Abstract
We identified a member of the Arabidopsis NRT1/PTR FAMILY (NPF), AtNPF4.6, as an abscisic acid (ABA) transporter, AIT1. AtNPF4.6 was originally characterized as a low-affinity nitrate transporter NRT1.2. We hypothesized that the competition between nitrate and ABA as substrates for AtNPF4.6 might be involved in the interactions between nitrate and ABA signaling. However, the ABA transport activity of AtNPF4.6 was not inhibited by an excess amount of nitrate. In addition, the npf4.6 mutant was less sensitive to ABA than the wild type during germination irrespective of nitrate concentrations in the media. Furthermore, nitrate promoted germination of both wild type and npf4.6 in the presence of ABA. These results do not support the idea of a physiological linkage between nitrate and ABA signals through AtNPF4.6.
Keywords: abscisic acid, nitrate, NRT1/PTR FAMILY (NPF), plant hormone, transporter
Abscisic acid (ABA) is a plant hormone that plays crucial roles in many aspects of plant life cycles, including the regulation of seed dormancy and germination, as well as plant responses to abiotic and biotic stress.1,2 So far, a number of genes involved in the biosynthesis, catabolism, perception, and signal transduction of ABA have been identified.3-7 These studies demonstrated that endogenous ABA levels as well as sensitivities (or responsiveness) of plants to ABA are important for the physiological responses. Recently, 2 ATP-binding cassette (ABC)-type ABA transporters, AtABCG25 and AtABCG40, were identified in Arabidopsis.8,9 AtABCG25 mediates ABA export from cells whereas AtABCG40 mediates ABA import into cells. Mutants defective in the transporters showed altered responses to ABA in terms of seed germination and/or stomatal aperture, suggesting that ABA transport is an important element in the regulation of ABA-mediated physiological processes.
By using a modified yeast two-hybrid screening system with the ABA receptor PYR/PYL/RCAR and PP2C protein phosphatases, we identified the third ABA transporter [ABA-IMPORTING TRANSPORTER (AIT) 1], a member of the NRT1/PTR FAMILY (NPF), NPF4.6.10,11 Our study suggests that NPF4.6 functions as an ABA importer,11 although we cannot exclude the possibility that it is a bidirectional transporter as reported for some NPF proteins.12,13 Interestingly, AIT1 is identical to the previously characterized low-affinity nitrate transporter NRT1.2.14 Km values of AtNPF4.6 for nitrate and ABA were approximately 5.9mM and 5μM, respectively, suggesting that ABA is the more efficient substrate of the transporter. However, nitrate concentrations in soils and plants are much higher than endogenous ABA levels.15,16 Thus it is still possible that AtNPF4.6 functions as a transporter for both nitrate and ABA.
Interactions between nitrate and ABA signals have been suggested.17-19 It has been suggested that the interactions occur at the level of gene expression.20 However, as AtNPF4.6 transports both nitrate and ABA, it is also possible that the competition between nitrate and ABA as substrates for AtNPF4.6 is involved in the interactions of the 2 signals. An example for such an interaction is the auxin (indole-3-acetic acid; IAA) transport via AtNPF6.3/NRT1.1/CHL1 where competition between nitrate and IAA as substrates for the transporter affects lateral root development in Arabidopsis.21 To evaluate the possibility, we first tested whether nitrate affects the ABA transport activity of AtNPF4.6 (Fig. 1). We used a modified yeast two-hybrid system with the ABI1 PP2C protein phosphatase fused to the GAL4 activation domain (AD; AD-ABI1) and the PYR1 ABA receptor fused to the GAL4 binding domain (BD; BD-PYR1) to detect ABA transport activities of AtNPF4.6.11 If an ABA importer is expressed in the yeast cells, the interaction between AD-ABI1 and BD-PYR1 is induced at lower ABA concentrations in the medium compared with cells without transporters. ABA-dependent interactions between AD-ABI1 and BD-PYR1 were detected as the expression of the marker gene lacZ determined by a β-gal assay with o-nitrophenyl β-D-galactopyranoside as the substrate. Because AtNPF4.1 (AIT3) transports ABA and gibberellin [GA (GA3)],11 we performed a control experiment using ABA and GA3 as competitive substrates for AtNPF4.1 (Fig. 1A). As expected, β-gal activities in the cells expressing AtNPF4.1 in the presence of 10 nM ABA was inhibited by addition of 10 μM or higher concentrations of GA3, indicating that AtNPF4.1-mediated ABA uptake into the cells was inhibited competitively by GA3. On the other hand, consistent with our previous observation that AtNPF4.6 did not transport GA3,11 AtNPF4.6-mediated ABA uptake was not inhibited under the same conditions. These results demonstrated that this assay allowed the detection of the competition between ABA and other substrates. Next, we determined the effects of nitrate on ABA uptake through AtNPF4.6 (Fig. 1B). Because nitrate exists in soils and plant tissues at much higher concentrations than ABA,15,16 we added up to 100 mM nitrate against 10 nM of ABA as a competitor. However, we could not detect any inhibitory effect of nitrate on ABA transport activities of AtNPF4.6 (and AtNPF4.1).
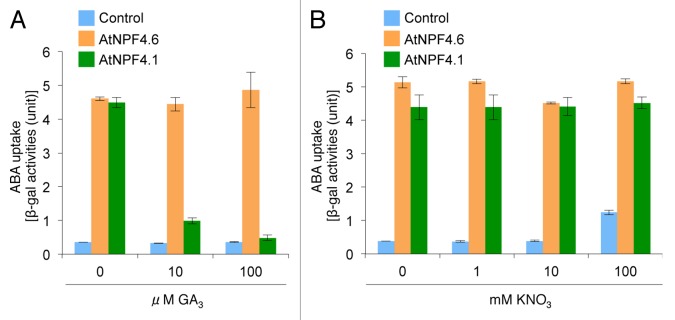
Figure 1. Effects of nitrate on ABA transport activities of AtNPF4.6. ABA transport activities of AtNPF4.6 and AtNPF4.1 were determined by a modified yeast two-hybrid system as described previously.11AtNPF4.6, AtNPF4.1, or the empty vector (Control) were introduced into yeast cells containing AD-ABI1 and BD-PYR1, and ABA-dependent interactions between AD-ABI1 and BD-PYR1 in the presence of 10 nM ABA were detected as the expression of lacZ as determined by a β-gal assay with o-nitrophenyl β-D-galactopyranoside as the substrate. (A) 0, 10, or 100 μM GA3 was added as a competitor of ABA. (B) 0, 1, 10, or 100 mM nitrate (KNO3) was added as a competitor of ABA. The assay was performed as described previously10 except that yeast cells were cultured in synthetic dextrose (SD) medium without Leu, Trp, and Ura, instead of yeast extract-peptone-dextrose (YPD) medium. The β-gal activities detected under these conditions were relatively high. Differences in the ABA transport activities of AtNPF4.6 and AtNPF4.1 were small, probably due to saturated ABA uptake or β-gal reactions. Values are means ± SD of 3 biological replicates.
We previously showed that the npf4.6 (ait1) mutant was less sensitive to exogenous ABA than the wild type during germination.11 On the other hand, AtNPF4.6 did not transport the ABA agonist pyrabactin, and the sensitivity of npf4.6 to pyrabactin was comparable to that of the wild type.11 These findings indicate that npf4.6 is defective in ABA transport rather than ABA signaling. However, it is unclear from these experiments whether nitrate competes with ABA as a substrate of AtNPF4.6. Other NPF members, together with AtNPF4.6, may be involved in the interactions between nitrate and ABA signals. We have shown that at least for members of NPF (AtNPF4.6/NRT1.2/AIT1, AtNPF4.5/AIT2, AtNPF4.1/AIT3, and AtNPF4.2/AIT4) exhibit ABA transport activity.11 However, among mutants defective in AtNPF4.6, AtNPF4.5, and AtNPF4.1, only npf4.6 showed an ABA-insensitive phenotype during germination. We were unable to obtain mutants defective in AtNPF4.2. However, based on the relatively low ABA transport activity of AtNPF4.2,11 its contribution to the physiological processes would be minor at best. Therefore, we investigated the effect of exogenously applied nitrate and ABA on seed germination of wild type and npf4.6 (Fig. 2). As expected from previous observations of nitrate-induced germination of dormant Arabidopsis seeds,22 nitrate promoted the germination and/or post-germination growth of non-dormant (after-ripened and stratified) wild-type seeds in the presence of 0.8 μM ABA, suggesting that nitrate negatively regulates the effect of ABA. If this phenomenon was caused by the inhibition of ABA transport through AtNPF4.6 by nitrate, npf4.6 should not respond to nitrate because AtNPF4.6 is not functional in the mutant irrespective of nitrate availability. However, germination of npf4.6 in the presence of ABA was promoted by nitrate similarly to that of the wild type (Fig. 2). Also, if AtNPF4.6 was involved in the nitrate-mediated promotion of seed germination in the presence of ABA, the ABA insensitive germination phenotype of npf4.6 should be masked at high nitrate concentrations. However, npf4.6 remained ABA-insensitive even in the presence of 10 mM nitrate, a saturating nitrate concentration with respect to the promotion of germination in the presence of ABA (Fig. 2). Thus, it appears that the ABA-insensitive phenotype of npf4.6 is independent of the function of AtNPF4.6 as a nitrate transporter. Taken together, our experiments do not support the idea that AtNPF4.6 mediates interactions between nitrate and ABA signals.
Figure 2. Germination of wild type and npf4.6 in the presence of nitrate and ABA. Seeds of wild type (Col-0 accession) and npf4.6 (ait1–1) were surface-sterilized in a solution containing 5% (vol/vol) NaClO and 0.05% (vol/vol) Tween 20, rinsed with water, and sown on 0.8% (w/vol) agar plate (pH 6.5) containing 0.5 × Murashige and Skoog salts, MES (0.5 g/l), ABA (0, 0.2, 0.5, or 0.8 μM), and nitrate (KNO3; 0, 1, or 10 mM). Stock solutions of ABA (1,000 × concentration) were prepared in DMSO and added to the media after autoclaving. Ten and 9 mM KCl was added to the media containing 0 and 1 mM KNO3, respectively, to maintain equal osmotic pressure among the treatments. After sowing, plates were incubated at 4 °C in the dark for 3 d. Germination rates were scored at 0, 1, 2, 3, and 4 d after transferring the plates to light conditions at 22 °C. Germination was determined as greening of cotyledons. Values are means ± SD of 3 biological replicates.
Glossary
Abbreviations:
- ABA
abscisic acid
- GA
gibberellin
- IAA
indole-3-acetic acid
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1.Zeevaart JAD, Creelman RA. Metabolism and physiology of abscisic acid. Annu Rev Plant Physiol Plant Mol Biol. 1988;39:439–73. doi: 10.1146/annurev.pp.39.060188.002255. [DOI] [Google Scholar]
- 2.Lee SC, Luan S. ABA signal transduction at the crossroad of biotic and abiotic stress responses. Plant Cell Environ. 2012;35:53–60. doi: 10.1111/j.1365-3040.2011.02426.x. [DOI] [PubMed] [Google Scholar]
- 3.Finkelstein RR, Rock CD. Abscisic Acid biosynthesis and response. Arabidopsis Book. 2002;1:e0058. doi: 10.1199/tab.0058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nambara E, Marion-Poll A. Abscisic acid biosynthesis and catabolism. Annu Rev Plant Biol. 2005;56:165–85. doi: 10.1146/annurev.arplant.56.032604.144046. [DOI] [PubMed] [Google Scholar]
- 5.Cutler SR, Rodriguez PL, Finkelstein RR, Abrams SR. Abscisic acid: emergence of a core signaling network. Annu Rev Plant Biol. 2010;61:651–79. doi: 10.1146/annurev-arplant-042809-112122. [DOI] [PubMed] [Google Scholar]
- 6.Hubbard KE. N. Nishimura N, Hitomi K, Getzoff ED, Schroeder JI. Early abscisic acid signal transduction mechanisms: newly discoverd components and newly emmmerging questions. Genes Dev. 2010;24:1695–1708. doi: 10.1101/gad.1953910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Seo M, Koshiba T. Transport of ABA from the site of biosynthesis to the site of action. J Plant Res. 2011;124:501–7. doi: 10.1007/s10265-011-0411-4. [DOI] [PubMed] [Google Scholar]
- 8.Kang J, Hwang JU, Lee M, Kim YY, Assmann SM, Martinoia E, Lee Y. PDR-type ABC transporter mediates cellular uptake of the phytohormone abscisic acid. Proc Natl Acad Sci U S A. 2010;107:2355–60. doi: 10.1073/pnas.0909222107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kuromori T, Miyaji T, Yabuuchi H, Shimizu H, Sugimoto E, Kamiya A, Moriyama Y, Shinozaki K. ABC transporter AtABCG25 is involved in abscisic acid transport and responses. Proc Natl Acad Sci U S A. 2010;107:2361–6. doi: 10.1073/pnas.0912516107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Léran S, Varala K, Boyer JC, Chiurazzi M, Crawford N, Daniel-Vedele F, David L, Dickstein R, Fernandez E, Forde B, et al. A unified nomenclature of NITRATE TRANSPORTER 1/PEPTIDE TRANSPORTER family members in plants. Trends Plant Sci. 2013 doi: 10.1016/j.tplants.2013.08.008. In press. [DOI] [PubMed] [Google Scholar]
- 11.Kanno Y, Hanada A, Chiba Y, Ichikawa T, Nakazawa M, Matsui M, Koshiba T, Kamiya Y, Seo M. Identification of an abscisic acid transporter by functional screening using the receptor complex as a sensor. Proc Natl Acad Sci U S A. 2012;109:9653–8. doi: 10.1073/pnas.1203567109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lin SH, Kuo HF, Canivenc G, Lin CS, Lepetit M, Hsu PK, Tillard P, Lin HL, Wang YY, Tsai CB, et al. Mutation of the Arabidopsis NRT1.5 nitrate transporter causes defective root-to-shoot nitrate transport. Plant Cell. 2008;20:2514–28. doi: 10.1105/tpc.108.060244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Léran S, Muños S, Brachet C, Tillard P, Gojon A, Lacombe B. The Arabidopsis NPF6.3/NRT1.1 is a bidirectional transporter involved in root-to-shoot nitrate translocation. Mol Plant. 2013 doi: 10.1093/mp/sst068. In press. [DOI] [PubMed] [Google Scholar]
- 14.Huang NC, Liu K-H, Lo HJ, Tsay YF. Cloning and functional characterization of an Arabidopsis nitrate transporter gene that encodes a constitutive component of low-affinity uptake. Plant Cell. 1999;11:1381–92. doi: 10.1105/tpc.11.8.1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Crawford NM, Glass ADM. Molecular and physiological aspects of nitrate uptake in plants. Trends Plant Sci. 1998;3:389–95. doi: 10.1016/S1360-1385(98)01311-9. [DOI] [Google Scholar]
- 16.Sauter A, Dietz KJ, Hartung W. A possible stress physiological role of abscisic acid conjugates in root-to-shoot signalling. Plant Cell Environ. 2002;25:223–8. doi: 10.1046/j.1365-3040.2002.00747.x. [DOI] [PubMed] [Google Scholar]
- 17.Rubio V, Bustos R, Irigoyen ML, Cardona-López X, Rojas-Triana M, Paz-Ares J. Plant hormones and nutrient signaling. Plant Mol Biol. 2009;69:361–73. doi: 10.1007/s11103-008-9380-y. [DOI] [PubMed] [Google Scholar]
- 18.Krouk G, Crawford NM, Coruzzi GM, Tsay YF. Nitrate signaling: adaptation to fluctuating environments. Curr Opin Plant Biol. 2010;13:266–73. doi: 10.1016/j.pbi.2009.12.003. [DOI] [PubMed] [Google Scholar]
- 19.Boursiac Y, Léran S, Corratgé-Faillie C, Gojon A, Krouk G, Lacombe B. ABA transport and transporters. Trends Plant Sci. 2013;18:325–33. doi: 10.1016/j.tplants.2013.01.007. [DOI] [PubMed] [Google Scholar]
- 20.Nero D, Krouk G, Tranchina D, Coruzzi GM. A system biology approach highlights a hormonal enhancer effect on regulation of genes in a nitrate responsive “biomodule”. BMC Syst Biol. 2009;3:59. doi: 10.1186/1752-0509-3-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Krouk G, Lacombe B, Bielach A, Perrine-Walker F, Malinska K, Mounier E, Hoyerova K, Tillard P, Leon S, Ljung K, et al. Nitrate-regulated auxin transport by NRT1.1 defines a mechanism for nutrient sensing in plants. Dev Cell. 2010;18:927–37. doi: 10.1016/j.devcel.2010.05.008. [DOI] [PubMed] [Google Scholar]
- 22.Alboresi A, Gestin C, Leydecker MT, Bedu M, Meyer C, Truong HN. Nitrate, a signal relieving seed dormancy in Arabidopsis. Plant Cell Environ. 2005;28:500–12. doi: 10.1111/j.1365-3040.2005.01292.x. [DOI] [PubMed] [Google Scholar]



