Abstract
J-proteins are co-chaperone components of the HSP70 system. J-proteins stimulate Hsp70ATPase activity, which is responsible for stabilizing the interaction of Hsp70 with client proteins. J-proteins are localized in various intracellular compartments including the cytoplasm, mitochondria and endoplasmic reticulum (ER). Five types of ER resident J-proteins (ERdjs) have been found in plants (P58, ERdj2, ERdj2A, ERdj3B and ERdj7). Rice OsERdj3A is located in the vacuole and protein storage vacuoles (PSV, PB-II) under conditions of ER stress. J-proteins that are localized to the vacuole or lysosome are not found in mammals and yeast, suggesting that the presence of OsERdj3A in the vacuole is plant-specific and one of the features unique to plant ERdjs. In this review, we summarize the current state of knowledge and recent research advancements regarding plant ERdjs, and compare mammalian and yeast ERdjs with plant ERdjs.
Keywords: BiP, endoplasmic reticulum, Hsp70, J-protein, rice seed, vacuole
Introduction
Protein folding often requires accessory machinery such as chaperones. The HSP70 molecular chaperone system consists of Hsp70 (DnaK), the co-chaperone Hsp40 (DnaJ), and a nucleotide exchange factor (NEF). Hsp70, the core chaperone of the HSP70 system, has a nucleotide-binding domain (NBD) and substrate-binding domain (SBD) (for a recent review, see1). ATP hydrolysis by the NBD is implicated in the regulation of substrate binding and dissociation; however, Hsp70 has very low ATPase activity, and Hsp40/DnaJ is required for stimulation of the ATPase activity of Hsp70. Hsp40/DnaJ, also termed a J-domain-containing protein (J-protein), contains a conserved J-domain with a signature length of approximately 70 amino acids that interacts with the NBD of Hsp70. During a cycle of ATP-dependent client-binding and release in the HSP70 system, the J-protein binds to an unfolded client protein, delivers it to Hsp70, and then stimulates the ATPase activity of Hsp70. After ATP hydrolysis on Hsp70, the J-protein dissociates from Hsp70 and NEF exchanges ADP for ATP, dissociating the client protein from Hsp70. J-proteins are involved in various cellular processes, including de novo protein folding, translocation of polypeptides across cellular membranes, and degradation of misfolded proteins.2 J-proteins are ubiquitously localized in mitochondria,2 endoplasmic reticulum (ER), and the cytosol. Mammals have 41 J-proteins, while the plant genomes of Arabidopsis and rice encode approximately 90 and 104 J-proteins, respectively.3,4 Six of the plant ERdjs are localized in the ER.5,6 The ER resident J-proteins are often termed ERj in mammalian systems; in this plant review, the ER J-proteins are denoted ERdj. The functions of most ERdjs in plants remain unclear; however, recent studies have revealed physiological roles for Arabidopsis ERdjs such as AtERdj2A, AtERdj3A and AtERdj3B.5,7,8 In addition, rice OsERdj3A is localized in vacuoles under ER stress conditions, indicating that plant ERdjs display distinct characteristics from those in mammals.6 This mini-review summarizes the current knowledge surrounding ERdjs in plants, as well as in mammals and yeast. In addition, plant ERdjs are compared with mammalian and yeast ERdjs to allow functional inference for plant ERdjs that are not yet fully characterized.
Gene Organization of ERdjs in Different Organisms
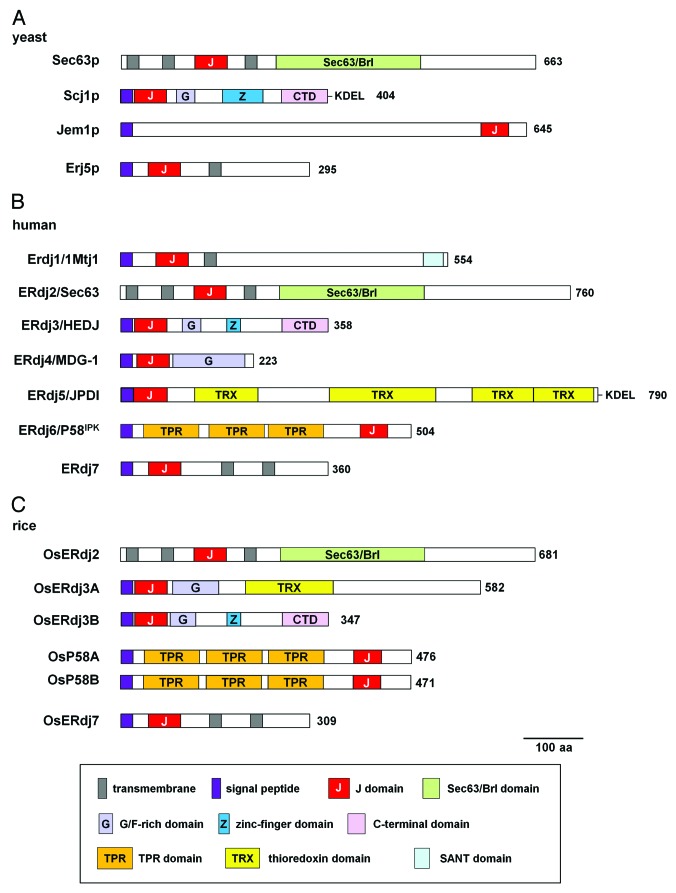
Gene organization of ERdjs varies between plants, animals, and yeast. Rice and Arabidopsis have six ERdjs: OsP58A, OsP58B, OsERdj2, OsERdj3A, OsERdj3B and OsERdj7 (Fig. 1); and AtP58IPK, AtERdj2A, AtERdj2B, AtERdj3A, AtERdj3B and AtERdj7 (At1g61770), respectively.5,6 Humans have seven ERdjs (ERdj1/Mtj1, ERdj2/Sec63, ERdj3/HEDJ, ERdj4/MDG-1, ERdj5/JPDI, ERdj6/P58IPK and ERdj7), and yeast has four ERdjs (Sec63p, Scj1p, Jem1p and Erj5p) (Fig. 1). Since Arabidopsis and rice ERdjs were designated according to their homology to mammalian ERdjs,5,6 plants have orthologs of mammalian ERdjs corresponding to ERdj2, ERdj3, ERdj6/P58IPK and ERdj7. While yeast nomenclature for ERdjs is not consistent with plant ERdj names, relationships are apparent between ERdjs from the different kingdoms.
Figure 1. Schematic representation of ER resident J-proteins in (A) yeast, (B) human and (C) rice. Domain structures of ER resident J-proteins from yeast, human and rice are shown. Arabidopsis ERdjs (AtP58IPK, AtERdj2A, AtERdj2B, AtERdj3A, AtERdj3B and AtERdj7) have the same domain organization of rice ERdjs, so rice ERdjs are shown as representatives of plant ERdjs. Human ERdjs (ERdj1/Mtj1, ERdj2/Sec63, ERdj3/HEDJ, ERdj4/MDG-1, ERdj5/JPDI, ERdj6/P58IPK and ERdj7) represent mammalian ERdjs. Gray, purple and red boxes indicate transmembrane regions, signal peptide sequences and J-domains, respectively.Auxiliary domains such as Sec63/Brl(light green), glycine and phenylalanine-rich (G/F-rich; light purple), zinc finger (ZF; blue), C-terminal domain (CTD; pink), tetratricopeptide repeat (TPR; orange), thioredoxin (TRX; yellow) and Swi3-Ada2-N-CoR-TFIIB (SANT; light blue) domains are indicated.
Although neither ArabidopsisAtERdj2A nor AtERdj2B were able to rescue the temperature-sensitive growth defect of the yeast sec63–1 mutant,5 the similarities of their domain structures suggests that plant ERdj2s are nevertheless likely to be orthologs of yeast Sec63p (Fig. 1). Plant ERdj3B shares domains with mammalian ERdj3 and yeast Scj1p such as a glycine/phenylalanine-rich (G/F-rich) domain, zinc finger domains, and a client protein-binding C-terminal domain.9 Furthermore, Arabidopsis AtERdj3B suppressed the growth defect of the yeast jem1/scj1 mutant in complementation experiments.5 Thus, plant ERdj3B is an ortholog of mammalian ERdj3 and yeast Scj1p. Mammalian and plant P58 have three copies of a tetratricopeptide repeat (TPR) domain, while this type of domain is absent from yeast Jem1p (Fig. 1). However, the growth defect of the yeast jem1/scj1 mutant was also suppressed by AtP58IPK, which is therefore proposed to be related to yeast Jem1p.5 Thus, ERdj2, ERdj3B and P58 are conserved between plants, yeast and mammals, respectively.
Both plants and mammals have ERdj7, but no corresponding gene is present in yeast. Alongside Sec63p, Jem1p and Scj1p, yeast has another ERdj: Erj5.10 Yeast Erj5 has a single transmembrane domain at the C-terminal region (Fig. 1), and loss of Erj5 impairs ER protein folding and leads to enhanced sensitivity to agents that cause ER stress in yeast.10 By contrast, mammalian ERdj7 and plant ERdj7 (AtERdj7 and OsErdj7) have two transmembrane domains at their C-terminal regions (Fig. 1). Expression of the yeast Erj5 is induced by ER stress,10 whereas expression of mammalian and plant ERdj7 are not affected by ER stress.5,6,11 Thus, mammalian and plant ERdj7 and yeast Erj5 may be distinct, and yeast Erj5 is likely to be a yeast-specific ERdj.
AtERdj3A can also suppress the growth defect of the yeast jem1/scj1 mutant, a proposed ortholog of yeast Scj1p.5 It is thus possible that plant ERdj3A is a paralog of ERdj3B, despite the lack of a zinc finger domain or CTD in ERdj3A (Fig. 1). However, orthologs of AtERdj3A are found only in plant genomes.5 Rice OsERdj3A localizes to the vacuole under ER-stressed conditions,6 but no mammalian or yeast ERdjs localize at the lysosome or vacuole. These observations suggest that OsERdj3A is a unique, plant-specific ERdj.
Mammals have several ERdjs not found in plants and yeast, including ERdj1, ERdj4 and ERdj5 (Fig. 1). ERdj1 is a type I transmembrane ERdj that associates with translating ribosomes through its cytosolic domain and inhibits further protein synthesis on ribosomes if BiP is not bound to its luminal J-domain.12 Therefore, ERdj1 serves as a checking system to ensure the presence of available BiP within the ER lumen for newly synthesized nascent polypeptides. ERdj4, which is required for degradation of misfolded proteins,13 is membrane-anchored via its uncleaved signal peptide, with the remaining protein located in the ER lumen.14,15 ERdj5 plays central roles in the degradation of misfolded proteins via its four thioredoxin-like domains (TRX) that contribute to reductase activity.16-18 As mentioned above, organization of ERdjs is different in mammals, yeast and plants (Fig. 1). Given this organizational difference, it remains unclear how plant ERdjs can substitute for the functions of mammalian ERdj1, ERdj4, and ERdj5. Further studies are thus necessary to understand the roles played by plant ERdjs in protein folding and in ER-associated degradation (ERAD) of unfolded and misfolded proteins.
ERdj genes are present in different copy numbers in Arabidopsis and rice. Arabidopsis has two copies of ERdj2 (AtERdj2A and AtERdj2B),5 while rice has only one copy, OsERdj2.6 Rice has two P58 gene copies (OsP58A and OsP58B),6 while Arabidopsis has only one copy.5 Expression of AtERdj2B is induced by ER stress, but AtERdj2A is constitutively expressed,5 rice OsP58A and OsP58B are also differentially impacted by ER stress; OsP58A is constitutively expressed while OsP58B is induced by ER stress.6 Thus, the duplicated genes encoding ERdjs might play distinct roles in different plants.
Possible Roles of ER Resident J-Proteins in Plants
In this section, we review the current state of knowledge regarding plant ERdjs. As there is minimal experimental evidence to support their functions, we speculate upon the possible roles of plant ERdjs based on the discoveries from their yeast and mammalian ERdj counterparts.
P58
Mammalian ERdj6/P58IPK was originally found as a cytosolic protein that is negatively regulated by phosphorylation of an ER stress sensor, PERK.19 P58IPK was later discovered to also be an ER resident protein that binds to unfolded proteins and acts as a co-chaperone for BiP.20,21 Although P58IPK binds to several unfolded proteins, modulation of P58IPK level did not affect degradation of the unfolded proteins.22 Binding of P58IPK to unfolded proteins is BiP-independent, and BiP enhances release of P58IPK from the unfolded proteins, implicating that P58IPK may reduce the burden of unfolded proteins by upregulating protein folding in the ER lumen.21
In plants, P58IPK was first identified as an interactor with a virus-encoded ligand, TMV-P50.23 P58IPK silencing in tobacco and knockout of AtP58IPKin Arabidopsis led to cell death upon infection with plant viruses, suggesting that plant P58IPK has similar cytoplasmic functions to mammalian P58IPK.23 However, Arabidopsis and rice P58s have been found localized in the ER, indicating that they are ER resident J-proteins.5,6 In addition, AtP58 and OsP58A&B, as well as OsBiP1, are detected under normal, non-stress conditions.5,6 OsP58A and OsP58B bind preferentially to OsBiP1 rather than other OsBiPs in rice protoplasts.6 Thus, together with OsBiP1, OsP58A and OsP58B may be involved in folding nascent polypeptides in the ER lumen.
ERdj2
Yeast Sec63p is involved in protein translocation across the ER by recruiting Kar2 (BiP), and thus providing a driving force for translocation.24,25 Sec63p, a transmembrane protein with a luminal J-domain and a cytosolic Sec63/Brl domain, is required for assembly of functional ER translocons.26 The J-domain of Sec63p, and not the Sec63/Brl domain, was recently shown to be required for ERAD of soluble proteins in yeast.27 Domain structures are similar between human Erdj2 and yeast Sec63p (Fig. 1); however, there is little evidence to support a role for protein translocation into the ER for mammalian ERdj2. Mutations in mammalian ERdj2 are linked to polycystic liver disease, an autosomal-dominant disorder characterized by the presence of multiple fluid filled cysts in the liver. This suggests that ERdj2 may contribute to cotranslational processing of proteins involved in biliary cell growth.28
Plant ERdj2s also have similar domain structures to the yeast and mammalian counterparts (Fig. 1). However, expression of Arabidopsis AtERdj2A and AtERdj2B did not suppress temperature-sensitive growth defects of the yeast sec63–1 mutant.5 It remains to be elucidated whether plant ERdj2 is a functional ortholog of yeast Sec63p. Insertional mutations of AtERdj2A, but not AtERdj2B, were lethal in Arabidopsis, and such aterdj2A mutants showed defects in pollen germination, implicating AtERdj2A involvement in protein secretion.5
Rice seed storage proteins are deposited into two different compartments. Protein body I (PB-I) contains several kinds of prolamins and is derived from ER, and protein body II (PB-II) is a storage vacuole that contains globulin and gluteilins.29,30 OsERdj2 accumulates gradually during endosperm development and is localized to the periphery of PB-I.6 OsBiP1 accumulates in a similar manner and is highly enriched at the periphery of PB-I through association with nascent prolamin polypeptide chains formed during translation; this indicates colocalization of OsERdj2 and OsBiP1.6,31,32 OsERdj2 may be involved in translocation of prolamin polypeptides during PB-Iformation. This may be similar to the role of Sec63p in yeast, which, together with Kar2 (BiP), plays crucial roles in protein import into the ER.
ERdj3B
Mammalian ERdj3/HEDJ and plant ERdj3B have a N-terminal domain, J-domain, G/F-rich domain, and an N-terminal zinc finger domain and C-terminal domain, indicating similar domain organization to yeast Scj1p (Fig. 1). Although the zinc finger domain is present in all these proteins, the types of zinc finger motifs differ: yeast Scj1p has four copies of the CXXCXGXG motif, whereas mammalian ERdj3 and plant ERdj3B have the CXC-X21-CXXC motif that is also found in yeast Ptr3p. Another difference is that yeast Scj1p uses a C-terminal KDEL sequence as the ER retention signal but this is absent in mammalian ERdj3 and plant ERdj3B. Mammalian ERdj3 forms a complex with unfolded proteins and multiple chaperones including BiP, PDI and GPR94.33 Complex formation is disrupted by treatment with a protein synthesis inhibitor, cycloheximide.33 ERdj3 associates directly with nascent unfolded proteins in an Hsp70-independent manner, then recruits Hsp70/BiP, and finally dissociates from the nascent unfolded proteins.14,34 These results indicate that ERdj3 serves to inhibit protein aggregation until BiP joins the complex.14 Recently, it was reported that ERdj3 is associated with the Sec61 translocon.35,36 Binding of ERdj3 to the Sec61 translocon may provide an elegant mechanism by which ERdj3 efficiently encounters newly synthesized peptides as they move through the Sec61 translocon. ERdj3 is fairly abundant in mammalian cell culture, at levels almost comparable to those of the Sec61 translocon.37 This would allow ERdj3 to survey aggregation-prone regions of newly synthesized peptides and bind to them to prevent their aggregation.
In plants, AtERdj3B forms a complex with BiP as well as stromal-derived factor 2 (SDF2).7 SDF2 is found in the ER and is required for the function of EFR, a pattern-recognition receptor involved in plant immunity.7 Aterdj3b mutants are susceptible to bacterial pathogens.7 The loss of SDF2 results in retention of the EFR receptor in the ER and then EFR degradation. These observations suggest that the AtERdj3B-BiP-SDF2 complex mediates proper accumulation of EFR at the plasma membrane and that complex-associated AtERdj3B and BiP serve to chaperone Hsp70.7 AtERdj3B is detected in seedlings during normal development,5 indicating an ongoing role for AtERdj3B. In rice, OsERdj3B binds preferentially to constitutive OsBiP1 rather than to ER stress inducible OsBiPs such as OsBiP4 and OsBiP5.6These suggest that, like mammalian ERdj3, plant ERdj3B may assist folding of nascent proteins under normal, non-stress, conditions. Yeast Scj1p is the most abundant of the five yeast ERdjs.38 While yeast scj1 mutants are viable, they grow slowly and exhibit constitutive ER stress.39 Furthermore, yeast Scj1 and BiP/Kar2p have been found in association with ERAD substrates.39 Because AtERdj3B and OsERdj3B are also induced by ER stress,5,6 it is possible that AtERdj3B and OsERdj3B also participate in protein degradation alongside their protein folding roles.
ERdj7
Mammalian ERdj7/DNAJC25 is a recently identified ERdj that has not yet been characterized in detail. ERdj7 is an abundant membrane protein that was observed as a subunit of a translocon, Sec61α, in canine pancreas microsomes.11 Canine ERdj7 has been shown to stimulate the ATPase activity of BiP in vitro, demonstrating a capacity to serve as a co-chaperone for BiP. Overexpression of human ERdj7 suppresses tumor growth; however, the molecular mechanisms of this suppression remain to be determined.40 The role of ERdj7 in plants is unclear, and further studies are necessary for functional elucidation.
Is ERdj3A a plant-specific ER resident protein?
It was recently demonstrated that rice OsERdj3A is induced by ER stress and is localized in protoplast vacuoles and vacuole-derived PB-II in seeds.6 No mammalian or yeast J-proteins have been identified that localize to the lysosome (mammals) or vacuole (yeast). Thus, plant ERdj3A is a unique ER resident J-protein and may have plant-specific functions. In this section, we describe the current state of knowledge for plant ERdj3A and its possible roles.
Plant ERdj3A structure is similar to ERdj3B, but ERdj3A lacks zinc finger and CTD domains (Fig. 1). Instead, plant ERdj3A has a domain at its C-terminal region that resembles a TRX domain (Fig. 1). Arabidopsis AtERdj3A has a second TRX domain that is similar to the C-terminal redox inactive domain in mammalian P5 PDI,8 but this region is not required for function and is not conserved between Arabidopsis and rice ERdj3A.6,8 Since mammalian ERdj5 has four TRX domains at the C terminus, it is possible that plant ERdj3As could be orthologous to mammalian ERdj5.16,17 Mammalian ERdj5 is involved in the reduction of BiP substrates and enhancement of their ability to be unfolded for retrotranslocation into cytoplasm. The reductase activity is dependent on cysteine residues in CXXC motifs found within the mammalian ERdj5 TRX domains.18 The TRX CXXC sequence motif is not present in either the Arabidopsis or the rice ERdj3A proteins, but not all PDI have CXXC sequences in their active sites.41 For example, PDILT (protein disulphide isomerase-like protein of the testis) has an incomplete active site that lacks the N-terminal cysteine residue, yet is able to form mixed disulfides with partners and substrates in vivo.42 Despite lacking the CXXC sequence motif, Arabidopsis AtERdj3A protein expressed in Escherichia coli cells retains reductase activity in vitro.8 AtERdj3A and OsERdj3A have other cysteines in their TRX domains, and it thus remains possible that plant ERdj3As may perform similar functions to mammalian ERdj5.
A dissociation (Ds) insertion mutant, Arabidopsis thermosensitive male 1 (tms1), has a male-sterile phenotype at higher temperature (30 °C), in which pollen growth is specifically reduced at higher temperature. Molecular cloning of the TMS1 gene revealed that it encodes AtERdj3A.8 Although aterdj2a mutants also have defective pollen growth,5 phenotypic differences are apparent between tsm1/aterdj3a and aterdj2a mutants. Pollen growth is severely reduced, even at normal temperature, in the aterdj2a mutant, which is therefore homozygous lethal.5 By contrast, pollen growth in the tsm1/aterdj3a plant is unaffected at normal temperatures.8 Since protein secretion defects often lead to inhibition of pollen germination and tube elongation, these results suggest that AtERdj2A and AtERdj3A are involved in protein secretion during pollen germination. The relative mutant severities suggest that AtERdj2A may have housekeeping roles in protein secretion in pollen, but that AtERdj3A may have a conditional role, becoming involved only when protein folding is severely damaged by higher temperatures. This temperature-sensitive role for AtERdj3A suggests the possibility that it is involved in refolding or degradation of misfolded proteins caused by higher temperatures. Consistent with this notion, expression of AtERdj3A and rice OsERdj3A is induced by ER stress conditions caused by heat, tunicamycin and DTT treatments.5,6,8 Furthermore, co-immunoprecipitation assays revealed that OsERdj3A preferentially binds to OsBiP5, which is specifically induced by ER-stressed conditions.6 Thus, plant ERdj3A is likely to have roles in the response to ER stress. OsERdj3A is localized in vacuoles under ER-stressed conditions,6 suggesting that plant ERdj3A may form a link between the ER and vacuoles, facilitating delivery of waste unfolded proteins. It was recently shown that ER stress induces ERAD by autophagy and that soluble ER marker proteins accumulate in the vacuole via autophagy in response to ER stress,43 suggesting that there is a disposal route for unfolded proteins produced under ER-stressed conditions. In mammalian cytoplasm, selective autophagy is mediated by several chaperones, including Hsc70 and BAG3, and is termed Chaperone-Assisted Selective autophagy (CASA).44 In a similar manner to this cytoplasmic CASA, OsERdj3A may mediate the selection of unfolded proteins prior to the formation of autophagosomes in the ER. However, the molecular mechanisms guiding elimination of aggregated ER proteins by autophagy are unclear, even in mammals. Further characterization of OsERdj3A may shed light on the ER protein quality control system, including the mechanisms leading to autophagy and vacuole disposal.
Perspectives
In contrast to mammalian systems, there is a paucity of information on the function of ERdjs in plants. Only a few proteins such as Arabidopsis AtERdj2A, AtERdj3A and AtERdj3B have been characterized, revealing their roles in protein secretion.5,7,8 Further studies are needed to fully understand plant ERdj functions. Of key importance is the identification of proteins targeted by the ER resident J-proteins. One useful strategy is proteomic analysis of mutant or RNAi-suppressed strains with diminished or altered ER resident J-proteins. Seed endosperm-specific RNAi suppression of ERdjs in rice is particularly promising for proteomic analysis because seed-specific RNAi suppression does not affect plant-wide physiological processes, avoiding lethality. In addition, rice seeds are relatively large in size and contain fewer protein types than other tissues, facilitating proteomic analysis. Rice endosperm produces a large amount of seed storage proteins as secretory proteins, allowing the effect of OsERdjs on protein secretion to be tested through endosperm-specific suppression.
Mammalian ERdj5, which has four thioredoxin-like domains and acts as a reductase to cleave disulphide bonds in unfolded proteins, has no ortholog in plants or yeast.17,18 Thus, it is unclear how plant cells handle unfolded proteins with disulphide bonds, as it is necessary to reduce these before the protein is dislocated back to the cytoplasm. In yeast, protein disulphide isomerase 1 (Pdi1) is necessary for the export of a mutant form of vacuolar carboxypeptidase Y (CPY*), which contains disulphide bonds and is a typically soluble ERAD substrate in yeast. This suggests that Pdi1 mediates reduction of disulphide in yeast in the absence of a mammalian ERdj5 ortholog.45 In plants, PDI may perform the disulphide bond reduction role for ERAD substrates. Alternatively, plant ERdj3A, which has a TRX domain, may act as a reductase in a similar manner to ERdj5. Further studies are required to understand how plant ERdj3A and/or other factors are involved in disulphide bond reduction of ERAD substrates.
The ER is a large factory that produces a range of secretory proteins, some of which are involved in plant immunity. Recent studies showed that components of the ER resident Hsp70 system such as AtBiP2 and AtERdj3B have important roles in plant immunity.7,46 Furthermore, the ER can be utilized for the production of foreign recombinant proteins with therapeutic or vaccine activities, particularly in rice seeds.47,48 Thus, further understanding of plant ER resident J-proteins will allow improvement of plant traits such as resistance against pathogens and efficient production of foreign recombinant proteins.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgment
This work was supported by Grants-in-Aid for Scientific Research to F.T. (# 25292220) from the Ministry of Education, Culture, Sports, Science, and Technology of Japan.
Glossary
Abbreviations:
- BiP
immunoglobulin binding protein
- ER
endoplasmic reticulum
- ERAD
ER-associated degradation
- Hsp
heat shock protein
References
- 1.Mayer MP. Hsp70 chaperone dynamics and molecular mechanism. Trends Biochem Sci. 2013;38:507–14. doi: 10.1016/j.tibs.2013.08.001. [DOI] [PubMed] [Google Scholar]
- 2.Kampinga HH, Craig EA. The HSP70 chaperone machinery: J proteins as drivers of functional specificity. Nat Rev Mol Cell Biol. 2010;11:579–92. doi: 10.1038/nrm2941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Miernyk JA. The J-domain proteins of Arabidopsis thaliana: an unexpectedly large and diverse family of chaperones. Cell Stress Chaperones. 2001;6:209–18. doi: 10.1379/1466-1268(2001)006<0209:TJDPOA>2.0.CO;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sarkar NK, Thapar U, Kundnani P, Panwar P, Grover A. Functional relevance of J-protein family of rice (Oryza sativa) Cell Stress Chaperones. 2013;18:321–31. doi: 10.1007/s12192-012-0384-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yamamoto M, Maruyama D, Endo T, Nishikawa S. Arabidopsis thaliana has a set of J proteins in the endoplasmic reticulum that are conserved from yeast to animals and plants. Plant Cell Physiol. 2008;49:1547–62. doi: 10.1093/pcp/pcn119. [DOI] [PubMed] [Google Scholar]
- 6.Ohta M, Wakasa Y, Takahashi H, Hayashi S, Kudo K, Takaiwa F. Analysis of rice ER-resident J-proteins reveals diversity and functional differentiation of the ER-resident Hsp70 system in plants. J Exp Bot. 2013;64:5429–41. doi: 10.1093/jxb/ert312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nekrasov V, Li J, Batoux M, Roux M, Chu ZH, Lacombe S, Rougon A, Bittel P, Kiss-Papp M, Chinchilla D, et al. Control of the pattern-recognition receptor EFR by an ER protein complex in plant immunity. EMBO J. 2009;28:3428–38. doi: 10.1038/emboj.2009.262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang KZ, Xia C, Liu XL, Dou XY, Wang W, Chen LQ, Zhang XQ, Xie LF, He L, Ma X, et al. A mutation in Thermosensitive Male Sterile 1, encoding a heat shock protein with DnaJ and PDI domains, leads to thermosensitive gametophytic male sterility in Arabidopsis. Plant J. 2009;57:870–82. doi: 10.1111/j.1365-313X.2008.03732.x. [DOI] [PubMed] [Google Scholar]
- 9.Goffin L, Georgopoulos C. Genetic and biochemical characterization of mutations affecting the carboxy-terminal domain of the Escherichia coli molecular chaperone DnaJ. Mol Microbiol. 1998;30:329–40. doi: 10.1046/j.1365-2958.1998.01067.x. [DOI] [PubMed] [Google Scholar]
- 10.Carla Famá M, Raden D, Zacchi N, Lemos DR, Robinson AS, Silberstein S. The Saccharomyces cerevisiae YFR041C/ERJ5 gene encoding a type I membrane protein with a J domain is required to preserve the folding capacity of the endoplasmic reticulum. Biochim Biophys Acta 2006; 1773: 232-42. [DOI] [PMC free article] [PubMed]
- 11.Zahedi RP, Völzing C, Schmitt A, Frien M, Jung M, Dudek J, Wortelkamp S, Sickmann A, Zimmermann R. Analysis of the membrane proteome of canine pancreatic rough microsomes identifies a novel Hsp40, termed ERj7. Proteomics. 2009;9:3463–73. doi: 10.1002/pmic.200800722. [DOI] [PubMed] [Google Scholar]
- 12.Dudek J, Greiner M, Müller A, Hendershot LM, Kopsch K, Nastainczyk W, Zimmermann R. ERj1p has a basic role in protein biogenesis at the endoplasmic reticulum. Nat Struct Mol Biol. 2005;12:1008–14. doi: 10.1038/nsmb1007. [DOI] [PubMed] [Google Scholar]
- 13.Dong M, Bridges JP, Apsley K, Xu Y, Weaver TE. ERdj4 and ERdj5 are required for endoplasmic reticulum-associated protein degradation of misfolded surfactant protein C. Mol Biol Cell. 2008;19:2620–30. doi: 10.1091/mbc.E07-07-0674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shen Y, Hendershot LM. ERdj3, a stress-inducible endoplasmic reticulum DnaJ homologue, serves as a cofactor for BiP’s interactions with unfolded substrates. Mol Biol Cell. 2005;16:40–50. doi: 10.1091/mbc.E04-05-0434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kurisu J, Honma A, Miyajima H, Kondo S, Okumura M, Imaizumi K. MDG1/ERdj4, an ER-resident DnaJ family member, suppresses cell death induced by ER stress. Genes Cells. 2003;8:189–202. doi: 10.1046/j.1365-2443.2003.00625.x. [DOI] [PubMed] [Google Scholar]
- 16.Cunnea PM, Miranda-Vizuete A, Bertoli G, Simmen T, Damdimopoulos AE, Hermann S, Leinonen S, Huikko MP, Gustafsson JA, Sitia R, et al. ERdj5, an endoplasmic reticulum (ER)-resident protein containing DnaJ and thioredoxin domains, is expressed in secretory cells or following ER stress. J Biol Chem. 2003;278:1059–66. doi: 10.1074/jbc.M206995200. [DOI] [PubMed] [Google Scholar]
- 17.Hosoda A, Kimata Y, Tsuru A, Kohno K. JPDI, a novel endoplasmic reticulum-resident protein containing both a BiP-interacting J-domain and thioredoxin-like motifs. J Biol Chem. 2003;278:2669–76. doi: 10.1074/jbc.M208346200. [DOI] [PubMed] [Google Scholar]
- 18.Ushioda R, Hoseki J, Araki K, Jansen G, Thomas DY, Nagata K. ERdj5 is required as a disulfide reductase for degradation of misfolded proteins in the ER. Science. 2008;321:569–72. doi: 10.1126/science.1159293. [DOI] [PubMed] [Google Scholar]
- 19.Yan W, Frank CL, Korth MJ, Sopher BL, Novoa I, Ron D, Katze MG. Control of PERK eIF2alpha kinase activity by the endoplasmic reticulum stress-induced molecular chaperone P58IPK. Proc Natl Acad Sci U S A. 2002;99:15920–5. doi: 10.1073/pnas.252341799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rutkowski DT, Kang SW, Goodman AG, Garrison JL, Taunton J, Katze MG, Kaufman RJ, Hegde RS. The role of p58IPK in protecting the stressed endoplasmic reticulum. Mol Biol Cell. 2007;18:3681–91. doi: 10.1091/mbc.E07-03-0272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Petrova K, Oyadomari S, Hendershot LM, Ron D. Regulated association of misfolded endoplasmic reticulum lumenal proteins with P58/DNAJc3. EMBO J. 2008;27:2862–72. doi: 10.1038/emboj.2008.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oyadomari S, Yun C, Fisher EA, Kreglinger N, Kreibich G, Oyadomari M, Harding HP, Goodman AG, Harant H, Garrison JL, et al. Cotranslocational degradation protects the stressed endoplasmic reticulum from protein overload. Cell. 2006;126:727–39. doi: 10.1016/j.cell.2006.06.051. [DOI] [PubMed] [Google Scholar]
- 23.Bilgin DD, Liu Y, Schiff M, Dinesh-Kumar SP. P58(IPK), a plant ortholog of double-stranded RNA-dependent protein kinase PKR inhibitor, functions in viral pathogenesis. Dev Cell. 2003;4:651–61. doi: 10.1016/S1534-5807(03)00125-4. [DOI] [PubMed] [Google Scholar]
- 24.Brodsky JL, Goeckeler J, Schekman R. BiP and Sec63p are required for both co- and posttranslational protein translocation into the yeast endoplasmic reticulum. Proc Natl Acad Sci U S A. 1995;92:9643–6. doi: 10.1073/pnas.92.21.9643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Matlack KE, Misselwitz B, Plath K, Rapoport TA. BiP acts as a molecular ratchet during posttranslational transport of prepro-alpha factor across the ER membrane. Cell. 1999;97:553–64. doi: 10.1016/S0092-8674(00)80767-9. [DOI] [PubMed] [Google Scholar]
- 26.Jermy AJ, Willer M, Davis E, Wilkinson BM, Stirling CJ. The Brl domain in Sec63p is required for assembly of functional endoplasmic reticulum translocons. J Biol Chem. 2006;281:7899–906. doi: 10.1074/jbc.M511402200. [DOI] [PubMed] [Google Scholar]
- 27.Servas C, Römisch K. The Sec63p J-domain is required for ERAD of soluble proteins in yeast. PLoS One. 2013;8:e82058. doi: 10.1371/journal.pone.0082058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zimmermann R, Müller L, Wullich B. Protein transport into the endoplasmic reticulum: mechanisms and pathologies. Trends Mol Med. 2006;12:567–73. doi: 10.1016/j.molmed.2006.10.004. [DOI] [PubMed] [Google Scholar]
- 29.Tanaka K, Sugimoto T, Ogawa M, Kasai Z. Isolation and characterization of two types of protein bodies in rice endosperm. Agric Biol Chem. 1980;44:1633–9. doi: 10.1271/bbb1961.44.1633. [DOI] [Google Scholar]
- 30.Krishnan HB, Franceschi VR, Okita TW. Immunochemical studies on the role of the Golgi complex in protein-body formation in rice seeds. Planta. 1986;169:471–80. doi: 10.1007/BF00392095. [DOI] [PubMed] [Google Scholar]
- 31.Li X, Wu Y, Zhang DZ, Gillikin JW, Boston RS, Franceschi VR, Okita TW. Rice prolamine protein body biogenesis: a BiP-mediated process. Science. 1993;262:1054–6. doi: 10.1126/science.8235623. [DOI] [PubMed] [Google Scholar]
- 32.Muench DG, Wu Y, Zhang Y, Li X, Boston RS, Okita TW. Molecular cloning, expression and subcellular localization of a BiP homolog from rice endosperm tissue. Plant Cell Physiol. 1997;38:404–12. doi: 10.1093/oxfordjournals.pcp.a029183. [DOI] [PubMed] [Google Scholar]
- 33.Meunier L, Usherwood YK, Chung KT, Hendershot LM. A subset of chaperones and folding enzymes form multiprotein complexes in endoplasmic reticulum to bind nascent proteins. Mol Biol Cell. 2002;13:4456–69. doi: 10.1091/mbc.E02-05-0311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jin Y, Awad W, Petrova K, Hendershot LM. Regulated release of ERdj3 from unfolded proteins by BiP. EMBO J. 2008;27:2873–82. doi: 10.1038/emboj.2008.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dejgaard K, Theberge JF, Heath-Engel H, Chevet E, Tremblay ML, Thomas DY. Organization of the Sec61 translocon, studied by high resolution native electrophoresis. J Proteome Res. 2010;9:1763–71. doi: 10.1021/pr900900x. [DOI] [PubMed] [Google Scholar]
- 36.Guo F, Snapp EL. ERdj3 regulates BiP occupancy in living cells. J Cell Sci. 2013;126:1429–39. doi: 10.1242/jcs.118182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schwanhäusser B, Busse D, Li N, Dittmar G, Schuchhardt J, Wolf J, Chen W, Selbach M. Global quantification of mammalian gene expression control. Nature. 2011;473:337–42. doi: 10.1038/nature10098. [DOI] [PubMed] [Google Scholar]
- 38.Jonikas MC, Collins SR, Denic V, Oh E, Quan EM, Schmid V, Weibezahn J, Schwappach B, Walter P, Weissman JS, et al. Comprehensive characterization of genes required for protein folding in the endoplasmic reticulum. Science. 2009;323:1693–7. doi: 10.1126/science.1167983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nishikawa SI, Fewell SW, Kato Y, Brodsky JL, Endo T. Molecular chaperones in the yeast endoplasmic reticulum maintain the solubility of proteins for retrotranslocation and degradation. J Cell Biol. 2001;153:1061–70. doi: 10.1083/jcb.153.5.1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu T, Jiang W, Han D, Yu L. DNAJC25 is downregulated in hepatocellular carcinoma and is a novel tumor suppressor gene. Oncol Lett. 2012;4:1274–80. doi: 10.3892/ol.2012.903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Appenzeller-Herzog C, Ellgaard L. The human PDI family: versatility packed into a single fold. Biochim Biophys Acta 2008; 1783:535-48. [DOI] [PubMed]
- 42.van Lith M, Hartigan N, Hatch J, Benham AM. PDILT, a divergent testis-specific protein disulfide isomerase with a non-classical SXXC motif that engages in disulfide-dependent interactions in the endoplasmic reticulum. J Biol Chem. 2005;280:1376–83. doi: 10.1074/jbc.M408651200. [DOI] [PubMed] [Google Scholar]
- 43.Liu Y, Burgos JS, Deng Y, Srivastava R, Howell SH, Bassham DC. Degradation of the endoplasmic reticulum by autophagy during endoplasmic reticulum stress in Arabidopsis. Plant Cell. 2012;24:4635–51. doi: 10.1105/tpc.112.101535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Arndt V, Dick N, Tawo R, Dreiseidler M, Wenzel D, Hesse M, Fürst DO, Saftig P, Saint R, Fleischmann BK, et al. Chaperone-assisted selective autophagy is essential for muscle maintenance. Curr Biol. 2010;20:143–8. doi: 10.1016/j.cub.2009.11.022. [DOI] [PubMed] [Google Scholar]
- 45.Gillece P, Luz JM, Lennarz WJ, de La Cruz FJ, Römisch K. Export of a cysteine-free misfolded secretory protein from the endoplasmic reticulum for degradation requires interaction with protein disulfide isomerase. J Cell Biol. 1999;147:1443–56. doi: 10.1083/jcb.147.7.1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang D, Weaver ND, Kesarwani M, Dong X. Induction of protein secretory pathway is required for systemic acquired resistance. Science. 2005;308:1036–40. doi: 10.1126/science.1108791. [DOI] [PubMed] [Google Scholar]
- 47.Takaiwa F, Takagi H, Hirose S, Wakasa Y. Endosperm tissue is good production platform for artificial recombinant proteins in transgenic rice. Plant Biotechnol J. 2007;5:84–92. doi: 10.1111/j.1467-7652.2006.00220.x. [DOI] [PubMed] [Google Scholar]
- 48.Takaiwa F. Update on the use of transgenic rice seeds in oral immunotherapy. Immunotherapy. 2013;5:301–12. doi: 10.2217/imt.13.4. [DOI] [PubMed] [Google Scholar]