Abstract
Continuous turnover of intracellular components by autophagy is necessary to preserve cellular homeostasis in all tissues. Despite recent advances in identifying autophagy-related genes and understanding the functions of autophagy in developmental and pathological conditions, so far, the role of autophagy in platelet, a specific anucleate cell type, is poorly understood. In this study, we showed that human platelets express the autophagy-related proteins ATG5, ATG7, and LC3. The same as in nucleated mammalian cells, autophagy was stimulated by cell starvation or the MTOR inhibitor rapamycin in a phosphatidylinositol 3-kinase (PtdIns3K)-dependent manner. Disruption of autophagic flux led to impairment of platelet aggregation and adhesion. Furthermore, Becn1 heterozygous knockout mice displayed a prolonged bleeding time and reduced platelet aggregation. These results suggest a potential role of autophagy in the regulation of platelet function, and imply that gene transcription may not be an essential prerequisite for adaptive autophagy.
Keywords: autophagy, autophagosome, platelet, thrombus, Beclin 1
Introduction
Autophagy is a conserved eukaryotic catabolic process that sequesters protein aggregates and damaged organelles into double-membrane vesicles for lysosomal degradation. Accumulating evidence has demonstrated that autophagy plays important roles in a variety of physiological and pathological processes such as maintenance of cellular homeostasis, adaptation to starvation, development, pathogen elimination, and tumor suppression.1 Autophagy consists of several sequential steps: induction, cargo recognition, autophagosome formation, autophagosome-lysosome fusion, and cargo breakdown followed by recycling of degradation products, all of which are coordinated by different sets of autophagy-related (ATG) proteins.2 Autophagosome formation is the central event in this pathway3 and includes 2 major steps: nucleation, mainly involving the class III phosphatidylinositol 3-kinase (PtdIns3K) complex; and phagophore elongation, mainly involving the microtubule-associated protein 1 light chain 3 (LC3)-conjugation system.2 LC3, a mammalian ortholog of yeast Atg8, is cleaved at its C terminus after synthesis to produce the cytosolic LC3-I form. During autophagy, LC3-I is converted to LC3-II, which is associated with the phagophore and autophagosome membrane by conjugation to phosphatidylethanolamine, and thus serves as a good autophagy marker.4 Extensive studies have identified many post-transcriptional regulatory mechanisms including phosphorylation, ubiquitination, and acetylation.5,6
Platelets are anucleate blood cells that play key roles in hemostasis, thrombosis, inflammation, immunity, and host defense.7 They are derived from the cytoplasm of megakaryocytes and live for 7 to 10 days in the bloodstream after production.8 Platelets have a highly organized cytoskeleton, unique sets of receptors, elaborate signaling pathways, and distinct membrane systems consisting of an open canalicular system, a dense tubular system, mitochondria, peroxisomes, lysosomes, and specialized secretory granules such as α-granules and dense granules.9,10 Although devoid of nuclei, platelets inherit a large reservoir of mRNA from the megakaryocytes, as well as the translational machinery,11 and therefore are capable of synthesizing new proteins12 and performing physiological functions.
In this study, we explored the impact of autophagy on platelet function. Our results showed that autophagic machinery is constitutively present in platelets. Autophagy runs constitutively and inductively in platelets, and is required for the maintenance of their fundamental functions.
Results
Expression of autophagy machinery in human platelets
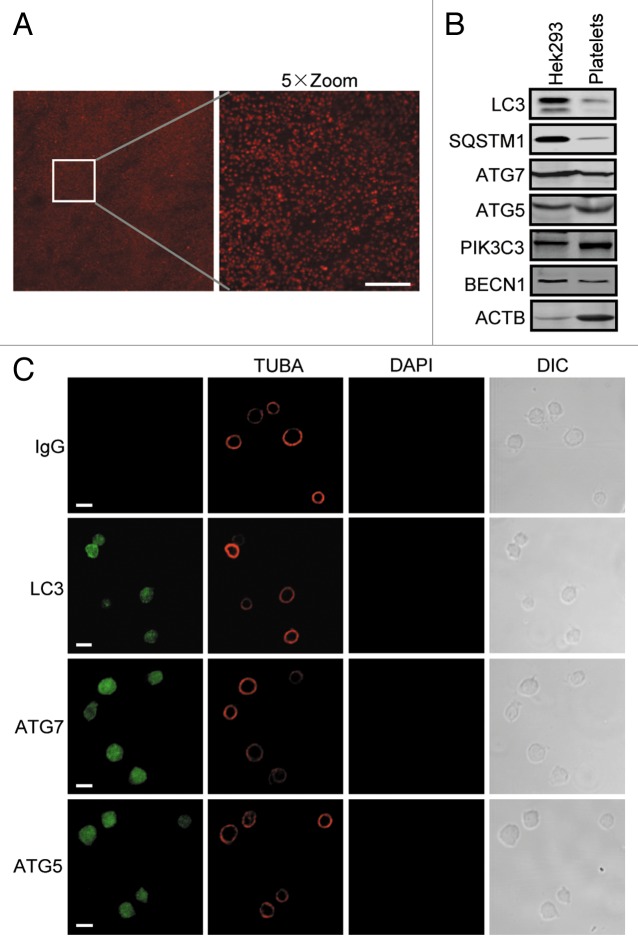
ATGs constitute the molecular basis of autophagy initiation and execution. We initially determined whether platelets possess autophagy modules by checking the expression of ATG5, LC3, and PIK3C3, the catalytic subunit of the class III PtdIns3K, which are the key components of the ATG12-conjugation system, the LC3-conjugation system and the class III PtdIns3K complex, respectively. Platelets were freshly prepared and immediately lysed or fixed for subsequent exploration to avoid unknown in vitro stimulation. Platelet purity assessment by microscopy analysis showed that the ratio of contaminating leukocytes was < 1/10,000 in the platelet suspension (Fig. 1A). Then the levels of LC3, SQSTM1/p62, ATG5, ATG7, BECN1/Beclin 1, and PIK3C3 were analyzed with specific antibodies by western blotting and immunostaining. Evidently, platelets expressed all the checked proteins, though they were relatively less abundant than in human embryonic kidney HEK293 cells (Fig. 1B). Immunostaining analysis demonstrated that, morphologically, LC3, ATG5, and ATG7 appeared to spread evenly in platelets (Fig. 1C). These observations support the notion that autophagy machinery is included in anucleate platelets.
Figure 1. Expression of autophagy machinery in human platelets. (A) Freshly isolated human platelets were stained with anti-TUBA antibody and DAPI, and the cells were imaged by confocal microscopy. Scale bar: 40 μm. (B) Lysates of HEK293 cells and freshly isolated platelets were subjected to western blot analysis with anti-LC3, SQSTM1, ATG5, ATG7, BECN1, or PIK3C3 antibody. Note that the ATG5 band is the ATG12–ATG5 conjugate and its molecular size is 55 kDa. (C) Freshly washed platelets were fixed and stained with anti-LC3, anti-ATG5, or anti-ATG7 with anti-TUBA antibodies. Then the cells were imaged by confocal microscopy. DAPI staining was used to exclude nucleated cell and the TUBA staining and DIC images display platelet morphology. Scale bars: 2 μm.
Induction of autophagy in human platelets
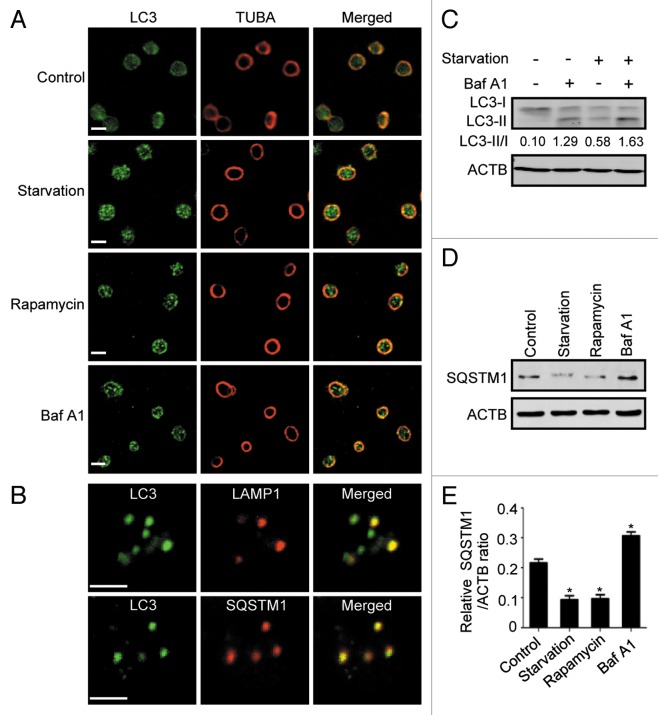
Next, we sought to determine whether autophagy is inducible in these special cells. Starvation and rapamycin are the most used triggers of autophagy, and functional mechanistic target of rapamycin (MTOR) has been found in platelets.13,14 Upon cell starvation and rapamycin treatment, LC3-positive puncta accumulated in the platelets (Fig. 2A), which phenocopied that induced by suppression of lysosomal degradation with the lysosome inhibitor bafilomycin A1 (Baf A1).15 These characteristics mimic those in nucleated cells in response to the same stimulation, implying an identity of autophagosomes with the punctate structures. We further observed the subcellular localization of the punctate structures by comparing it with that of lysosomes and SQSTM1 proteins. Protein SQSTM1 is a selective autophagic adaptor and itself is incorporated into autophagosomes and degraded along with other substrates by lysosomal hydrolyses.16 During starvation, LC3-positive puncta partially colocalized with either lysosomal marker LAMP1 or SQSTM1 (Fig. 2B), suggesting that the puncta were bona fide autophagosomes. Western blot analysis revealed that cell starvation or Baf A1 treatment resulted in the accumulation of LC3-II (Fig. 2C). In addition, compared with Baf A1 treatment, which caused accumulation of SQSTM1, cell starvation and rapamycin treatment led to a decrease in SQSTM1 level (Fig. 2D and E), further confirming that platelets have an effective autophagy pathway at both the constitutive and inductive levels.
Figure 2. Induction of autophagy in human platelets. (A) Human platelets were starved for 1.5 h or treated by rapamycin or Baf A1 in PRP for 2 h. They were then fixed and stained with anti-LC3 and imaged by confocal microcopy. The ring-like TUBA staining represented the typical morphology of resting platelets. (B) Platelets were starved and stained with anti-LC3 and anti-LAMP1 antibodies, or anti-LC3 and anti-SQSTM1 antibodies. (C) Platelets were starved with or without Baf A1 and subjected to western blotting using anti-LC3 antibody. The LC3-II to LC3-I ratio was evaluated by densitometric analysis. The results are representative data of 3 independent experiments. (D) Platelets were starved or treated with rapamycin or Baf A1 and the cellular SQSTM1 level was analyzed by western blot using SQSTM1 antibody. (E) Quantitative analysis of SQSTM1/ACTB ratio of (D). Results are shown as mean ± SEM from 3 independent experiments. *P < 0.05. Scale bars: 2 μm.
Autophagy in human platelets is class III PtdIns3K-dependent
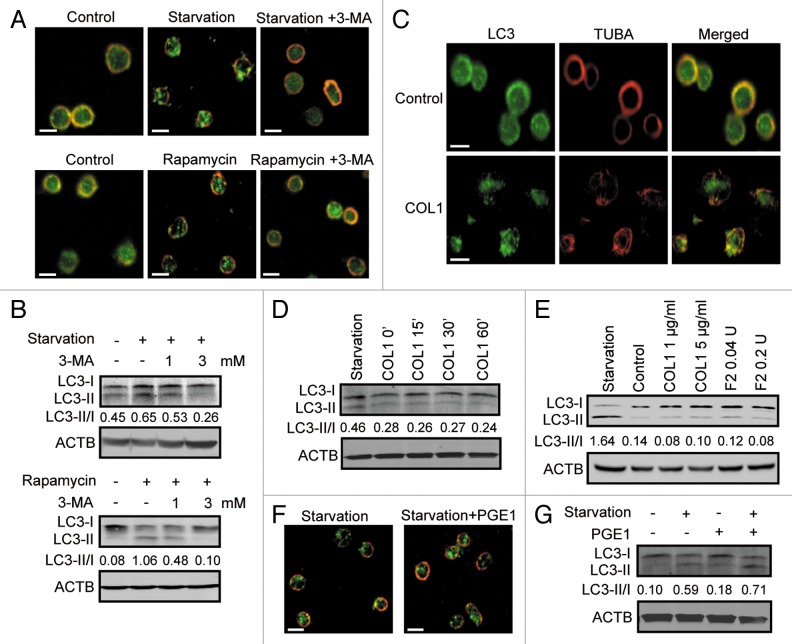
In nucleated cells, autophagy is tightly regulated by several signaling pathways in which the signal molecules MTOR and class III PtdIns3K occupy central positions.17,18 Induction of autophagy in platelets by rapamycin identified MTOR as a negative regulator of the event, so we then sought to determine whether class III PtdIns3K is essential to and operational in the process. Freshly purified human platelets were starved or treated with rapamycin with or without addition of 3-methyladenine (3-MA), a specific inhibitor of class III PtdIns3K.19 We found that autophagosome formation induced in platelets by either starvation or rapamycin was dramatically inhibited by addition of 3-MA (Fig. 3A). Consistently, the results from western blotting also demonstrated a clear suppression of LC3-II production by 3-MA (Fig. 3B), suggesting a crucial role of class III PtdIns3K in the autophagic process of platelets.
Figure 3. Autophagy in platelets is class III PtdIns3K activity-dependent. (A) Platelets were starved for 1.5 h or treated with rapamycin for 2 h in the presence or absence of 3 mM 3-MA. Then the platelets were fixed and immunostained using anti-LC3 antibody, and were imaged by confocal microcopy. (B) Platelets were starved or treated with rapamycin with or without 1 mM or 3 mM 3-MA. Then the platelets were analyzed by western blot using anti-LC3 antibody. (C) Platelets incubated with or without 1 μg/ml of COL1 for 30 min were labeled with anti-LC3 and anti-TUBA antibodies. (D and E) Platelets incubated with 1 μg/ml COL1 for the indicated durations (D) or different concentrations of COL1 or F2/thrombin for 10 min (E) were analyzed by western blot with anti-LC3 antibody. (F and G) Platelets starved with or without 300 nM PGE1 were labeled (F) or analyzed by western blot using anti-LC3 antibody (G). The LC3-II to LC3-I ratio was evaluated by densitometric analysis in all the western blot experiments. All the results are representative data of 3 independent experiments. Scale bars: 2 μm.
Platelet activation by stimuli initiates multiple intracellular signaling cascades to induce shape change, secretion, aggregation, and other events.20 During purification, platelets tend to be easily activated. To exclude that starvation- or rapamycin-stimulated autophagy was due to unwanted activation of the platelets, we then investigated the correlation between platelet activation and platelet autophagy. We revealed that activation of human platelets by either COL1 (collagen, type 1) or F2 [coagulation factor II (thrombin)] treatment, which acts through either tyrosine kinase-coupled receptors or G-protein-coupled receptors, caused neither the formation of autophagosomes nor the increase in LC3-II (Fig. 3C–E). The activation of the platelets by COL1 or F2/thrombin was confirmed by detection of the exposure of SELP [selectin P (granule membrane protein 140 kDa, antigen CD62)] on the cell surface (Fig. S1). In addition, when platelets were treated with prostaglandin E1 (PGE1), a commonly used platelet inhibitor that functions by increasing the intracellular level of cyclic AMP,21 starvation was still able to induce autophagy (Fig. 3F and G). Collectively, these data indicate that autophagy in platelets is independent of their activation.
Blocking autophagic degradation inhibits platelet aggregation and adhesion
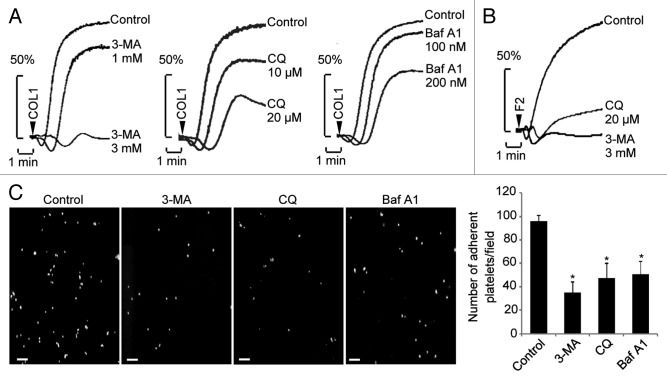
The existence of autophagy machinery and the inducibility of autophagy in platelets prompted us to investigate the physiological function of autophagic degradation in platelets. In Tyrode’s buffer-cultured platelets, preincubation with 3-MA dose-dependently inhibited the aggregation of platelets triggered by the physiological agonists COL1 and F2/thrombin (Fig. 4A and B). In addition, treatment with Baf A1 or chloroquine (CQ), commonly-used inhibitor of lysosomes,15 also evidently weakened COL1- or F2/thrombin-stimulated platelet aggregation (Fig. 4A and B). Similar results were obtained from platelets in platelet-rich plasma (PRP) (Fig. S2), indicating constitutive autophagy is required for platelet aggregation. Platelet adhesion was also checked upon disruption of autophagy. Perfusion of human blood over a reconstituted COL1-coated surface at a shear rate of 1000 s−1 led to significant platelet adhesion. We found, under this condition, that platelet adhesion was significantly inhibited by 3-MA, and to a milder degree, by CQ and Baf A1 (Fig. 4C). To exclude the possibility that the inhibition of platelet function by the chemicals was due to decreased cell viability, we analyzed phosphatidylserine exposure to detect apoptosis of the platelets. Very little ANXA5/annexin V staining was detected in the platelets treated with each of the chemicals used for autophagy blocking, when more than 85% of the platelets were positively stained for ANXA5 by ionomycin, a typical apoptosis inducer (Fig. S3A and S3B). Results from the [3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium, inner salt] (MTS) assay also confirmed that the platelet viability was not influenced by each of the chemicals with the same dose and treatment time used for autophagy blocking (Fig. S3C). Taken together, these results suggest a potential role of the autophagic degradation pathway in the regulation of platelet function.
Figure 4. Inhibition of autophagic degradation impairs aggregation and adhesion of platelets. (A and B) Washed platelets were pretreated with the indicated chemicals for 1.5 h, then COL1 (1 μg/ml) or F2/thrombin (0.04 U) was added and the aggregation was recorded using an aggregometer. (C) After treatment with the indicated chemicals for 1.5 h, calcein-labeled platelets were added to washed red blood cells and the reconstituted blood was perfused over COL1-coated coverslips. Adherent platelets were scored under a fluorescence microscope. Values are shown as mean ± SEM from 6 visual fields of duplicate experiments. *P < 0.05. Scale bars: 20 μm.
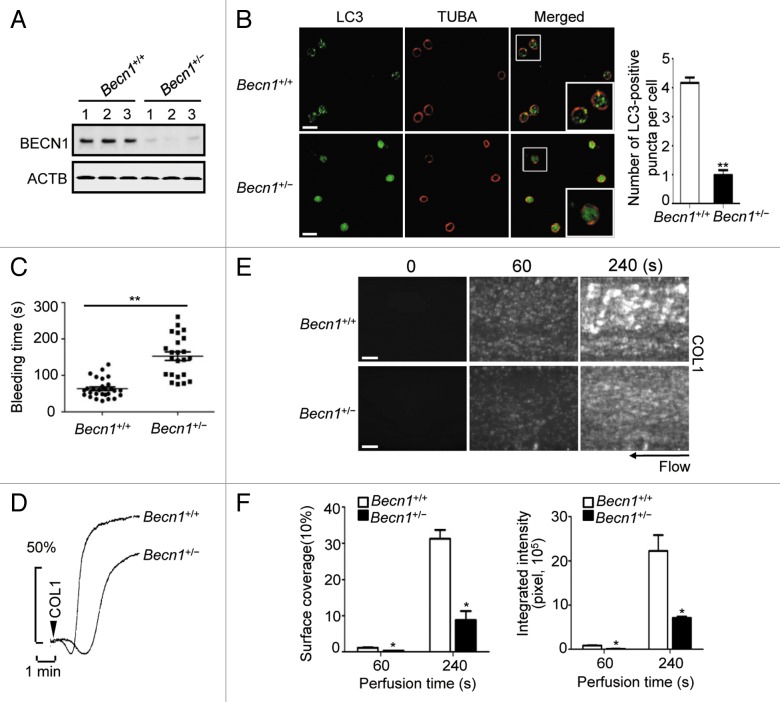
Impaired platelet function in mice with Becn1 heterozygous disruption
As a part of a class III PtdIns3K complex, BECN1 is required for autophagy by mediating the localization of other autophagy proteins to phagophores.22,23 Becn1 heterozygous knockout mice have shown reduced autophagy in several tissues and the development of tumorigenesis.24 To further define the relevance of autophagy to the physiological functions of platelets and rule out the potential off-target effect of the chemical inhibitors, we checked the function of platelets from Becn1+/− mice. First, we checked BECN1 expression level in platelets from Becn1+/+ and Becn1+/− mice. As expected, the BECN1 expression in Becn1+/− platelets was much lower compared with that in Becn1+/+ mice (Fig. 5A). We found that starvation-induced LC3-positive puncta were much less in Becn1+/− platelets than in Becn1+/+ platelets (Fig. 5B), indicating a reduced autophagosome formation in the Becn1 heterozygous deletion platelets. However, the LC3-I to LC3-II conversion was not dramatically affected by cell starvation (Fig. S4A and S4B), supporting a previous suggestion that BECN1 is required for LC3 association with autophagic membranes but not for the LC3 lipidation.23,25 To further verify autophagy in Becn1+/− platelets, we examined the protein level of SQSTM1. Apparently, the basal level of SQSTM1 in Becn1+/− platelets was higher than Becn1+/+ platelets, and SQSTM1 maintained at a high level after cell starvation in Becn1+/− platelets (Fig. S4A and S4C). These data indicated that autophagy is suppressed in platelets of mice with heterozygous Becn1 deletion.
Figure 5. Platelet function in mice with Becn1 heterozygous disruption. (A) The expression level of BECN1 was analyzed in platelets isolated from 3 individual Becn1+/+ or Becn1+/− mice by western blot using specific anti-BECN1 antibody. (B) Fresh platelets form Becn1+/+ or Becn1+/− mice were starved, fixed, and labeled with anti-LC3 and anti-TUBA. Values are shown as number of LC3-positive puncta per cell from 3 independent experiments. **P < 0.01. Scale bars: 2 μm. (C) Quantification of the tail-bleeding time of Becn1+/+ mice and their Becn1+/− littermates. Values are mean ± SEM **P < 0.01. (D) Fresh platelets from Becn1+/+ or Becn1+/− mice were analyzed for aggregation response upon 1 μg/ml COL1 stimulation. (E) The whole blood with mepacrine-labeled platelets from Becn1+/+ or Becn1+/− mice was perfused over COL1-coated microfluidic channels. Platelet adhesion and thrombus formation were visualized and imaged by fluorescence microscopy. Scale bars: 20 μm. (F) Quantification of platelet adhesion and thrombus formation in (E). Results are expressed as the mean percentage of surface coverage (left panels) and total integrated fluorescence intensity (right panels) (n = 6 per group). *P < 0.05.
We then used the Becn1+/+ and Becn1+/− mouse littermates to measure the tail bleeding time and the platelet aggregation response. As expected, Becn1+/− mice showed clearly a longer bleeding time compared with that of the wild-type mice (Fig. 5C), and the platelet aggregation stimulated by COL1 was reduced evidently in Becn1+/− mice (Fig. 5D). We further assayed the adhesion dynamics of platelets to the immobilized-COL1 surface under blood flow condition. We found that the primary attachment rate of Becn1+/− platelets was lower than the wild-type platelets, and when most of the adherent Becn1+/+ platelets developed into aggregates, adherent Becn1+/− platelets remained solitary (Fig. 5E and F), indicating an impaired thrombus growth on COL1. Together, these results suggest in vivo the critical role of autophagy in platelet function.
Discussion
In this study, we have demonstrated that platelets express ATG proteins and autophagy can be induced in platelets. As in nucleated cells, autophagy in platelets is mediated by class III PtdIns3K activity and signals initiated by MTOR inhibition. Our data suggest that lysosome-dependent cellular degradation is an essential pathway for platelets to maintain their physiological functions.
It seems so far that all the events involved in autophagic degradation, including the formation of autophagosomes and autophagosome-lysosome fusion, occur in the cytoplasm, while the function of the nucleus in autophagy remains unclear. Although ATG proteins are abundantly expressed in eukaryotic cells, several studies have described transcriptional alteration of some ATG proteins during autophagy.26,27 Nevertheless, it has been shown that autophagy can be stimulated in artificially enucleated cells.28,29 Using platelets as an anucleate cell model, our data suggested that gene transcription may not be an essential prerequisite for autophagy, at least for adaptive autophagy that can be induced within a short time. However, basal or constitutive autophagy, which operates continuously at a low level, or long-term autophagy induction, might rely on extensive transcriptions.
Accumulating evidence has shown that platelets can synthesize new proteins constitutively or in an activation-dependent manner.12 However, little is known about how misfolded or damaged proteins or organelles are eliminated in these anucleate cells. It is necessary that unneeded proteins be cleared by proteasomal and/or autophagic degradation pathway(s) for the maintenance of intracellular homeostasis, especially in the platelet, a cell-type that is highly responsive to environmental cues. In spite of the fact that proteasomes are present in platelets,30,31 our results indicate that the autophagic degradation pathway plays an important role in platelet function. It is possible that, as in nucleated cells, the proteasome system in platelets digests soluble and short-lived proteins while the autophagy pathway mainly degrades long-lived proteins and large protein aggregates. Nevertheless, due to the difficulty of gene manipulation in platelets, in our study, depression of autophagy was achieved mostly by use of specific inhibitors. In addition, BECN1 may not be specific to autophagy and the defect observed in Becn1+/− mice may be caused by a defect other than autophagy. Therefore, utilizing hematopoietic stem cell-specific atg knockout mice and samples from clinical patients would provide further evidence for the role of autophagy in platelet function.
The mechanism underlying the finding that inactivation of autophagy inhibits platelet aggregation and adhesion remains to be elucidated. Despite the lack of direct data in platelets, it has been estimated in liver cells that ~1% to 1.5% of cellular proteins are catabolized per hour by autophagy, even under nutrient-rich conditions.32 Thus, one possibility is that upon blockade of autophagic degradation, abnormal or damaged components accumulate inside platelets, homeostasis is disrupted, and platelet activation signals are blunted. Preincubating the platelets with the inhibitors for a relatively short time clearly suppressed autophagic degradation, implying a more direct pathway may be involved. In fact, based on the regulatory function of autophagy in WNT signaling,33 PTK2/FAK signaling,34 and Drosophila hemocyte cortical remodeling,35 it has been proposed that autophagy acts as a means to dampen signaling activities in response to changing cues by sequestering or degrading ubiquitinated signaling components.35 Given that some kinases in platelet are known to undergo ubiquitination,36 and the extent of platelet activation is ultimately determined by the balance of activating and inactivating signals,37 autophagic clearance of certain potentially ubiquitinated proteins that antagonize platelet activation may represent a direct mechanism for the inhibition of platelets by autophagy disruption.
In conclusion, we provide the first evidence that autophagy occurs in anucleate platelets and defective autophagy causes impaired platelet aggregation and adhesion. Our findings provide new substances for the regulation of platelet function and potential mechanism for some of the platelet-related hemorrhagic diseases. Improvement of the function of the platelets by targeting autophagic machinery including the lysosomes may provide promising choices for the treatment of these diseases. More importantly, development of specific autophagy inhibitor of platelets may become a novel strategy for antithrombotic therapy.
Materials and Methods
Reagents and antibodies
Rapamycin (Sigma, R8781), bafilomycin A1 (Baf A1) (Sigma, B1793), chloroquine (CQ) (Sigma, C6628), 3-methyladenine (3-MA) (Sigma, M9281), F2/thrombin (Sigma, T6884), prostaglandin E1 (PGE1) (Sigma, P5515), ionomycin (Sigma, I3909) and mepacrine (quinacrine dihydrochloride) (Sigma, Q3251) were from Sigma Aldrich. COL1/collagen, type 1 (Chrono-Log Corp, 385) was from Chrono-Log Corporation. Protein G beads (Santa Cruz, sc2002) were from Santa Cruz Biotechnology. In all the experiments, rapamycin, Baf A1, and CQ were used at 200 nM, 200 nM and 20 μM, respectively, unless otherwise indicated.
The following antibodies were used: rabbit polyclonal anti-LC3 (Sigma, L7643), anti-ATG7 (Sigma, A2856) and mouse monoclonal anti-ACTB (Sigma, A5316), anti-TUBA (Sigma, T9026); rabbit polyclonal anti-ATG5 (Novus Biologicals, NB110-53818); rabbit monoclonal anti-PIK3C3 (Cell Signaling, 3811); mouse monoclonal anti-SQSTM1 (Santa Cruz, sc28359), rabbit polyclonal anti-SQSTM1 (Santa Cruz, sc25575), rabbit polyclonal anti-BECN1 (Proteintech, 11306-1-AP), mouse monoclonal anti-LAMP1 (Santa Cruz, sc20011), FITC-conjugated CD62P (BD Biosciences, 550866), FITC-conjugated ANXA5/annexin V (BD Biosciences, 556547) and FITC-conjugated mouse IgG1 (BD Biosciences, 553443). Alexa Fluor 488- and 546-tagged secondary antibodies for immunostaining were from Molecular Probes. The secondary antibodies for western blots were donkey anti-rabbit IRDye800CW (LI-COR Biosciences, 926-32213) and donkey anti-mouse IRDye680 (LI-COR Biosciences, 926-32222) from LI-COR Biosciences.
Human platelet isolation and culture
All studies were approved by Zhejiang University Institutional Review Committee and informed consent was obtained. Whole blood from healthy donors was drawn into BD vacutainers (BD Biosciences, 301746) containing sodium citrate and centrifuged at 150 × g for 20 min to collect PRP. Without disturbing the buffy coat, the upper 3/4 of PRP was carefully removed to new tubes, diluted 3-fold with acid-citrate-dextrose (ACD, 75 mM sodium citrate, 39 mM citric acid, and 135 mM dextrose, pH 6.5), and centrifuged at 800 × g for 10 min. The platelet pellet fraction was resuspended in platelet-poor plasma (PPP) for immunostaning experiments and in M199 medium (Gibco, 12340-030) for western blot experiments.
Mouse platelet isolation and tail bleeding measurement
Animal experiments were approved by Review Committee of Zhejiang University School of Medicine and were in compliance with Institutional Guidelines of the ISTH. Twenty four Becn1+/− mice24 (8 to 12 wk old) and their Becn1+/+ littermates were anesthetized (400 mg/kg chloraldurat). Whole blood from the inferior vena cava was collected into ACD and centrifuged at 180 × g for 10 min to obtain PRP. Then PRP was diluted in ACD containing 1 U/ml apyrase and centrifuged at 800 × g for 10 min. The platelet pellet was resuspended in Tyrode’s buffer [137 mM NaCl, 12 mM NaHCO3, 2 mM KCl, 0.34 mM Na2HPO4, 1 mM MgCl2, 5.5 mM glucose, 5 mM HEPES, and 0.35% bovine serum albumin (BSA; MP, 0218054980)].
Tail bleeding times were determined as previously described.38 Tails of Becn1+/− and Becn1+/+ mice after anesthesia were transected 5 mm from the tip with a scalpel blade. The bleeding end was immersed in saline prewarmed at 37 °C, and the time to bleeding cessation (for more than 10 s) was recorded.
Autophagy induction by starvation or rapamycin treatment
For starvation, the platelets were resuspended in starvation medium (140 mM NaCl, 1 mM CaCl2, 1 mM MgCl2, 3 mM glucose, 20 mM HEPES and 0.35% BSA, pH 7.4) and incubated at 37 °C for 1.5 h. For rapamycin-induced autophagy, platelets were treated with 200 nM rapamycin in designated medium at 37 °C for 2 h.
Western blot
Western blotting was performed as described previously39 with minor modifications. In brief, proteins from lysed platelets were resolved on SDS polyacrylamide gels. The proteins were then transferred to a polyvinylidene difluoride membrane. After blocking with 3% (w/v) BSA in TBST (150 mM NaCl, 10 mM TRIS-HCl, pH 7.5 and 0.1% Tween 20), the membrane was stained with corresponding primary antibodies. The specific bands were analyzed using an Odyssey infrared imaging system (LI-COR Biosciences) after incubated with the corresponding secondary antibodies, goat anti-rabbit IRDye 800CW and goat anti-mouse IRDye 680 (LI-COR Biosciences). Signal intensity was quantified using NIH ImageJ.
Immunostaining and confocal microscopy
For immunostaining, platelets attached on Poly-L-Lysine-coated coverslip were fixed with precooled methanol for 10 min. To strengthen the association of the platelets to the coverslip, at the last 3 min of fixation, the coverslip was spun at 500 × g. After washing twice in phosphate-buffered saline (PBS; 135 mM NaCl, 4.7 mM KCl, 10 mM Na2HPO4, 2 mM NaH2PO4, pH 7.4), platelets were blocked with 10% FCS in PBS. The platelets were then incubated with appropriate primary and secondary antibodies in 0.15% saponin (Sigma, 8047-15-2) as indicated in the legends.
Confocal images were taken in multitracking mode on a LSM510 Meta laser-scanning confocal microscope (Carl Zeiss) with a plan apochromat 100×/1.4 NA oil immersion lens (optical slice thickness 0.5 μm). Images were analyzed with Zeiss LSM Image Examiner Software.
Flow cytometry
Human platelets (1 × 106/reaction) were suspended in modified Tyrode’s buffer (134 mM NaCl, 2.9 mM KCl, 0.34 mM Na2HPO4, 12 mM NaHCO3, 20 mM HEPES, 1.0 mM MgCl2, 5.0 mM glucose, pH 7.35) containing 2.0 mM CaCl2. After treatment with the indicated chemicals at 37 °C without stirring, the platelets were incubated for 10 min with FITC-conjugated CD62P, FITC-conjugated ANXA5/annexin V, or FITC-conjugated mouse IgG1. The platelets were analyzed on a FACScan flow cytometer (Beckman Coulter) and gated by light scatter.
MTS assay of platelet viability
[3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium, inner salt] (MTS) assay (Promega, G358A) was performed as described previously.40 Briefly, 10 µl of PRP containing 1.5 × 107 platelets or PPP was diluted with PBS to 100 µl in 96-well plates. After addition of 20 µl of MTS, the platelet suspension was incubated for 4 h at 37 °C, and the reaction was read in a kinetic microplate reader (Molecular Devices) at 490 nm. The results were calculated by subtracting the OD of the PPP from the OD of the PRP samples.
Platelet aggregation
For aggregation analysis, platelets were suspended in Tyrode’s buffer. After treatment with chemicals at 37 °C, 5 mM CaCl2 was added, and aggregation was stimulated by designated agonists and measured using a Chrono-log 700 aggregometer (Chrono-Log Corp). During aggregation recording, platelets were stirred at 900 rpm.
Platelet adhesion
Adhesion analysis was performed as previously described with minor modifications.41 Briefly, glass coverslips were cleaned in dichromic acid, rinsed in dH2O, and coated with COL1 at a density of 0.08 μg/cm2 overnight at 4 °C. The coverslip was assembled into the flow chamber as the lower surface. A Master-2 flex L/S peristaltic pump (Cole-Parmer) was used to aspirate blood through the flow chamber.
Platelets resuspended in Tyrode’s buffer were pretreated at 37 °C with the indicated chemicals for 1.5 h and calcein (10 μg/ml; Molecular Probes, C3099) was added for the last 20 min. Then the platelet suspension was mixed with washed red blood cells to form reconstituted blood at a platelet concentration of 200 × 105/ml and a hematocrit of 45%. Reconstituted blood was perfused over the coverslip for 5 min at a shear rate of 1,000/s. The perfusion was live-monitored under a TE-2000S fluorescence microscope equipped with a DS-2MBWc-U1 CCD camera (Nikon). After the termination of perfusion, the flow chamber was washed with PBS for 2 min to remove nonadherent cells. And the adherent platelets were counted using Image-Pro Plus software.
Ex vivo flow-based thrombus formation assay
Thrombus formation was evaluated by a whole-blood perfusion assay utilizing a Bioflux-200 microfluidic well-plate (Fluxion Biosciences) and performed as previously described.42 The microfluidic devices were coated with 100 μg/ml COL1 overnight at 4 °C. Subsequently, the devices were rinsed with an excessive PBS and blocked with 0.5% BSA in PBS for 30 min. Mepacrine (2 μM, 30 min at 37 °C) was used to label platelets in whole blood. Labeled blood was then perfused through the COL1-coated microcapillaries at a wall shear rate of 40 dynes/cm2, followed by washing at the same shear rate with PBS. Images of platelet adhesion and thrombus formation were acquired by epifluorescence microscopy in real-time. Thrombus formation was determined as the mean percentage of total area covered by thrombi and as the mean integrated fluorescence intensity per micrometer squared. Image analysis was performed using Metamorph software (Universal Imaging).
Statistics
All the statistical data were presented as mean ± SEM. Statistical significance of the differences was determined using the Student t test. P < 0.05 was considered to be statistically significant.
Supplementary Material
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We are grateful to Nannan Wu,Yaosen Wu, and the Imaging Center of Zhejiang University School of Medicine for technical assistance and Cunji Gao for helpful discussion. This study was supported by the National Basic Research Program of China (2011CB910100 and 2013CB910200), the National Natural Science Foundation of China (30971429 and 31171288).
Glossary
Abbreviations:
- 3-MA
3-methyladenine
- ACTB
actin, beta
- ATG
autophagy-related
- Baf A1
bafilomycin A1
- BECN1
Beclin 1, autophagy-related
- BSA
bovine serum albumin
- COL1
collagen, type 1
- CQ
chloroquine
- F2
coagulation factor II (thrombin)
- MAP1LC3 (LC3)
microtubule-associated protein 1 light chain 3
- MTOR
mechanistic target of rapamycin
- PGE1
prostaglandin E1
- PIK3C3
phosphatidylinositol 3-kinase, catalytic subunit type 3
- PRP
platelet-rich plasma
- PtdIns3K
phosphatidylinositol 3-kinase
- SQSTM1/p62
sequestosome 1
- TUBA
tubulin, alpha
References
- 1.Mizushima N. The pleiotropic role of autophagy: from protein metabolism to bactericide. Cell Death Differ. 2005;12(Suppl 2):1535–41. doi: 10.1038/sj.cdd.4401728. [DOI] [PubMed] [Google Scholar]
- 2.Mizushima N, Yoshimori T, Levine B. Methods in mammalian autophagy research. Cell. 2010;140:313–26. doi: 10.1016/j.cell.2010.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xie Z, Klionsky DJ. Autophagosome formation: core machinery and adaptations. Nat Cell Biol. 2007;9:1102–9. doi: 10.1038/ncb1007-1102. [DOI] [PubMed] [Google Scholar]
- 4.Kabeya Y, Mizushima N, Yamamoto A, Oshitani-Okamoto S, Ohsumi Y, Yoshimori T. LC3, GABARAP and GATE16 localize to autophagosomal membrane depending on form-II formation. J Cell Sci. 2004;117:2805–12. doi: 10.1242/jcs.01131. [DOI] [PubMed] [Google Scholar]
- 5.Gozuacik D, Kimchi A. Autophagy and cell death. Curr Top Dev Biol. 2007;78:217–45. doi: 10.1016/S0070-2153(06)78006-1. [DOI] [PubMed] [Google Scholar]
- 6.Lee IH, Cao L, Mostoslavsky R, Lombard DB, Liu J, Bruns NE, Tsokos M, Alt FW, Finkel T. A role for the NAD-dependent deacetylase Sirt1 in the regulation of autophagy. Proc Natl Acad Sci U S A. 2008;105:3374–9. doi: 10.1073/pnas.0712145105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Spinelli SL, Maggirwar SB, Blumberg N, Phipps RP. Nuclear emancipation: a platelet tour de force. Sci Signal. 2010;3:pe37. doi: 10.1126/scisignal.3144pe37. [DOI] [PubMed] [Google Scholar]
- 8.Leeksma CH, Cohen JA. Determination of the life of human blood platelets using labelled diisopropylfluorophosphanate. Nature. 1955;175:552–3. doi: 10.1038/175552b0. [DOI] [PubMed] [Google Scholar]
- 9.Weyrich AS, Lindemann S, Tolley ND, Kraiss LW, Dixon DA, Mahoney TM, Prescott SP, McIntyre TM, Zimmerman GA. Change in protein phenotype without a nucleus: translational control in platelets. Semin Thromb Hemost. 2004;30:491–8. doi: 10.1055/s-2004-833484. [DOI] [PubMed] [Google Scholar]
- 10.White JG. Ultrastructural studies of the gray platelet syndrome. Am J Pathol. 1979;95:445–62. [PMC free article] [PubMed] [Google Scholar]
- 11.Lindemann S, Tolley ND, Dixon DA, McIntyre TM, Prescott SM, Zimmerman GA, Weyrich AS. Activated platelets mediate inflammatory signaling by regulated interleukin 1beta synthesis. J Cell Biol. 2001;154:485–90. doi: 10.1083/jcb.200105058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Weyrich AS, Schwertz H, Kraiss LW, Zimmerman GA. Protein synthesis by platelets: historical and new perspectives. J Thromb Haemost. 2009;7:241–6. doi: 10.1111/j.1538-7836.2008.03211.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Weyrich AS, Denis MM, Schwertz H, Tolley ND, Foulks J, Spencer E, Kraiss LW, Albertine KH, McIntyre TM, Zimmerman GA. mTOR-dependent synthesis of Bcl-3 controls the retraction of fibrin clots by activated human platelets. Blood. 2007;109:1975–83. doi: 10.1182/blood-2006-08-042192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aslan JE, Tormoen GW, Loren CP, Pang J, McCarty OJ. S6K1 and mTOR regulate Rac1-driven platelet activation and aggregation. Blood. 2011;118:3129–36. doi: 10.1182/blood-2011-02-331579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Klionsky DJ, Abeliovich H, Agostinis P, Agrawal DK, Aliev G, Askew DS, Baba M, Baehrecke EH, Bahr BA, Ballabio A, et al. Guidelines for the use and interpretation of assays for monitoring autophagy in higher eukaryotes. Autophagy. 2008;4:151–75. doi: 10.4161/auto.5338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Komatsu M, Waguri S, Koike M, Sou YS, Ueno T, Hara T, Mizushima N, Iwata J, Ezaki J, Murata S, et al. Homeostatic levels of p62 control cytoplasmic inclusion body formation in autophagy-deficient mice. Cell. 2007;131:1149–63. doi: 10.1016/j.cell.2007.10.035. [DOI] [PubMed] [Google Scholar]
- 17.Ravikumar B, Vacher C, Berger Z, Davies JE, Luo S, Oroz LG, Scaravilli F, Easton DF, Duden R, O’Kane CJ, et al. Inhibition of mTOR induces autophagy and reduces toxicity of polyglutamine expansions in fly and mouse models of Huntington disease. Nat Genet. 2004;36:585–95. doi: 10.1038/ng1362. [DOI] [PubMed] [Google Scholar]
- 18.Tassa A, Roux MP, Attaix D, Bechet DM. Class III phosphoinositide 3-kinase--Beclin1 complex mediates the amino acid-dependent regulation of autophagy in C2C12 myotubes. Biochem J. 2003;376:577–86. doi: 10.1042/BJ20030826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Seglen PO, Gordon PB. 3-Methyladenine: specific inhibitor of autophagic/lysosomal protein degradation in isolated rat hepatocytes. Proc Natl Acad Sci U S A. 1982;79:1889–92. doi: 10.1073/pnas.79.6.1889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li Z, Delaney MK, O’Brien KA, Du X. Signaling during platelet adhesion and activation. Arterioscler Thromb Vasc Biol. 2010;30:2341–9. doi: 10.1161/ATVBAHA.110.207522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Roberts W, Magwenzi S, Aburima A, Naseem KM. Thrombospondin-1 induces platelet activation through CD36-dependent inhibition of the cAMP/protein kinase A signaling cascade. Blood. 2010;116:4297–306. doi: 10.1182/blood-2010-01-265561. [DOI] [PubMed] [Google Scholar]
- 22.Liang XH, Kleeman LK, Jiang HH, Gordon G, Goldman JE, Berry G, Herman B, Levine B. Protection against fatal Sindbis virus encephalitis by beclin, a novel Bcl-2-interacting protein. J Virol. 1998;72:8586–96. doi: 10.1128/jvi.72.11.8586-8596.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Suzuki K, Kirisako T, Kamada Y, Mizushima N, Noda T, Ohsumi Y. The pre-autophagosomal structure organized by concerted functions of APG genes is essential for autophagosome formation. EMBO J. 2001;20:5971–81. doi: 10.1093/emboj/20.21.5971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Qu X, Yu J, Bhagat G, Furuya N, Hibshoosh H, Troxel A, Rosen J, Eskelinen EL, Mizushima N, Ohsumi Y, et al. Promotion of tumorigenesis by heterozygous disruption of the beclin 1 autophagy gene. J Clin Invest. 2003;112:1809–20. doi: 10.1172/JCI20039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Matsui Y, Takagi H, Qu X, Abdellatif M, Sakoda H, Asano T, Levine B, Sadoshima J. Distinct roles of autophagy in the heart during ischemia and reperfusion: roles of AMP-activated protein kinase and Beclin 1 in mediating autophagy. Circ Res. 2007;100:914–22. doi: 10.1161/01.RES.0000261924.76669.36. [DOI] [PubMed] [Google Scholar]
- 26.Zhao J, Brault JJ, Schild A, Cao P, Sandri M, Schiaffino S, Lecker SH, Goldberg AL. FoxO3 coordinately activates protein degradation by the autophagic/lysosomal and proteasomal pathways in atrophying muscle cells. Cell Metab. 2007;6:472–83. doi: 10.1016/j.cmet.2007.11.004. [DOI] [PubMed] [Google Scholar]
- 27.Settembre C, Di Malta C, Polito VA, Garcia Arencibia M, Vetrini F, Erdin S, Erdin SU, Huynh T, Medina D, Colella P, et al. TFEB links autophagy to lysosomal biogenesis. Science. 2011;332:1429–33. doi: 10.1126/science.1204592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tasdemir E, Maiuri MC, Galluzzi L, Vitale I, Djavaheri-Mergny M, D’Amelio M, Criollo A, Morselli E, Zhu C, Harper F, et al. Regulation of autophagy by cytoplasmic p53. Nat Cell Biol. 2008;10:676–87. doi: 10.1038/ncb1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Morselli E, Mariño G, Bennetzen MV, Eisenberg T, Megalou E, Schroeder S, Cabrera S, Bénit P, Rustin P, Criollo A, et al. Spermidine and resveratrol induce autophagy by distinct pathways converging on the acetylproteome. J Cell Biol. 2011;192:615–29. doi: 10.1083/jcb.201008167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yukawa M, Sakon M, Kambayashi J, Shiba E, Kawasaki T, Uemura Y, Murata K, Tanaka T, Nakayama T, Shibata H, et al. Purification and characterization of endogenous protein activator of human platelet proteasome. J Biochem. 1993;114:317–23. doi: 10.1093/oxfordjournals.jbchem.a124174. [DOI] [PubMed] [Google Scholar]
- 31.Ostrowska H, Ostrowska JK, Worowski K, Radziwon P. Human platelet 20S proteasome: inhibition of its chymotrypsin-like activity and identification of the proteasome activator PA28. A preliminary report. Platelets. 2003;14:151–7. doi: 10.1080/0953710031000092802. [DOI] [PubMed] [Google Scholar]
- 32.Mizushima N, Komatsu M. Autophagy: renovation of cells and tissues. Cell. 2011;147:728–41. doi: 10.1016/j.cell.2011.10.026. [DOI] [PubMed] [Google Scholar]
- 33.Gao C, Cao W, Bao L, Zuo W, Xie G, Cai T, Fu W, Zhang J, Wu W, Zhang X, et al. Autophagy negatively regulates Wnt signalling by promoting Dishevelled degradation. Nat Cell Biol. 2010;12:781–90. doi: 10.1038/ncb2082. [DOI] [PubMed] [Google Scholar]
- 34.Sandilands E, Serrels B, McEwan DG, Morton JP, Macagno JP, McLeod K, Stevens C, Brunton VG, Langdon WY, Vidal M, et al. Autophagic targeting of Src promotes cancer cell survival following reduced FAK signalling. Nat Cell Biol. 2012;14:51–60. doi: 10.1038/ncb2386. [DOI] [PubMed] [Google Scholar]
- 35.Kadandale P, Stender JD, Glass CK, Kiger AA. Conserved role for autophagy in Rho1-mediated cortical remodeling and blood cell recruitment. Proc Natl Acad Sci U S A. 2010;107:10502–7. doi: 10.1073/pnas.0914168107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dangelmaier CA, Quinter PG, Jin J, Tsygankov AY, Kunapuli SP, Daniel JL. Rapid ubiquitination of Syk following GPVI activation in platelets. Blood. 2005;105:3918–24. doi: 10.1182/blood-2004-09-3689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ravetch JV, Lanier LL. Immune inhibitory receptors. Science. 2000;290:84–9. doi: 10.1126/science.290.5489.84. [DOI] [PubMed] [Google Scholar]
- 38.Sambrano GR, Weiss EJ, Zheng YW, Huang W, Coughlin SR. Role of thrombin signalling in platelets in haemostasis and thrombosis. Nature. 2001;413:74–8. doi: 10.1038/35092573. [DOI] [PubMed] [Google Scholar]
- 39.Liu W, Kato M, Akhand AA, Hayakawa A, Suzuki H, Miyata T, Kurokawa K, Hotta Y, Ishikawa N, Nakashima I. 4-hydroxynonenal induces a cellular redox status-related activation of the caspase cascade for apoptotic cell death. J Cell Sci. 2000;113:635–41. doi: 10.1242/jcs.113.4.635. [DOI] [PubMed] [Google Scholar]
- 40.Picker SM, Schneider V, Oustianskaia L, Gathof BS. Cell viability during platelet storage in correlation to cellular metabolism after different pathogen reduction technologies. Transfusion. 2009;49:2311–8. doi: 10.1111/j.1537-2995.2009.02316.x. [DOI] [PubMed] [Google Scholar]
- 41.Savage B, Saldívar E, Ruggeri ZM. Initiation of platelet adhesion by arrest onto fibrinogen or translocation on von Willebrand factor. Cell. 1996;84:289–97. doi: 10.1016/S0092-8674(00)80983-6. [DOI] [PubMed] [Google Scholar]
- 42.Gutierrez E, Petrich BG, Shattil SJ, Ginsberg MH, Groisman A, Kasirer-Friede A. Microfluidic devices for studies of shear-dependent platelet adhesion. Lab Chip. 2008;8:1486–95. doi: 10.1039/b804795b. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.