of SOD1 mutant mice. Autophagy. 2008;4:290–3. doi: 10.4161/auto.5524. [DOI] [PubMed] [Google Scholar]
- 7.Sasaki S. Autophagy in spinal cord motor neurons in sporadic amyotrophic lateral sclerosis. J Neuropathol Exp Neurol. 2011;70:349–59. doi: 10.1097/NEN.0b013e3182160690. [DOI] [PubMed] [Google Scholar]
- 8.Morimoto N, Nagai M, Ohta Y, Miyazaki K, Kurata T, Morimoto M, Murakami T, Takehisa Y, Ikeda Y, Kamiya T, et al. Increased autophagy in transgenic mice with a G93A mutant SOD1 gene. Brain Res. 2007;1167:112–7. doi: 10.1016/j.brainres.2007.06.045. [DOI] [PubMed] [Google Scholar]
- 9.Cheung ZH, Ip NY. Autophagy deregulation in neurodegenerative diseases - recent advances and future perspectives. J Neurochem. 2011;118:317–25. doi: 10.1111/j.1471-4159.2011.07314.x. [DOI] [PubMed] [Google Scholar]
- 10.Wong E, Cuervo AM. Autophagy gone awry in neurodegenerative diseases. Nat Neurosci. 2010;13:805–11. doi: 10.1038/nn.2575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Banerjee R, Beal MF, Thomas B. Autophagy in neurodegenerative disorders: pathogenic roles and therapeutic implications. Trends Neurosci. 2010;33:541–9. doi: 10.1016/j.tins.2010.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nixon RA, Wegiel J, Kumar A, Yu WH, Peterhoff C, Cataldo A, Cuervo AM. Extensive involvement of autophagy in Alzheimer disease: an immuno-electron microscopy study. J Neuropathol Exp Neurol. 2005;64:113–22. doi: 10.1093/jnen/64.2.113. [DOI] [PubMed] [Google Scholar]
- 13.Chen Y, Klionsky DJ. The regulation of autophagy - unanswered questions. J Cell Sci. 2011;124:161–70. doi: 10.1242/jcs.064576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang X, Li L, Chen S, Yang D, Wang Y, Zhang X, Wang Z, Le W. Rapamycin treatment augments motor neuron degeneration in SOD1(G93A) mouse model of amyotrophic lateral sclerosis. Autophagy. 2011;7:412–25. doi: 10.4161/auto.7.4.14541. [DOI] [PubMed] [Google Scholar]
- 15.Nassif M, Hetz C. Targeting autophagy in ALS: a complex mission. Autophagy. 2011;7:450–3. doi: 10.4161/auto.7.4.14700. [DOI] [PubMed] [Google Scholar]
- 16.Bhattacharya A, Bokov A, Muller FL, Jernigan AL, Maslin K, Diaz V, Richardson A, Van Remmen H. Dietary restriction but not rapamycin extends disease onset and survival of the H46R/H48Q mouse model of ALS. Neurobiol Aging. 2012;33:1829–32. doi: 10.1016/j.neurobiolaging.2011.06.002. [DOI] [PubMed] [Google Scholar]
- 17.Ching JK, Weihl CC. Rapamycin-induced autophagy aggravates pathology and weakness in a mouse model of VCP-associated myopathy. Autophagy. 2013;9:799–800. doi: 10.4161/auto.23958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sarkar S, Floto RA, Berger Z, Imarisio S, Cordenier A, Pasco M, Cook LJ, Rubinsztein DC. Lithium induces autophagy by inhibiting inositol monophosphatase. J Cell Biol. 2005;170:1101–11. doi: 10.1083/jcb.200504035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Morrison KE, Dhariwal S, Hornabrook R, Savage L, Burn DJ, Khoo TK, Kelly J, Murphy CL, Al-Chalabi A, Dougherty A, et al. UKMND-LiCALS Study Group Lithium in patients with amyotrophic lateral sclerosis (LiCALS): a phase 3 multicentre, randomised, double-blind, placebo-controlled trial. Lancet Neurol. 2013;12:339–45. doi: 10.1016/S1474-4422(13)70037-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fornai F, Longone P, Cafaro L, Kastsiuchenka O, Ferrucci M, Manca ML, Lazzeri G, Spalloni A, Bellio N, Lenzi P, et al. Lithium delays progression of amyotrophic lateral sclerosis. Proc Natl Acad Sci U S A. 2008;105:2052–7. doi: 10.1073/pnas.0708022105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pizzasegola C, Caron I, Daleno C, Ronchi A, Minoia C, Carrì MT, Bendotti C. Treatment with lithium carbonate does not improve disease progression in two different strains of SOD1 mutant mice. Amyotroph Lateral Scler. 2009;10:221–8. doi: 10.1080/17482960902803440. [DOI] [PubMed] [Google Scholar]
- 22.Verstraete E, Veldink JH, Huisman MH, Draak T, Uijtendaal EV, van der Kooi AJ, Schelhaas HJ, de Visser M, van der Tweel I, van den Berg LH. Lithium lacks effect on survival in amyotrophic lateral sclerosis: a phase IIb randomised sequential trial. J Neurol Neurosurg Psychiatry. 2012;83:557–64. doi: 10.1136/jnnp-2011-302021. [DOI] [PubMed] [Google Scholar]
- 23.Elbein AD, Pan YT, Pastuszak I, Carroll D. New insights on trehalose: a multifunctional molecule. Glycobiology. 2003;13:17R–27R. doi: 10.1093/glycob/cwg047. [DOI] [PubMed] [Google Scholar]
- 24.Schaeffer V, Lavenir I, Ozcelik S, Tolnay M, Winkler DT, Goedert M. Stimulation of autophagy reduces neurodegeneration in a mouse model of human tauopathy. Brain. 2012;135:2169–77. doi: 10.1093/brain/aws143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sarkar S, Davies JE, Huang Z, Tunnacliffe A, Rubinsztein DC. Trehalose, a novel mTOR-independent autophagy enhancer, accelerates the clearance of mutant huntingtin and alpha-synuclein. J Biol Chem. 2007;282:5641–52. doi: 10.1074/jbc.M609532200. [DOI] [PubMed] [Google Scholar]
- 26.Wooley CM, Sher RB, Kale A, Frankel WN, Cox GA, Seburn KL. Gait analysis detects early changes in transgenic SOD1(G93A) mice. Muscle Nerve. 2005;32:43–50. doi: 10.1002/mus.20228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sarkar S, Ravikumar B, Floto RA, Rubinsztein DC. Rapamycin and mTOR-independent autophagy inducers ameliorate toxicity of polyglutamine-expanded huntingtin and related proteinopathies. Cell Death Differ. 2009;16:46–56. doi: 10.1038/cdd.2008.110. [DOI] [PubMed] [Google Scholar]
- 28.Blokhuis AM, Groen EJ, Koppers M, van den Berg LH, Pasterkamp RJ. Protein aggregation in amyotrophic lateral sclerosis. Acta Neuropathol. 2013;125:777–94. doi: 10.1007/s00401-013-1125-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen S, Zhang X, Song L, Le W. Autophagy dysregulation in amyotrophic lateral sclerosis. Brain Pathol. 2012;22:110–6. doi: 10.1111/j.1750-3639.2011.00546.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dobrowolny G, Aucello M, Rizzuto E, Beccafico S, Mammucari C, Boncompagni S, Belia S, Wannenes F, Nicoletti C, Del Prete Z, et al. Skeletal muscle is a primary target of SOD1G93A-mediated toxicity. Cell Metab. 2008;8:425–36. doi: 10.1016/j.cmet.2008.09.002. [DOI] [PubMed] [Google Scholar]
- 31.Bernardini C, Censi F, Lattanzi W, Barba M, Calcagnini G, Giuliani A, Tasca G, Sabatelli M, Ricci E, Michetti F. Mitochondrial network genes in the skeletal muscle of amyotrophic lateral sclerosis patients. PLoS One. 2013;8:e57739. doi: 10.1371/journal.pone.0057739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Davies JE, Sarkar S, Rubinsztein DC. Trehalose reduces aggregate formation and delays pathology in a transgenic mouse model of oculopharyngeal muscular dystrophy. Hum Mol Genet. 2006;15:23–31. doi: 10.1093/hmg/ddi422. [DOI] [PubMed] [Google Scholar]
- 33.Mizushima N. Autophagy: process and function. Genes Dev. 2007;21:2861–73. doi: 10.1101/gad.1599207. [DOI] [PubMed] [Google Scholar]
- 34.Tashiro Y, Urushitani M, Inoue H, Koike M, Uchiyama Y, Komatsu M, Tanaka K, Yamazaki M, Abe M, Misawa H, et al. Motor neuron-specific disruption of proteasomes, but not autophagy, replicates amyotrophic lateral sclerosis. J Biol Chem. 2012;287:42984–94. doi: 10.1074/jbc.M112.417600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gal J, Ström AL, Kwinter DM, Kilty R, Zhang J, Shi P, Fu W, Wooten MW, Zhu H. Sequestosome 1/p62 links familial ALS mutant SOD1 to LC3 via an ubiquitin-independent mechanism. J Neurochem. 2009;111:1062–73. doi: 10.1111/j.1471-4159.2009.06388.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Klionsky DJ, Abdalla FC, Abeliovich H, Abraham RT, Acevedo-Arozena A, Adeli K, Agholme L, Agnello M, Agostinis P, Aguirre-Ghiso JA, et al. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy. 2012;8:445–544. doi: 10.4161/auto.19496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mizushima N, Yoshimori T, Levine B. Methods in mammalian autophagy research. Cell. 2010;140:313–26. doi: 10.1016/j.cell.2010.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Castillo K, Nassif M, Valenzuela V, Rojas F, Matus S, Mercado G, Court FA, van Zundert B, Hetz C. Trehalose delays the progression of amyotrophic lateral sclerosis by enhancing autophagy in motoneurons. Autophagy. 2013;9:1308–20. doi: 10.4161/auto.25188. [DOI] [PubMed] [Google Scholar]
- 39.Muller FL, Liu Y, Jernigan A, Borchelt D, Richardson A, Van Remmen H. MnSOD deficiency has a differential effect on disease progression in two different ALS mutant mouse models. Muscle Nerve. 2008;38:1173–83. doi: 10.1002/mus.21049. [DOI] [PubMed] [Google Scholar]
- 40.Yamamoto M, Suzuki SO, Himeno M. The effects of dynein inhibition on the autophagic pathway in glioma cells. Neuropathology. 2010;30:1–6. doi: 10.1111/j.1440-1789.2009.01034.x. [DOI] [PubMed] [Google Scholar]
- 41.Filimonenko M, Stuffers S, Raiborg C, Yamamoto A, Malerød L, Fisher EM, Isaacs A, Brech A, Stenmark H, Simonsen A. Functional multivesicular bodies are required for autophagic clearance of protein aggregates associated with neurodegenerative disease. J Cell Biol. 2007;179:485–500. doi: 10.1083/jcb.200702115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Deng HX, Chen W, Hong ST, Boycott KM, Gorrie GH, Siddique N, Yang Y, Fecto F, Shi Y, Zhai H, et al. Mutations in UBQLN2 cause dominant X-linked juvenile and adult-onset ALS and ALS/dementia. Nature. 2011;477:211–5. doi: 10.1038/nature10353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Laird FM, Farah MH, Ackerley S, Hoke A, Maragakis N, Rothstein JD, Griffin J, Price DL, Martin LJ, Wong PC. Motor neuron disease occurring in a mutant dynactin mouse model is characterized by defects in vesicular trafficking. J Neurosci. 2008;28:1997–2005. doi: 10.1523/JNEUROSCI.4231-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Webb JL, Ravikumar B, Rubinsztein DC. Microtubule disruption inhibits autophagosome-lysosome fusion: implications for studying the roles of aggresomes in polyglutamine diseases. Int J Biochem Cell Biol. 2004;36:2541–50. doi: 10.1016/j.biocel.2004.02.003. [DOI] [PubMed] [Google Scholar]
- 45.Zhang F, Ström AL, Fukada K, Lee S, Hayward LJ, Zhu H. Interaction between familial amyotrophic lateral sclerosis (ALS)-linked SOD1 mutants and the dynein complex. J Biol Chem. 2007;282:16691–9. doi: 10.1074/jbc.M609743200. [DOI] [PubMed] [Google Scholar]
- 46.Furuta N, Fujita N, Noda T, Yoshimori T, Amano A. Combinational soluble N-ethylmaleimide-sensitive factor attachment protein receptor proteins VAMP8 and Vti1b mediate fusion of antimicrobial and canonical autophagosomes with lysosomes. Mol Biol Cell. 2010;21:1001–10. doi: 10.1091/mbc.E09-08-0693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pankiv S, Alemu EA, Brech A, Bruun JA, Lamark T, Overvatn A, Bjørkøy G, Johansen T. FYCO1 is a Rab7 effector that binds to LC3 and PI3P to mediate microtubule plus end-directed vesicle transport. J Cell Biol. 2010;188:253–69. doi: 10.1083/jcb.200907015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wurmser AE, Sato TK, Emr SD. New component of the vacuolar class C-Vps complex couples nucleotide exchange on the Ypt7 GTPase to SNARE-dependent docking and fusion. J Cell Biol. 2000;151:551–62. doi: 10.1083/jcb.151.3.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lee JH, Yu WH, Kumar A, Lee S, Mohan PS, Peterhoff CM, Wolfe DM, Martinez-Vicente M, Massey AC, Sovak G, et al. Lysosomal proteolysis and autophagy require presenilin 1 and are disrupted by Alzheimer-related PS1 mutations. Cell. 2010;141:1146–58. doi: 10.1016/j.cell.2010.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Federico A, Cardaioli E, Da Pozzo P, Formichi P, Gallus GN, Radi E. Mitochondria, oxidative stress and neurodegeneration. J Neurol Sci. 2012;322:254–62. doi: 10.1016/j.jns.2012.05.030. [DOI] [PubMed] [Google Scholar]
- 51.Crowe JH. Trehalose as a “chemical chaperone”: fact and fantasy. Adv Exp Med Biol. 2007;594:143–58. doi: 10.1007/978-0-387-39975-1_13. [DOI] [PubMed] [Google Scholar]
- 52.Atkin JD, Farg MA, Walker AK, McLean C, Tomas D, Horne MK. Endoplasmic reticulum stress and induction of the unfolded protein response in human sporadic amyotrophic lateral sclerosis. Neurobiol Dis. 2008;30:400–7. doi: 10.1016/j.nbd.2008.02.009. [DOI] [PubMed] [Google Scholar]
- 53.Nagata T, Ilieva H, Murakami T, Shiote M, Narai H, Ohta Y, Hayashi T, Shoji M, Abe K. Increased ER stress during motor neuron degeneration in a transgenic mouse model of amyotrophic lateral sclerosis. Neurol Res. 2007;29:767–71. doi: 10.1179/016164107X229803. [DOI] [PubMed] [Google Scholar]
- 54.Xu Z, Chen S, Li X, Luo G, Li L, Le W. Neuroprotective effects of (-)-epigallocatechin-3-gallate in a transgenic mouse model of amyotrophic lateral sclerosis. Neurochem Res. 2006;31:1263–9. doi: 10.1007/s11064-006-9166-z. [DOI] [PubMed] [Google Scholar]
- 55.Koh SH, Kim Y, Kim HY, Hwang S, Lee CH, Kim SH. Inhibition of glycogen synthase kinase-3 suppresses the onset of symptoms and disease progression of G93A-SOD1 mouse model of ALS. Exp Neurol. 2007;205:336–46. doi: 10.1016/j.expneurol.2007.03.004. [DOI] [PubMed] [Google Scholar]
- 56.Manabe Y, Nagano I, Gazi MS, Murakami T, Shiote M, Shoji M, Kitagawa H, Abe K. Glial cell line-derived neurotrophic factor protein prevents motor neuron loss of transgenic model mice for amyotrophic lateral sclerosis. Neurol Res. 2003;25:195–200. doi: 10.1179/016164103101201193. [DOI] [PubMed] [Google Scholar]
- 57.Zhang X, Chen S, Li L, Wang Q, Le W. Folic acid protects motor neurons against the increased homocysteine, inflammation and apoptosis in SOD1 G93A transgenic mice. Neuropharmacology. 2008;54:1112–9. doi: 10.1016/j.neuropharm.2008.02.020. [DOI] [PubMed] [Google Scholar]
- 58.Misawa H, Nakata K, Matsuura J, Moriwaki Y, Kawashima K, Shimizu T, Shirasawa T, Takahashi R. Conditional knockout of Mn superoxide dismutase in postnatal motor neurons reveals resistance to mitochondrial generated superoxide radicals. Neurobiol Dis. 2006;23:169–77. doi: 10.1016/j.nbd.2006.02.014. [DOI] [PubMed] [Google Scholar]
- 59.Gurney ME, Pu H, Chiu AY, Dal Canto MC, Polchow CY, Alexander DD, Caliendo J, Hentati A, Kwon YW, Deng HX, et al. Motor neuron degeneration in mice that express a human Cu,Zn superoxide dismutase mutation. Science. 1994;264:1772–5. doi: 10.1126/science.8209258. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.