Abstract
Helicases are molecular motor proteins that perform a variety of cellular functions including transcription, translation, DNA replication and repair, RNA maturation, ribosome synthesis, nuclear export and splicing processes. The p68 is an evolutionarily conserved protein which plays pivotal roles in all aspect RNA metabolism processes. It is well established that helicases provides abiotic stress adaptation in plants but analysis of cis-regulatory elements present in the upstream regions is still infancy. Here we report isolation and functional characterization of the promoter of a DEAD-box helicase Psp68 in response to abiotic stress and hormonal regulation. The promoter of Psp68 was isolated by gene walking PCR from pea genomic DNA library constructed in BD genome walker kit. In silico analysis revealed that promoter of Psp68 contained a TATA, a CAAT motif and also harbors some important stress and hormone associated cis regulatory elements, including E-box, AGAAA, GATA-box, ACGT, GAAAA and GTCTC. Functional analyses were performed by Agrobacterium-mediated transient assay in tobacco leaves. Very high level of GUS activity was observed in agroinfiltrated tobacco leaves by the construct carrying the Psp68 promoter::GUS, subjected to abiotic stress and exogenous hormonal treatments. Stress-inducible nature of Psp68 promoter opens possibility for the study of the gene regulation under stress condition. Therefore, may be useful in the field of agriculture and biotechnology.
Keywords: Agro-infiltration, Psp68, cis-regulatory elements, helicase, promoter, transient assay
Introduction
p68, a member of DEAD-box helicase family is highly conserved in eukaryotes and involved in almost all RNA metabolic processes. Recently, a number of studies showed that helicases are not only involved in many cellular processes including plant growth and development1-7 but also provides stress tolerance in transgenic plants.3,7-13 The appropriate regulation of gene expression is important for all cellular processes, in which transcriptional control is primarily concerned with improved survival. Mostly, genes are expressed in transgenic plants under the control of promoter, a DNA sequences required for appropriate spatial and temporal expression pattern. The most widely used promoter for expression of transgenes is CaMV 35S promoter (cauliflower mosaic virus) but sometime it may causes some undesirable effect in plants such as gene silencing, delayed growth, dwarfism and low yield.14-18
So, inducible and tissue-specific promoters are required to study the gene regulatory networks in plant.19,20 Cis-acting regulatory elements present in promoter sequence may function as molecular switch by controlling transcriptional regulation of gene activities. Previously, it was reported that promoter of helicases contained stress responsive cis-elements21-24 but the isolation of stress-inducible and tissue-specific promoters25,26 is still interest in the fiend of molecular breeding, biotechnology and agriculture.
In this study, we have isolated and functionally characterized the promoter of Psp68 in response to abiotic and hormonal treatment by Agrobacterium-mediated transient assay. In silico analysis also identified that the promoter of Psp68 harbored multiple stress responsive cis-acting elements. Transient assay showed that promoter of Psp68 drives high levels of GUS expression under abiotic stress and hormonal treatment. Therefore, this promoter could be used for the study of the spatio-temporal expression pattern and development of stress tolerant transgenic crops in the future.
Results
Isolation of the promoter of Psp68
Pea genomic DNA library was prepared by digesting genomic DNA with different restriction enzymes (EcoRV, DraI, PvuII and SspI) in 4 separate tubes to generate blunt ends of genomic DNA. The digested genomic DNA was purified and further ligated into BD genome walker kit. The primary PCR was done by using AP1 as forward (5′–GTAATACGAC TCACTATAGG GC–3′) primer and gene specific reverse primer R3 (5′–CCTCGCATTC TCTTCCTCGT A–3′). Four DNA genomic libraries were used as a template for the first PCR. The PCR products were resolved on a 1% agarose gel and a smear was observed in all the 4 libraries. Secondary PCR was done using primary PCR product as template (1: 10 dilution) with AP2 (5′-ACTATAGGGC ACGCGTGGT-3′) as forward and R2 (5′-AGAAGAGTTG GAGTGA-GGGTACG-3′) as a reverse primer. The PCR products were resolved on 1% agarose gel but only library 4 gave a band size of 750 bp. After the nested PCR, the specific band was purified and cloned into pGEM-T vector (data not shown) and sequenced. After sequencing, 531bp upstream regions was successfully isolated and verified by finding the overlapping regions at the 3′ end of Psp68 gene.
In silico analysis of Psp68 promoter
To identify the transcription start site, putative TATA box and CCAAT box, the promoter sequence of Psp68 were analyzed by using Plant Prom Database. The promoter region has a TATA (TACAAA, consensus TATAAA) and CCAAT box at position –81 and –117bp respectively (Fig. 1). To identify the cis-regulatory elements, present in the Psp68 promoter, the sequence was analyzed using PLANTCARE and PLACE databases (Fig. 1). Various cis-acting elements including E-box, AGAAA, GATA-box, dehydration and salt responsive elements (ACGT and GAAAA) and auxin response factor (GTCTC) were identified in the promoter sequence (Fig. 1). The additional cis-acting elements presents among others are 8 transcriptional activators elements (NGATT, GANTTNC, MACCWAMC and CTGACY), 4 mesophyll-specific gene expression elements (YACT), 6 pollen specific activator elements (GTGA and AGAAA), 10 (AAAGAT, CTCTT, AAAGAT and CTCTT) nodule specificity regulatory elements, one light-activated (ACTTTG), and one WRKY transcription factors (TGAC) element (Table 1). A complete list of all predicted cis-elements present in the Psp68 promoter was shown in Table 1.
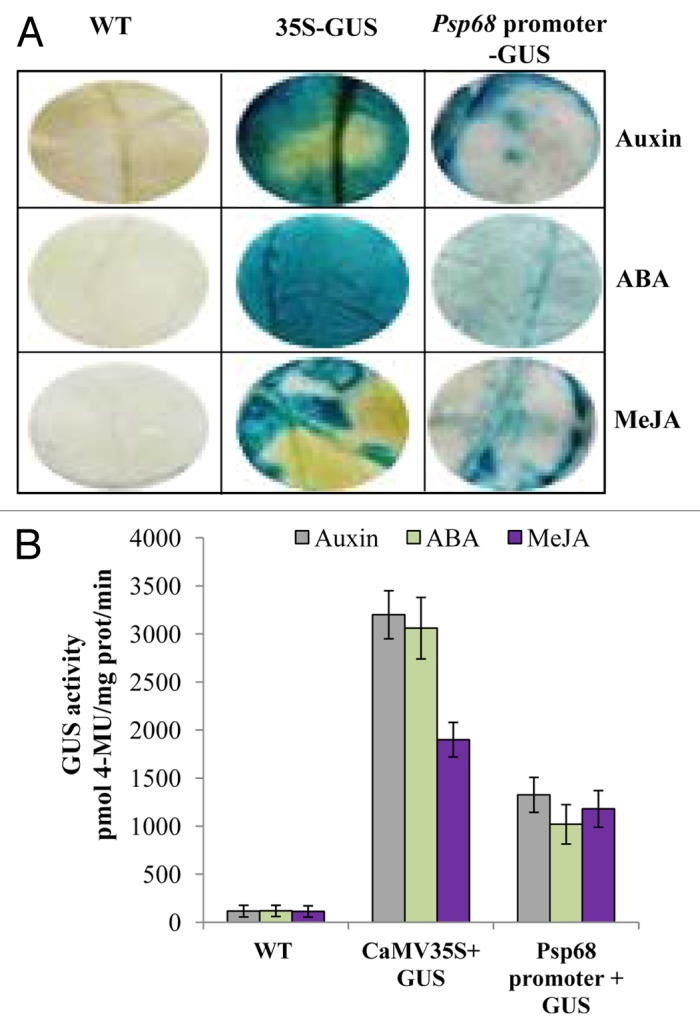
Figure 1.Psp68 promoter sequence. A schematic representation showing various cis-elements present in the upstream region of Psp68 gene as determined by PLACE program. TATA-box and CAAT sequences and various cis-acting elements are shown in different color.
Table 1. Prediction of cis-regulatory elements of PsP68 promoters using PLACE database.
| Element name and number | Sequence | Function |
|---|---|---|
| ARR1AT (5) EECCRCAH1 (1) MYBPLANT (1) BOXNTCHN48 (1) |
NGATT GANTTNC MACCWAMC CTGACY | Transcriptional activators |
| ACGTATERD1 (2) | ACGT | Responsive to dehydration |
| ARFAT (1) SURECOREATSULTR11 (3) |
TGTCTC GAGAC |
Auxin response factor |
| CAATBOX1 (3) GATABOX (5) NTBBF1ARROLB1 (1) |
CAAT GATA ACTTTA |
Responsible for the tissue specific promoter activity. Tissue-specific and auxin-regulated expression |
| CACTFTPPCA1 (4) | YACT | Elements for mesophyll-specific gene expression |
| CAREOSREP1 (1) | CAACTC | Gibberellin-upregulated proteinase expression |
| CCAATBOX1 (1) | CCAAT | Enhanced expression of chimaeric heat shock |
| DOFCOREZM (5) | AAAG | Transcription factors |
| GTGANTG10 (3) POLLEN1LELAT52 (3) |
GTGA AGAAA |
Late pollen gene Responsible for pollen specific activation |
| INTRONLOWER (1) | TGCAGG | Catalog of splice junction |
| MYBCOREATCYCB1 (1) | AACGG | Activator of reporter gene |
| MYBPZM (1) | CCWACC | Controls phlobaphene pigmentation |
| NODCON1GM (2) NODCON2GM (3) OSE1ROOTNODULE (2) OSE2ROOTNODULE (3) |
AAAGAT CTCTT AAAGAT CTCTT |
Nodule specificity of cis-acting regulatory elements. Activated in the infected cells of root nodules |
| SEBFCONSSTPR10A (1) | YTGTCWC | Potato silencing element binding factor |
| SORLIP2AT (1) | GGGCC | Involved in the network of phytochrome A-regulated gene expression |
| TAAAGSTKST1 (1) | TAAAG | Transcription factors in guard cell-specific gene expression |
| TBOXATGAPB (1) | ACTTTG | Light-activated gene transcription |
| WBOXNTERF3 (1) | TGACY | Rapid and transient activation of transcription of the ERF3 gene by wounding in tobacco leaves |
| WRKY71OS (1) | TGAC | Early nuclear events in plant defense signaling: rapid gene activation by WRKY transcription factors |
Cloning of Psp68 promoter in binary vector and Agrobacterium transformation
The PCR amplified Psp68 promoter fragment was first cloned into pGEMT easy vector. It was then released by BamHI and HindIII restriction digestion and further cloned in pCAMBIA-1391Z (promoter less vector) binary vector in the same restriction site. Cloning of the Psp68 promoter was confirmed by colony PCR and restriction analysis (data not shown). The fusion construct containing Psp68 promoter-GUS (β-glucuronidase) in pCAMBIA-1391Z was further transformed Agrobacterium tumefaciens strain (LBA4404) and verified by colony PCR using promoter specific primers.
Regulation of Psp68 promoter activities
To compare the regulation of Psp68 promoter activity, we used transient expression by agro infiltration in the tobacco leaves.27This method was selected to avoid long-time regeneration protocol. The Psp68 promoter was fused with GUS reporter gene in pCAMBIA-1391Z vector and infected into the leaves of tobacco by Agrobacterium infiltration. The CaMV35S promoter fused with GUS and WT tobacco plants used was as positive and negative control respectively, in order to determine Psp68 promoter activity.
Activities of Psp68 promoter in tobacco leaves
To check whether the isolated 5/-flanking region of Psp68 genes have promoter activity, the constructs containing promoter of Psp68::GUS and CaMV35S::GUS were agroinfiltrated into the leaves tobacco. Both the constructs drove strong levels of GUS expression but maximum GUS gene expression was driven by the CaMV35S promoter (Fig. 2). No GUS expression was observed for the negative control. These results indicated that the promoter sequence isolated from the upstream of the Psp68 gene was functional in tobacco leaves.
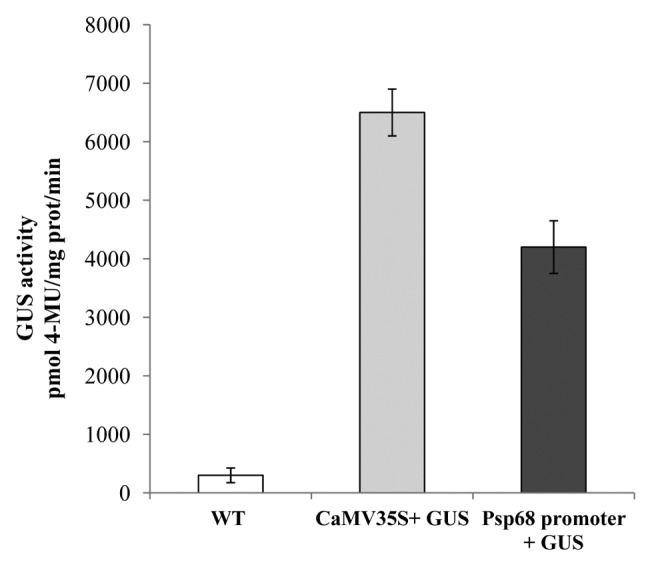
Figure 2. Transient expression of Psp68 promoters in agroinfiltrated tobacco leaves. GUS activity was determined 48h after infiltration of tobacco leaves with Agrobacterium (OD0.7) containing promoter::GUS and CaMV35S::GUS constructs, or WT (negative control). Data represent the mean and SD of 4 independent experiments.
Abiotic stress-induced activities of Psp68 promoter
The impact of abiotic stress (salt, PEG and cold) on the activities of Psp68 promoter was verified by transient assay in the leaves tobacco (Fig. 3). Abiotic stress treatment applied on agroinfiltrated leaves increases the expression of GUS activity (Fig. 3A). The effect of abiotic stress was varied for the Psp68 promoter; GUS activity increased ~15-folds in response to salt stress but upon PEG and cold treatment the expression was increased ~20 and ~11-folds respectively (Fig. 3B). The CaMV 35S promoter also displayed high GUS activity levels. These results indicated that the promoter of Psp68 is a stress inducible promoter.
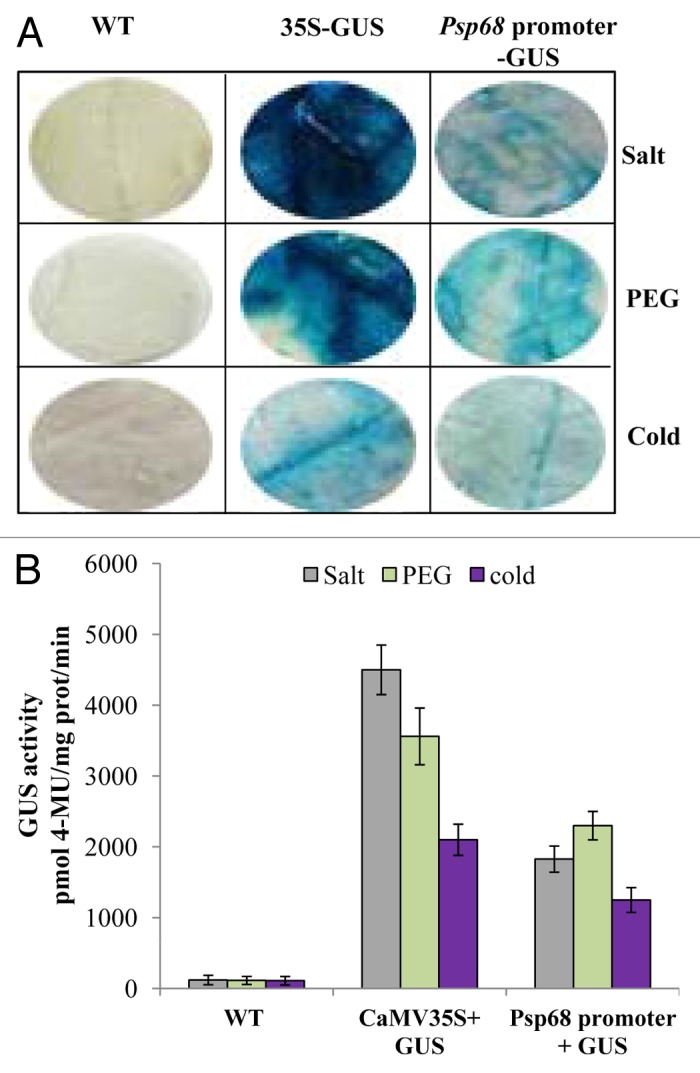
Figure 3.Psp68 promoter-GUS analysis in response to abiotic stress. (A) GUS activity in agroinfiltrated leaves. GUS was detected in X-Gluc solution followed by stress treatment. (B) Effect of abiotic stress on the transient expression of Psp68 promoter in agroinfiltrated tobacco leaves. Two days after infiltration with A. tumefaciens (OD 0.7) containing either Psp68 promoter::GUS or CaMV35S::GUS constructs, tobacco leaves were sprayed with 200 mM NaCl, 20% PEG. For cold treatment infiltrated leaves were kept on 4 °C. After treatment, all the samples were used for quantification assay. Infiltrated leaves of WT without treatment (water) used as negative control. Data represent the mean and SD of 4 independent experiments
Hormone-induced GUS Activity
In the agroinfiltrated leaves of the tobacco, the Psp68 promoter construct showed GUS positive expression in response to hormonal (Auxin, ABA and MeJA) treatments (Fig. 4). High GUS expression was observed in response to Auxin and MeJA followed by ABA treatment (Fig. 4A). To quantify the GUS expression, equal amounts of protein was isolated from agroinfiltrated leaves and assayed for fluorescence. GUS activity increased ~11, ~8 and ~10-folds respectively by application of Auxin, ABA and MeJA treatments (Fig. 4B). The variation in the activity may be due to present of hormone-induced cis-acting elements in different position of Psp68 promoter.
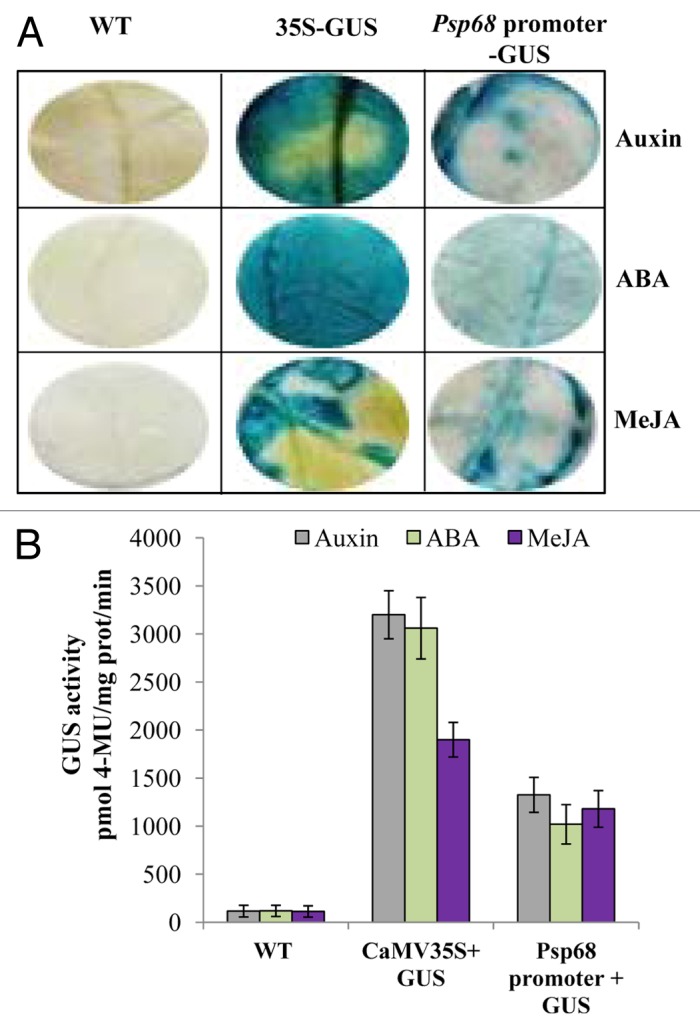
Figure 4.Psp68 promoter-GUS analysis in response to hormonal treatment. (A) GUS activity in agroinfiltrated leaves. GUS was detected in X-Gluc solution followed by hormone treatment. (B) Effect of abiotic stress on the transient expression of Psp68 promoter in agroinfiltrated tobacco leaves. Two days after infiltration with A. tumefaciens (OD0.7) containing either Psp68 promoter::GUS or CaMV35S::GUS constructs, tobacco leaves were sprayed with 100 µM Auxin, 100 µM ABA and 10 µM MeJA and used for quantification assay. Water treated infiltrated leaves of WT used as negative control. Data represent the mean and SD of 4 independent experiments.
Discussion
Cis- regulatory elements present in the promoters of stress-responsive gene controlled many essential biological processes including abiotic stress responses, hormone responses and developmental processes. In plants, a number of cis-regulatory elements have shown to be essential for the transcription of stress-responsive genes.28,29 A recent chromatin immunoprecipitation study identified that ETHYLENE RESPONSE FACTOR1 bind with stress-specific GCC or DRE/CRT elements and upregulates specific suites of genes in response to abiotic stresses.30 Drought and salt stress lead to increase ABA accumulation which may triggers adaptive responses.31 The presence of either a single ABRE or multiple ABREs is sufficient to confer ABA-mediated osmotic stress.32 The significance of a few cis- regulatory elements (G-box and ABREs) combinations have also been showed that stress-responsive genes are regulated by multiple transcription factors.33,34 Therefore, to understand the regulatory gene networks in stress-responsive cascades, functional analyses of cis-acting elements is desirable.
The promoter of Psp68 contain canonical E-box element which is critical for p68 promoter activity.35 E-box motifs can be recognized by Myc-Max heterodimers that are known to function in the regulation of many growth regulating genes. E-box motif is found in other DEAD-box proteins in human Ddx5, mouse MrDb (Myc-regulated DEAD-box protein) and Drosophila.36,37 The p68 function as transcription coactivators by binding with CBP, the CREB-binding protein.38 The CBP bridges the CRE/CREB complex to components of the basal transcription apparatus and it is possible that p68 directly influence its own transcription. Furthermore, the promoter of Psp68 contains overrepresentation of different transcriptional activators elements. These findings indicated that transcription of Psp68 might be highly complex and developmentally regulated. Several putative cis-regulatory elements associated with tissue-specific expression (GATA and CAAT motifs), pollen specific activator elements (GTGA and AGAAA motifs), mesophyll (YACT motif) and guard cell-specific (TAAAG motif) gene expression elements and nodule specific regulatory elements (AAAGAT, CTCTT, AAAGAT and CTCTT motifs) were identified in Psp68 promoter sequences. Apparent enrichment of these tissue-specific expression regulatory elements indicates the involvement of Psp68 gene in wide range of cellular process but need to validate.
The expression of Psp68 was induced by abiotic stress. Salt (GAAAAA) and dehydration (ACGT-box) responsive cis-acting elements were identified by computational analysis. The promoter of Psp68 is able to drive GUS expression in agroinfiltrated leaves of tobacco challenged with NaCl, PEG and cold stress. The presence of the GT-1 like element (5/-GAAAAA-3/) in the upstream region of Psp68 gene might be responsible salinity specific expression. Earlier report showed that in response to salinity stress, the GA sequence in GT-1 cis regulatory element bind to nuclear factor(s)39 resulting salinity stress tolerance. In this study, GUS activity was also observed in response to PEG and ABA treatments. The existence of ACGT-box and ABRE elements might support the above statement. In response to dehydration, ABA levels increased. It has been reported that most dehydration-inducible genes are also induced by ABA40,41 and ABA is known be involved in dehydration-inducible gene expression in land plants.40-42 The promoter sequence of Psp68 also contain cold responsive element like CCGAC (DRE). Earlier, the DRE/CRT and ABRE elements found together in the promoters of many well-studied cold-regulated genes in Arabidopsis43, 44 which is consistent with a role for the ABA regulation of cold-induced genes.
Auxin is a major plant hormone, required for many developmental processes in plant45 including root formation,46 apical dominance47 and growth-related tropisms.48 The transcriptional response to auxin is mediated by the auxin responsive cis-regulatory elements present in the upstream region of auxin responsive genes.49 We have identified 5 auxin responsive cis-regulatory elements in Psp68 promoter sequence. Furthermore, high GUS expression was observed in the agroinfiltrated tobacco leaves upon treatment with auxin. This indicated that Psp68 gene may play an important role in auxin-mediating signal transduction pathways. Although conserved similar sequence were observed in the promoter of many auxin responsive genes,50,51 it remains need to be tested the functional significance of these conserved sequences. Jasmonates are another growth regulators52 important for plant biotic and abiotic stress responses.53-55 Either GCC or G-box elements are required for MeJA-inducible expression of different genes. A number of studies have been identified these elements in a variety of plant gene promoters and their role in response to light, anaerobiosis, and various phytohormones.56,57 We found 7 GCC motifs in the promoter of Psp68 and promoter:: GUS analysis also detected very high level of GUS expression under MeJA treatment, indicating a positive regulatory role of Psp68 gene toward abiotic stress tolerance.
The promoter of Psp68, drive the expression reporter gene in response to abiotic stress and hormonal treatments. The Psp68 promoter contains dehydration, salt, cold, auxin ABA and MeJA related cis-elements, which may regulate the expression of this gene. Therefore, Psp68 promoter could be used as a new and powerful candidate for the study of tissue specific and stress responsive transgene expression in crop plants.
Materials and Methods
Isolation of Psp68 promoter: gene walking by PCR
Pea genomic DNA was isolated by a previously described method58 and ~5 µg genomic DNA was digested overnight at 37 °C with 4 blunt end cutting restriction enzymes: DraI, EcoRI, PvuII and StuI/SmaI independently. Following digestion, the genomic DNA was purified by phenol-chloroform precipitation and each pool of DNA fragments was ligated to the BD Genome Walker Adaptor as per the manufacturer’s instruction. The primer AP1corresponding to gene-specific primer R3 was used for primary PCR reactions. The 50 µl reactions mixture contain 0.4 µl stock diluted DNA, 0.2 µM of each primer and 0.5 µl Advantage® 2 Polymerase mix (Clontech, USA) with the following conditions: 35 cycles, 94 °C 45s, 64 °C 30s, 72 °C 1 min. The primary PCR were then diluted in 50 fold for prior to the nested-PCR reaction using the primer AP2 in combination with R2 in the same cycle conditions. The bands of interest were separated by electrophoresis, purified and cloned into pGEMT vector and sequenced.
In silico analyses of promoter sequence
Homologies to sequence were searched in Basic local alignment search tools (BLASTN and BLASTX) and was aligned using the ClustalW program. The prediction of transcriptional start site, TATA-box and CAAT-box were done using Plant Prom Database.59 Putative cis-acting elements were identified by using Plant CARE Database (http://bioinformatics.psb.-ugent.be/webtools-/plantcare/html/).
Plasmid constructions
The Psp68 promoter was amplified from Psp68 promoter-pGEMT clone, using the primers by introducing BamHI and HindIII restriction sites. The amplified bands were run on 1% agarose gel, cut, eluted and again ligated into pGEMT-easy cloning vector. The clone was verified by colony PCR and restriction digestion analysis with BamHI and HindIII enzymes. The Psp68 promoter was further cloned in pCAMBIA-1391Z in the same restriction site. The colonies were checked by PCR, followed by restriction analysis with BamHI and HindIII restriction enzymes. The Psp68 promoter cloned in pCAMBIA-1391Z vector was again transformed in Agrobacterium tumefaciens (LBA4404) and confirmed by colony PCR using Psp68 promoter specific primers.
Agrobacterium-mediated transient assays
Agroinfiltration assays were performed by a previously described method earlier. 27 The Psp68 promoter, transform in Agrobacterium tumefaciens (LBA4404) were grown in LB medium containing 50 µg/ml rifampicin, 50 µg/ml kanamycin and incubated overnight at 28 °C. The cells were harvested by centrifugation at 3,0009 g for 15 min and further resuspended in infiltration media (10 mM MgSO4, 200 µM acetosyringone, 20 mM MES pH 5.6). Fully expanded leaves of tobacco (Nicotiana tobaccum cv USA) plants grown in greenhouse at 22 °C were agroinfiltrated by using a 1-ml syringe. After 48h, infiltrated leaf discs were collected.
Stress treatments
For salinity and drought stress, tobacco leaves were agroinfiltration with 200 mM NaCl and 20% PEG solution or water as a control and then collected after 24h. For cold treatment, infiltrated leaf discs kept on 4 °C and collected after 24h. For hormonal stress, tobacco leaves were agroinfiltration with 10 μM naphthalene acetic acid (auxin), 100 μM ABA and 10 μM MeJA respectively or water as a control and then collected after 24h.
GUS activity detection
The leaf discs were incubated overnight at 37 °C in GUS assay solution containing 1 mg/ml X-Gluc, 5 mM potassium ferrocyanide, 5 mM potassium ferricyanide, 0.2% Triton X-100 in 100 mM sodium phosphate buffer (pH 7.4) followed by washing with 70% ethanol solution till the chlorophyll cleared.
GUS activity quantification
β-Glucuronidase activity was quantified by fluorometric GUS assays. Agroinfiltrated leaves were homogenized in 1 ml extraction buffer containing 10 mM EDTA, 50 mM NaH2PO4 pH 7, 0.1% sodium lauryl sarcosine, 10 mM β-mercaptoethanol and 0.1% Triton X-100. The homogenate was centrifuged at 12,000 rpm for 15 min at 4 °C and finally supernatant was collected. The concentration of protein was measured by Bradford method60 by using bovine serum albumin (BSA) as a standard. GUS activity was performed by earlier described method61 and expressed as picomoles of 4-MU (methylumbelliferone) per minute per milligram of protein.
Contributions
MSAB performed the research, analyzed data and written the manuscript, KMKH performed the experiments and helped in writing the manuscript, NT designed research, analyzed the data and written the manuscript.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
The research of NT’s laboratory is partially supported by Department of Biotechnology (DBT) and Department of Science and Technology (DST), Government of India. M.S.A.B. and K.M.K.H. are the recipients of Arturo Falaschi International Centre for Genetic Engineering and Biotechnology pre-doctoral fellowships.
References
- 1.Tuteja N. Plant cell and viral helicases: essential enzymes for nucleic acid transactions. Crit Rev Plant Sci. 2000;19:449–78. doi: 10.1080/07352689.2000.10131825. [DOI] [Google Scholar]
- 2.Tuteja N. Plant DNA helicases: the long unwinding road. J Exp Bot. 2003;54:2201–14. doi: 10.1093/jxb/erg246. [DOI] [PubMed] [Google Scholar]
- 3.Gong Z, Dong CH, Lee H, Zhu J, Xiong L, Gong D, Stevenson B, Zhu JK. A DEAD box RNA helicase is essential for mRNA export and important for development and stress responses in Arabidopsis. Plant Cell. 2005;17:256–67. doi: 10.1105/tpc.104.027557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Owttrim GW. RNA helicases and abiotic stress. Nucleic Acids Res. 2006;34:3220–30. doi: 10.1093/nar/gkl408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cordin O, Banroques J, Tanner NK, Linder P. The DEAD-box protein family of RNA helicases. Gene. 2006;367:17–37. doi: 10.1016/j.gene.2005.10.019. [DOI] [PubMed] [Google Scholar]
- 6.Kammel C, Thomaier M, Sørensen BB, Schubert T, Längst G, Grasser M, Grasser KD. Arabidopsis DEAD-box RNA helicase UAP56 interacts with both RNA and DNA as well as with mRNA export factors. PLoS One. 2013;8:e60644. doi: 10.1371/journal.pone.0060644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guan Q, Wu J, Zhang Y, Jiang C, Liu R, Chai C, Zhu J. A DEAD box RNA helicase is critical for pre-mRNA splicing, cold-responsive gene regulation, and cold tolerance in Arabidopsis. Plant Cell. 2013;25:342–56. doi: 10.1105/tpc.112.108340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sanan-Mishra N, Pham XH, Sopory SK, Tuteja N. Pea DNA helicase 45 overexpression in tobacco confers high salinity tolerance without affecting yield. Proc Natl Acad Sci U S A. 2005;102:509–14. doi: 10.1073/pnas.0406485102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vashisht AA, Pradhan A, Tuteja R, Tuteja N. Cold- and salinity stress-induced bipolar pea DNA helicase 47 is involved in protein synthesis and stimulated by phosphorylation with protein kinase C. Plant J. 2005;44:76–87. doi: 10.1111/j.1365-313X.2005.02511.x. [DOI] [PubMed] [Google Scholar]
- 10.Li D, Liu H, Zhang H, Wang X, Song F. OsBIRH1, a DEAD-box RNA helicase with functions in modulating defence responses against pathogen infection and oxidative stress. J Exp Bot. 2008;59:2133–46. doi: 10.1093/jxb/ern072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu HH, Liu J, Fan SL, Song MZ, Han XL, Liu F, Shen FF. Molecular cloning and characterization of a salinity stress-induced gene encoding DEAD-box helicase from the halophyte Apocynum venetum. J Exp Bot. 2008;59:633–44. doi: 10.1093/jxb/erm355. [DOI] [PubMed] [Google Scholar]
- 12.Luo Y, Liu YB, Dong YX, Gao XQ, Zhang XS. Expression of a putative alfalfa helicase increases tolerance to abiotic stress in Arabidopsis by enhancing the capacities for ROS scavenging and osmotic adjustment. J Plant Physiol. 2009;166:385–94. doi: 10.1016/j.jplph.2008.06.018. [DOI] [PubMed] [Google Scholar]
- 13.Tuteja N, Sahoo RK, Garg B, Tuteja R. OsSUV3 dual helicase functions in salinity stress tolerance by maintaining photosynthesis and antioxidant machinery in rice (Oryza sativa L. cv. IR64) Plant J. 2013;76:115–27. doi: 10.1111/tpj.12277. [DOI] [PubMed] [Google Scholar]
- 14.Ow DW, DE Wet JR, Helinski DR, Howell SH, Wood KV, Deluca M. Transient and stable expression of the firefly luciferase gene in plant cells and transgenic plants. Science. 1986;234:856–9. doi: 10.1126/science.234.4778.856. [DOI] [PubMed] [Google Scholar]
- 15.Vaucheret H, Béclin C, Elmayan T, Feuerbach F, Godon C, Morel JB, Mourrain P, Palauqui JC, Vernhettes S. Transgene-induced gene silencing in plants. Plant J. 1998;16:651–9. doi: 10.1046/j.1365-313x.1998.00337.x. [DOI] [PubMed] [Google Scholar]
- 16.Youm JW, Jeon JH, Choi D, Yi SY, Joung H, Kim HS. Ectopic expression of pepper CaPF1 in potato enhances multiple stresses tolerance and delays initiation of in vitro tuberization. Planta. 2008;228:701–8. doi: 10.1007/s00425-008-0782-5. [DOI] [PubMed] [Google Scholar]
- 17.Achard P, Gong F, Cheminant S, Alioua M, Hedden P, Genschik P. The cold-inducible CBF1 factor-dependent signaling pathway modulates the accumulation of the growth-repressing DELLA proteins via its effect on gibberellin metabolism. Plant Cell. 2008;20:2117–29. doi: 10.1105/tpc.108.058941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kanneganti V, Gupta AK. Overexpression of OsiSAP8, a member of stress associated protein (SAP) gene family of rice confers tolerance to salt, drought and cold stress in transgenic tobacco and rice. Plant Mol Biol. 2008;66:445–62. doi: 10.1007/s11103-007-9284-2. [DOI] [PubMed] [Google Scholar]
- Oettgen P. Transcriptional regulation of vascular development. Circ Res. 2001;89:380–8. doi: 10.1161/hh1701.095958. [DOI] [PubMed] [Google Scholar]
- 20.Huda KM, Banu MS, Pathi KM, Tuteja N. Reproductive organ and vascular specific promoter of the rice plasma membrane Ca2+ATPase mediates environmental stress responses in plants. PLoS One. 2013;8:e57803. doi: 10.1371/journal.pone.0057803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tran NQ, Dang HQ, Tuteja R, Tuteja N. A single subunit MCM6 from pea forms homohexamer and functions as DNA helicase. Plant Mol Biol. 2010;74:327–36. doi: 10.1007/s11103-010-9675-7. [DOI] [PubMed] [Google Scholar]
- 22.Dang HQ, Tran NQ, Gill SS, Tuteja R, Tuteja N. A single subunit MCM6 from pea promotes salinity stress tolerance without affecting yield. Plant Mol Biol. 2011;76:19–34. doi: 10.1007/s11103-011-9758-0. a. [DOI] [PubMed] [Google Scholar]
- 23.Dang HQ, Tran NQ, Tuteja R, Tuteja N. Promoter of a salinity and cold stress-induced MCM6 DNA helicase from pea. Plant Signal Behav. 2011;6:1006–8. doi: 10.4161/psb.6.7.15502. b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tajrishi MM, Tuteja N. Isolation and in silico analysis of promoter of a high salinity stress-regulated pea DNA helicase 45. Plant Signal Behav. 2011;6:1447–50. doi: 10.4161/psb.6.10.17106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Song P, Allen RD. Identification of a cotton fiber-specific acyl carrier protein cDNA by differential display. Biochim Biophys Acta. 1997;1351:305–12. doi: 10.1016/S0167-4781(96)00218-7. [DOI] [PubMed] [Google Scholar]
- 26.Song P, Heinen JL, Burns TH, Allen RD. Expression of a promoter from a fiber-specific acyl carrier protein gene in transgenic cotton plants. Proc. Beltwide Cotton Prod. Res. Conf; 1998 Jan 5 – Jan 9; San Diego, CA. Memphis, TN: Natl. Cotton Counc. Am.; 1998 [Google Scholar]
- Yang Y, Li R, Qi M. In vivo analysis of plant promoters and transcription factors by agroinfiltration of tobacco leaves. Plant J. 2000;22:543–51. doi: 10.1046/j.1365-313x.2000.00760.x. [DOI] [PubMed] [Google Scholar]
- 28.Machens F, Becker M, Umrath F, Hehl R. Identification of a novel type of WRKY transcription factor binding site in elicitor-responsive cis-sequences from Arabidopsis thaliana. Plant Mol Biol. 2014;84:371–85. doi: 10.1007/s11103-013-0136-y. [DOI] [PubMed] [Google Scholar]
- 29.Yadav DK, Shukla D, Tuteja N. Isolation, in silico characterization, localization and expression analysis of abiotic stress-responsive rice G-protein β subunit (RGB1) Plant Signal Behav. 2014;9 doi: 10.4161/psb.28890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cheng MC, Liao PM, Kuo WW, Lin TP. The Arabidopsis ETHYLENE RESPONSE FACTOR1 regulates abiotic stress-responsive gene expression by binding to different cis-acting elements in response to different stress signals. Plant Physiol. 2013;162:1566–82. doi: 10.1104/pp.113.221911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fukao T, Yeung E, Bailey-Serres J. The submergence tolerance regulator SUB1A mediates crosstalk between submergence and drought tolerance in rice. Plant Cell. 2011;23:412–27. doi: 10.1105/tpc.110.080325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim JS, Mizoi J, Yoshida T, Fujita Y, Nakajima J, Ohori T, Todaka D, Nakashima K, Hirayama T, Shinozaki K, et al. An ABRE promoter sequence is involved in osmotic stress-responsive expression of the DREB2A gene, which encodes a transcription factor regulating drought-inducible genes in Arabidopsis. Plant Cell Physiol. 2011;52:2136–46. doi: 10.1093/pcp/pcr143. [DOI] [PubMed] [Google Scholar]
- 33.Liu N, Ding Y, Fromm M. Avramova1 Z. Different gene-specific mechanisms determine the ‘revised-response’ memory transcription patterns of a subset of A. thaliana dehydration stress responding genes. Nucleic Acids Res. 2014 doi: 10.1093/nar/gku220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang X, Yan Y, Li Y, Chu X, Wu C, Guo X. GhWRKY40, a multiple stress-responsive cotton WRKY gene, plays an important role in the wounding response and enhances susceptibility to Ralstonia solanacearum infection in transgenic Nicotiana benthamiana. PLoS One. 2014;9:e93577. doi: 10.1371/journal.pone.0093577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rössler OG, Hloch P, Schütz N, Weitzenegger T, Stahl H. Structure and expression of the human p68 RNA helicase gene. Nucleic Acids Res. 2000;28:932–9. doi: 10.1093/nar/28.4.932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Grandori C, Mac J, Siëbelt F, Ayer DE, Eisenman RN. Myc-Max heterodimers activate a DEAD box gene and interact with multiple E box-related sites in vivo. EMBO J. 1996;15:4344–57. [PMC free article] [PubMed] [Google Scholar]
- 37.Zaffran S, Chartier A, Gallant P, Astier M, Arquier N, Doherty D, Gratecos D, Sémériva M. A Drosophila RNA helicase gene, pitchoune, is required for cell growth and proliferation and is a potential target of d-Myc. Development. 1998;125:3571–84. doi: 10.1242/dev.125.18.3571. [DOI] [PubMed] [Google Scholar]
- 38.Endoh H, Maruyama K, Masuhiro Y, Kobayashi Y, Goto M, Tai H, Yanagisawa J, Metzger D, Hashimoto S, Kato S. Purification and identification of p68 RNA helicase acting as a transcriptional coactivator specific for the activation function 1 of human estrogen receptor alpha. Mol Cell Biol. 1999;19:5363–72. doi: 10.1128/mcb.19.8.5363. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 39.Park HC, Kim ML, Kang YH, Jeon JM, Yoo JH, Kim MC, Park CY, Jeong JC, Moon BC, Lee JH, et al. Pathogen- and NaCl-induced expression of the SCaM-4 promoter is mediated in part by a GT-1 box that interacts with a GT-1-like transcription factor. Plant Physiol. 2004;135:2150–61. doi: 10.1104/pp.104.041442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chandler PM, Robertson M. Gene-expression regulated by abscisic-acid and its relation to stress tolerance. Annu Rev Plant Physiol Plant Mol Biol. 1994;45:113–41. doi: 10.1146/annurev.pp.45.060194.000553. [DOI] [Google Scholar]
- 41.Shinozaki K, Yamaguchi-Shinozaki K, Seki M. Regulatory network of gene expression in the drought and cold stress responses. Curr Opin Plant Biol. 2003;6:410–7. doi: 10.1016/S1369-5266(03)00092-X. [DOI] [PubMed] [Google Scholar]
- 42.Tougane K, Komatsu K, Bhyan SB, Sakata Y, Ishizaki K, Yamato KT, Kohchi T, Takezawa D. Evolutionarily conserved regulatory mechanisms of abscisic acid signaling in land plants: characterization of ABSCISIC ACID INSENSITIVE1-like type 2C protein phosphatase in the liverwort Marchantia polymorpha. Plant Physiol. 2010;152:1529–43. doi: 10.1104/pp.110.153387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Baker SS, Wilhelm KS, Thomashow MF. The 5′-region of Arabidopsis thaliana cor15a has cis-acting elements that confer cold-, drought- and ABA-regulated gene expression. Plant Mol Biol. 1994;24:701–13. doi: 10.1007/BF00029852. [DOI] [PubMed] [Google Scholar]
- 44.Yamaguchi-Shinozaki K, Shinozaki K. A novel cis-acting element in an Arabidopsis gene is involved in responsiveness to drought, low-temperature, or high-salt stress. Plant Cell. 1994;6:251–64. doi: 10.1105/tpc.6.2.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Möller B, Weijers D. Auxin control of embryo patterning. Cold Spring Harb Perspect Biol. 2009;1:a001545. doi: 10.1101/cshperspect.a001545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bennett T, Scheres B. Root development-two meristems for the price of one? Curr Top Dev Biol. 2010;91:67–102. doi: 10.1016/S0070-2153(10)91003-X. [DOI] [PubMed] [Google Scholar]
- 47.Leyser O. The fall and rise of apical dominance. Curr Opin Genet Dev. 2005;15:468–71. doi: 10.1016/j.gde.2005.06.010. [DOI] [PubMed] [Google Scholar]
- 48.Muday GK. Auxins and tropisms. J Plant Growth Regul. 2001;20:226–43. doi: 10.1007/s003440010027. [DOI] [PubMed] [Google Scholar]
- 49.Ulmasov T, Hagen G, Guilfoyle TJ. Dimerization and DNA binding of auxin response factors. Plant J. 1999;19:309–19. doi: 10.1046/j.1365-313X.1999.00538.x. [DOI] [PubMed] [Google Scholar]
- 50.Takahashi Y, Ishida S, Nagata T. Function and modulation of expression of auxin-regulated genes. Int Rev Cytol. 1994;152:109–44. doi: 10.1016/S0074-7696(08)62555-3. [DOI] [PubMed] [Google Scholar]
- 51.van der Zaal EJ, Droog FN, Boot CJ, Hensgens LA, Hoge JH, Schilperoort RA, Libbenga KR. Promoters of auxin-induced genes from tobacco can lead to auxin-inducible and root tip-specific expression. Plant Mol Biol. 1991;16:983–98. doi: 10.1007/BF00016071. [DOI] [PubMed] [Google Scholar]
- 52.Seo JS, Koo YJ, Jung C, Yeu SY, Song JT, Kim JK, Choi Y, Lee JS, Do Choi Y. Identification of a novel jasmonate-responsive element in the AtJMT promoter and its binding protein for AtJMT repression. PLoS One. 2013;8:e55482. doi: 10.1371/journal.pone.0055482. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 53.Howe GA, Jander G. Plant immunity to insect herbivores. Annu Rev Plant Biol. 2008;59:41–66. doi: 10.1146/annurev.arplant.59.032607.092825. [DOI] [PubMed] [Google Scholar]
- 54.Browse J. Jasmonate passes muster: a receptor and targets for the defense hormone. Annu Rev Plant Biol. 2009;60:183–205. doi: 10.1146/annurev.arplant.043008.092007. [DOI] [PubMed] [Google Scholar]
- 55.Koo AJ, Howe GA. The wound hormone jasmonate. Phytochemistry. 2009;70:1571–80. doi: 10.1016/j.phytochem.2009.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ishige F, Takaichi M, Foster R, Chua NH, Oeda K. A G-box motif (GCCACGTGCC) tetramer confers high-level constitutive expression in dicot and monocot plants. Plant J. 1999;18:443–8. doi: 10.1046/j.1365-313X.1999.00456.x. [DOI] [Google Scholar]
- 57.Sibéril Y, Doireau P, Gantet P. Plant bZIP G-box binding factors. Modular structure and activation mechanisms. Eur J Biochem. 2001;268:5655–66. doi: 10.1046/j.0014-2956.2001.02552.x. [DOI] [PubMed] [Google Scholar]
- 58.Murray MG, Thompson WF. Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 1980;8:4321–5. doi: 10.1093/nar/8.19.4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shahmuradov IA, Gammerman AJ, Hancock JM, Bramley PM, Solovyev VV. PlantProm: a database of plant promoter sequences. Nucleic Acids Res. 2003;31:114–7. doi: 10.1093/nar/gkg041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–54. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 61.Jefferson RA. Assaying chimeric genes in plants: the GUS gene fusion system. Plant Mol Biol. 1987;21:121–31. [Google Scholar]


