Abstract
Flavonoids are plant secondary polyphenolic metabolites and fulfil many vital biological functions, offering a valuable metabolic and genetic model for studying transcriptional control of gene expression. Arabidopsis thaliana mainly accumulates 3 types of flavonoids, including flavonols, anthocyanins, and proanthocyanidins (PAs). Flavonoid biosynthesis involves a multitude of well-characterized enzymatic and regulatory proteins. Three R2R3-MYB proteins (MYB11, MYB12, and MYB111) control flavonol biosynthesis via activating the early biosynthetic steps, whereas the production of anthocyanins and PAs requires the MYB-bHLH-WD40 (MBW) complex to activate the late biosynthetic genes. Additional regulators of flavonoid biosynthesis have recently come to light, which interact with R2R3-MYBs or bHLHs to organize or disrupt the formation of the MBW complex, leading to enhanced or compromised flavonoid production. This mini-review gives an overview of how these novel players modulate flavonoid metabolism and thus plant developmental processes and further proposes a fine-tuning mechanism to complete the complex regulatory network controlling flavonoid biosynthesis.
Keywords: flavonoids, MYBL2, SPL9, TT1, TCP3, MBW complexes, Arabidopsis thaliania
Introduction
Flavonoids constitute a large family of plant secondary polyphenolic metabolites categorized into 6 major subgroups according to their molecular structures, including chalcones, flavones, flavonols, flavandiols, anthocyanins, and proanthocyanidins (PAs; also called condensed tannins). These natural endogenous compounds are widely distributed in higher plants and fulfill a variety of important biological functions. Besides genetically contributing to Mendel’s elucidation of inheritance in peas (Pisum sativum), McClintock’s discovery of transposable elements in maize (Zea mays), and genetic dissection of co-suppression phenomena in petunia (Petunia hybrida), flavonoids also provide plants with beautiful pigmentation to protect against UV radiation and attract pollinators and seed dispersers.1 In addition, flavonoids exert potential beneficial effects on human health and play vital roles in plant-microbe interactions and defense response as antimicrobial agents.1
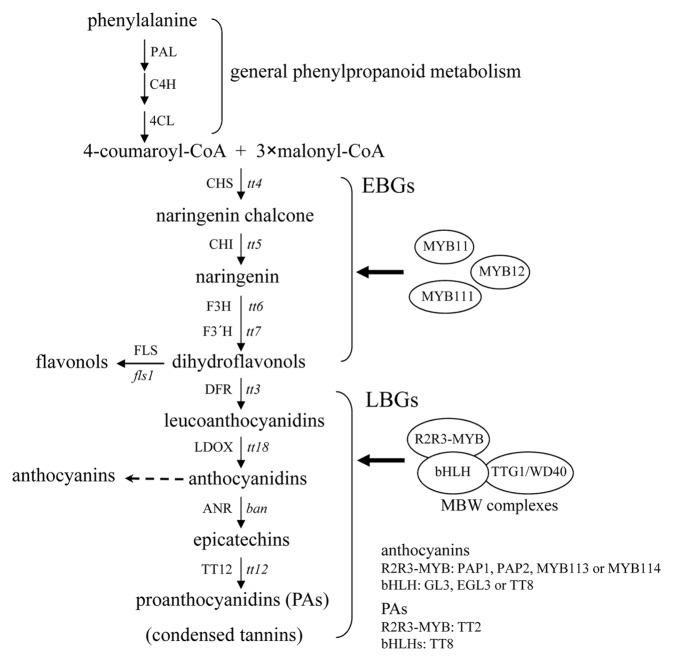
Arabidopsis (Arabidopsis thaliana) plants produce 3 major groups of flavonoids, including colorless to pale yellow flavonols in vegetative tissues and seeds, red to purple anthocyanins in vegetative organs, as well as colorless PAs in seeds, which turn brown upon oxidative reactions during seed desiccation.1 Flavonoid biosynthesis is largely modulated at the transcriptional level and starts with the general phenylpropanoid metabolism involving phenylalanine ammonia-lyase (PAL), cinnamate 4-hydroxylase (C4H), and 4-coumarate:CoA ligase (4CL) (Fig. 1). Genetic and biochemical dissection of the flavonoid biosynthetic pathway reveals that TRANSPARENT TESTA4 (TT4), TT5, TT6, and TT7 encode chalcone synthase (CHS), chalcone isomerase (CHI), flavanone 3-hydroxylase (F3H), and flavanone 3′-hydroxylase (F3′H), respectively. Successive reactions catalyzed by these structural enzymes generate dihydroflavonols, the last common precursors for the biosynthesis of flavonols, anthocyanins, and PAs (Fig. 1).2-4 Dihydroflavonols are then oxidized by flavonol synthase (FLS) to produce flavonols, such as quercetins and kaempferols. These early biosynthetic steps are transcriptionally regulated by the 3 closely related R2R3-MYB proteins MYB11, MYB12, and MYB111, which activate the early flavonoid biosynthetic genes CHS, CHI, F3H, and FLS1 (EBGs; Fig. 1).5,6 Intriguingly, the early flavonoid biosynthetic steps are even discovered in the bryophytes (Physcomitrella patens and Marchantia polymorpha), implying that flavonoids might have evolved to act first as chemical messengers and then UV sunscreens.7 Dihydroflavonol 4-reductase (DFR) encoded by the late flavonoid biosynthetic gene DFR (TT3) reduces dihydroflavonols to leucoanthocyanidins and further downstream catalytic enzymes, including leukoanthocyanidin dioxygenase (LDOX), anthocyanidin reductase (ANR, encoded by BANYULS or BAN) and TT12, eventually result in the production of anthocyanins and PAs (Fig. 1).2-4 Several of the late flavonoid biosynthetic genes (LBGs) are activated by the MYB-bHLH-WD40 (MBW) ternary transcriptional complex comprising 3 classes of regulatory proteins, including R2R3-MYBs, bHLHs, and TRANSPARENT TESTA GLABROUS1 (TTG1; also termed WD40) (Fig. 1).2-4 Anthocyanin production is transcriptionally regulated by the MBW complex composed of TTG1, one R2R3-MYB protein from PRODUCTION OF ANTHOCYANIN PIGMENTS (PAP1), PAP2, MYB113, or MYB114, as well as one bHLH protein from TT8, GLABROUS3 (GL3), or ENHANCER OF GLABRA3 (EGL3) (Fig. 1).4 Seed-specific accumulation of PAs requires the activity of the MBW complex consisting of TT2, TT8, and TTG1 (Fig. 1).2,8 Similar MBW complexes also control various aspects of epidermal cell patterning, such as the development of trichomes and root-hairless cells as well as the biosynthesis of seed mucilage.9 These epidermal cell-specific MBW complexes comprise TTG1, one R2R3-MYB protein from GL1 or WEREWOLF (WER) and one bHLH protein from GL3 or EGL3.9
Figure 1. The biosynthetic pathway for flavonols, anthocyanins, and PAs in Arabidopsis. This pathway starts with the general phenylpropanoid metabolism and subsequent steps are catalyzed by a series of structural enzymes leading to the biosynthesis of 3 final end products, including flavonols, anthocyanins, and PAs. The early biosynthetic genes (EBGs) are activated by 3 functionally redundant R2R3-MYB proteins (MYB11, MYB12, and MYB111), whereas the expression of the late biosynthetic genes (LBGs) requires the transcriptional activation activity of the R2R3-MYB/bHLH/WD40 (MBW) complex. Enzymes are denoted in uppercase and corresponding genetic loci are indicated in italic lowercase letters. PAL, phenylalanine ammonia-lyase; C4H, cinnamate 4-hydroxylase; 4CL, 4-coumarate:CoA ligase; CHS, chalcone synthase; CHI, chalcone isomerase; F3H, flavanone 3-hydroxylase; F3′H, flavanone 3′-hydroxylase; DFR, dihydroflavonol 4-reductase; LDOX, leucoanthocyanidin dioxygenase; ANR, anthocyanidin reductase; tt, transparent testa; ban, banyuls.
As described above, the flavonoid biosynthetic pathway in Arabidopsis involves a plethora of functionally well-known catalytic or regulatory proteins. Additional regulators of flavonoid production have recently emerged, some of which participate in the flavonoid pathway via directly interacting with the component of the MBW complex.3,4,6,10-13 These modulators belong to different families of transcription factors, including the R3-MYB protein MYBL2, the miR156-targeted SQUAMOSA PROMOTER BINDING PROTEIN-LIKE9 (SPL9), the WIP-type zinc finger protein TT1, and the class II CIN-TCP protein TCP3.3,4,10-12 Here, I summarize the current knowledge of how these novel players regulate flavonoid biosynthesis and thus plant developmental processes and further put forward a fine-adjusting mechanism to complete the complex regulatory network involved in flavonoid production.
MYBL2 inhibits the activity of the MBW complex and negatively regulates anthocyanin biosynthesis
MYB proteins make up the largest transcription factor family in Arabidopsis and most of its members belong to plant-specific R2R3-MYBs.14 A group of R3-MYBs participates in the modulation of epidermal cell fates, acting as inhibitors of the MBW complex GL1-GL3/EGL3-TTG1.14-18 Five of these closely related R3-MYBs, namely CPC (CAPRICE), TRY (TRIPTYCHON), ETC1 (ENHANCER OF CAPRICE AND TRIPTYCHON1), ETC2, and ETC3, are capable of interacting with the bHLH proteins GL3, EGL3, and TT8 to counteract the transcriptional activity of the MBW complex by sequestering its bHLH component.16,17 Another related R3-MYB protein, TCL1 (TRICHOMELESS1), can be recruited to the cis-acting regulatory elements of GL1 to inhibit its transcription and thus negatively regulates trichome formation.18
MYBL2 encodes a more distantly related small R3-MYB protein and its ectopic expression in Arabidopsis leaves prevents trichome initiation, implicating that MYBL2 exerts a similar regulatory function as other small R3-MYBs in the determination of epidermal cell fates.10,11,13 In contrast, the seed-specific expression of MYBL2 or other small R3-MYB genes (CPC, TRY, ETC1, ETC2, and ETC3) under the control of the TT8 promoter demonstrates that MYBL2 does not function redundantly with other small R3-MYBs and is the only small R3-MYB protein interfering with the flavonoid pathway.10,11 The loss of MYBL2 activity in the mybl2 null mutant does not affect the biosynthesis of flavonols and PAs in seeds or vegetative tissues, but results in anthocyanin hyperaccumulation and heightened expression of structural and regulatory anthocyanin genes, including DFR, LDOX, GL3, TT8, and PAP1.10,11 Transient expression analysis using the DRF promoter as a target activated by regulatory proteins (TT2, PAP1, TT8, and EGL3) provides evidence implying that MYBL2 directly inhibits the activity of the MBW complex.10,11 Consistently, MYBL2 fails to associate directly with the DRF promoter and interacts with the bHLH proteins GL3, EGL3, and TT8 in yeast cells.10,11 Besides, the expression of MYBL2 is not only developmentally controlled but also regulated by environmental stimuli, such as light intensity.10,11 These observations together indicate that MYBL2 interacts with the bHLHs by competing with R2R3-MYBs to prevent the formation of the MBW complex and thus negatively regulates anthocyanin production in response to developmental and environmental stimuli. Intriguingly, the R3-MYB protein PhMYBx from petunia operates as an inhibitor of anthocyanin biosynthesis via sequestering the bHLH protein PhAN1 into inactive complexes, indicative for a conserved regulatory mechanism among dicots.19
SPL9 negatively regulates anthocyanin accumulation via destabilizing the MBW complex
The Arabidopsis genome encodes 17 SQUAMOSA PROMOTER BINDING PROTEIN-LIKE (SPL) proteins that are classified into 2 groups.20 The first group comprises only 5 members (SPL1, SPL7, SPL12, SPL14, and SPL16) and the remaining 12 SPL proteins fall into the second group. With the exception of SPL8, all members of the second group are targeted by miR156/157 for posttranscriptional regulation.20,21 These plant-specific SPB-box proteins are known to influence a large array of developmental processes in Arabidopsis, such as promoting the juvenile-to-adult phase transition by activating miR172, inducing flowering via activating LEAFY and the MADS box genes, inhibiting trichome initiation on floral organs through activating TCL1, as well as concertedly controlling male fertility.21-24 Recently, the miR156/157-targeted SPLs have been shown to affect metabolic fluxes through the branched flavonoid biosynthetic pathway.4
Dihydroflavonols serve as common precursors of flavonols and anthocyanins and can be oxidized either by FLS to form flavonols or reduced by DFR to produce leucoanthocyanidins in the first dedicated step for anthocyanin biosynthesis (Fig. 1). Substrate competition between FLS and DFR controls metabolic flow in the flavonoid biosynthetic pathway. In Arabidopsis stems, anthocyanins accumulate in an acropetal manner, with the highest level at the junction between the stem and the rosette leaves. This pattern of anthocyanin distribution is controlled by miR156/157-targeted SPL genes.4 In the tissues with a high anthocyanin concentration, high levels of miR156/157 reduce the SPL activity and thus elevate the expression of anthocyanin biosynthetic genes.4 As a result, dihydroflavonols are directed into the anthocyanin branch. Contrarily, along the elongating stem of plants, gradually reduced miR156/157 levels result in progressively increased expression of SPL genes during plant development.4 As SPL levels rise, the expression of anthocyanin biosynthetic genes is more and more inhibited, leading to increased flavonol production by FLS.4 Enhanced flavonol accumulation is also observed in the overexpressors of SPL9 or SPL10.4 Therefore, an antagonistic relationship exists between anthocyanin and flavonol biosynthesis in Arabidopsis stems. An increase in the ratio of flavonols to anthocyanins advances side by side with the transition from the formation of leaves to the formation of flowers on Arabidopsis shoots. At least one of the miR156/157 targets, SPL9, competes with TT8 for binding to anthocyanin-specific R2R3-MYBs (PAP1 and MYB113) to disrupt the MBW complex and thus negatively regulates anthocyanin biosynthesis via directly preventing the expression of anthocyanin biosynthetic genes (F3′H, DFR, BAN, and UGT75C1).4 The molecular mechanism by which SPL9 controls the transcription of anthocyanin biosynthetic genes bears a striking similarity to that of MYBL2.
TT1 interacts with TT2 and positively regulates PA accumulation in endothelial cells of the seed coat
Development of the seed coat (also termed the testa) involves a complex process whereby the integuments of the ovule differentiate into 5 layers of specialized cells: 2 layers in the outer integument and 3 layers in the inner integument. During early stages of seed development, the innermost layer of the testa (also called the endothelium) and the seed base (micropyle and chalaza) synthesize the precursors of PAs.25 These colorless precursors polymerize and oxidize to condensed tannins at later phases of seed development, giving a brown color to mature seeds.25 Three regulatory proteins (TT2, TT8, and TTG1) form the MBW complex to regulate PA biosynthesis via activating the expression of LBGs (DFR, LDOX, BAN, and TT12).8,26,27 TT2 expression is confined to PA-producing cells in the seed coat, whereas TT8 and TTG1 are expressed in both seeds and vegetative tissues.26 Specific accumulation of PAs in the testa and at the seed base is conferred by the spatio-temporally restricted expression pattern of TT2.26
TT1 encodes a WIP-type zinc finger protein that together with 5 other WIP proteins belongs to the subgroup A1d of the Arabidopsis zinc finger proteins.28 Functional characterization of 3 WIPs (WIP2/NTT, WIP6/DOT5, and CmWIP1) reveals the involvement of this class of proteins in plant developmental processes.29,30TT1 is expressed in the endothelium of the seed coat and is required for the competence of endothelial cells to synthesize condensed tannins.31TT16 encodes the ARABIDOPSIS BSISTER (ABS) MADS domain protein and also exhibits endothelium-specific expression in the testa.2 Although the endothelium layer of the tt1 and tt16 seeds lacks PA accumulation, the chalazal-micropylar area of these mutant seeds still produces PAs.2,3 These data suggest that PA biosynthesis at the seed base is not controlled by TT1 and TT16. TT1 is able to interact with TT2 but not with TT8 or TTG1 and ectopic expression of TT2 partially restores PA production in the tt1 and tt16 endothelial cells.3 These findings together indicate that TT1 may organize the formation of the PA-specific MBW complex via associating with TT2 and that the full activity of this complex requires the participation of TT1.
TCP3 enhances flavonoid biosynthesis and negatively regulates auxin response via interacting with R2R3-MYBs
The plant-specific TCP family of transcription factors is represented by the first 3 characterized members, TEOSINTE BRANCHED1 in maize (Zea mays), CYCLOIDEA (CYC) in Antirrhinum majus, and PROLIFERATING CELL NUCLEAR ANTIGEN FACTOR in rice (Oryza sativa).32 Among the 24 TCP proteins encoded by the Arabidopsis genome, 13 are classified as members of the class I/TCP-P and the other 11 TCPs fall into the class II/TCP-C, which is further subdivided into the CYC/TB1 and CINCINNATA (CIN) subgroups.32,33 TCP proteins have been demonstrated to control a multitude of plant biological processes, such as floral symmetry, plant architecture, the morphogenesis of lateral organs, leaf senescence, and defense response.34-44 In Arabidopsis, several TCP proteins are known to participate in developmental processes via regulating the biosynthetic pathways of plant hormones.43-46 TCP1 directly activates DWARF4 expression to promote brassinosteroid (BR) biosynthesis, thereby resulting in the longitudinal elongation of leaves and stems.45,46 The miRJAW-targeted CIN-TCPs (TCP2–4, TCP10, and TCP24) operate redundantly to regulate leaf senescence and defense response via directly activating LIPOXYGENASE2 (LOX2) expression to modulate jasmonic acid (JA) biosynthesis.43,44
Combinatorial analysis of transgenic plants expressing the miRJAW-resistant mTCP3 and the dominant-negative TCP3SRDX has recently revealed a novel regulatory role for TCP3 in enhancing flavonoid biosynthesis.12 Seedlings and seeds of mTCP3 plants hyperaccumulate flavonols, anthocyanins and PAs, whereas slightly reduced levels of these compounds are detected in TCP3SRDX plants.12 R2R3-MYBs not only activate the transcription of EBGs but also participate in the formation of the MBW complex to control the expression of LBGs.8,14,27,47 TCP3 interacts with R2R3-MYBs but neither with bHLHs nor with TTG1 and significantly strengthens the transcriptional activation capacity of TT8-bound R2R3-MYBs.12 Heterodimerization of TCP3 with MYB12 or MYB111 likely upregulates EBGs and hence enhances flavonol production. The interactions of TCP3 with anthocyanin- and PA-specific R2R3-MYBs suggest that TCP3 acts in a manner similar to TT1 to promote flavonoid production via stabilizing the formation of the MBW complexes and thus stimulating the expression of LBGs. In agreement, mTCP3 expression activates many enzymatic and regulatory genes involved in the early (4CL, CHS/TT4, F3H/TT6) and late steps (DFR/TT3, LDOX/TT18, PAP1) of the flavonoid pathway and downregulation of these respective genes is also detected in TCP3SRDX plants.12 The failure to detect the interaction between TCP3 and GL1 is consistent with the lack of trichome phenotypes in transgenic plants, lending support to the specificity of TCP3 interactions with flavonoid R2R3-MYBs. Interestingly, TCP3 forms a heterodimer with MYBL2 in yeast cells and it is therefore also possible that TCP3 reinforces the formation of the MBW complex via binding to MYBL2 and thus freeing bHLHs.12
Additionally, transgenic plants expressing mTCP3 display altered leaf phyllotaxy, abnormal vasculature patterning, reduced apical dominance, impaired root development, and reduced organ size.12 These developmental abnormalities are reminiscent of auxin-deficient or -insensitive mutants.48-50 Microarray analysis of mTCP3 seedlings reveals many auxin-related genes with reduced expression levels, including the early auxin-responsive genes (GH3.17 and AUX/IAA29), the auxin transporter protein genes (PGP19, PGP1, and AUX1) and the auxin receptor gene TIR1.12 Contrarily, enhanced expression of several auxin-related genes (AUX/IAA29, PAR1, and PGP1) is detectable in TCP3SRDX plants.12 In conjunction with the data from studies of the auxin response reporter pDR5::GUS and the auxin efflux carrier pPIN1::PIN1-GFP, morphological defects and transcriptome data of transgenic plants strongly indicate that TCP3 negatively modulates auxin response, likely by weakening auxin transport capacity.
Endogenous flavonols, including quercetins and kaempferols, are considered to be negative regulators of cellular auxin efflux and polar auxin transport.51-55 Altering in planta flavonol levels via applying exogenous flavonols, activating or inactivating the flavonoid pathway interferes with auxin transport capacity and thus affects many auxin-related biological processes.51-56 Expression of mTCP3 in wild type plants strengthens flavonol production and causes auxin-related developmental abnormalities.12 The knockout mutant tt4–11 does not produce any class of flavonoids57,58 and is therefore utilized to unveil the regulatory hierarchy of TCP3 for these 2 biological processes. Phenotypic analysis of mTCP3/tt4–11 plants demonstrates that the absence of flavonoid biosynthesis in the tt4–11 mutant disables the auxin-related defects caused by expressing mTCP3.12 Given this observation, compromised auxin response observed in mTCP3 plants is thus due to heightened flavonol production. Taken together, these results indicate that TCP3 promotes flavonol accumulation via binding to R2R3-MYBs and thus reduces auxin response likely by impairing auxin transport capacity.
Conclusions and Perspectives
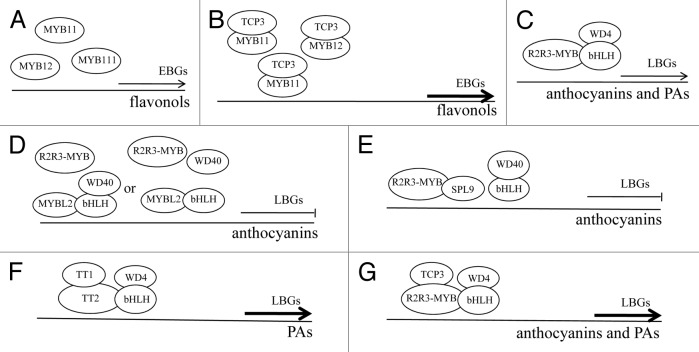
Based on the data presented above, a model is proposed to elucidate how the aforementioned fine-tuners control flavonoid biosynthesis (Fig. 2). Three closely related R2R3-MYBs (MYB11, MYB12, and MYB111) redundantly activate the transcription of EBGs (Fig. 2A). TCP3 interacts with these 3 R2R3-MYBs to stimulate the expression of EBGs and thus promotes flavonol production (Fig. 2B). The ternary MBW complex regulates the biosynthesis of anthocyanins and PAs via directly activating the late flavonoid biosynthetic steps (Fig. 2C). The negative fine-tuners of the flavonoid pathway disrupt the formation of the MBW complex via competitively interacting with bHLHs in the case of MYBL2 (Fig. 2D) or with R2R3-MYBs in the case of SPL9 (Fig. 2E). By contrast, the positive fine-tuners, including TT1 and TCP3, organize the formation of the MBW complex by synergistically associating with R2R3-MYBs (Fig. 2F, G). The counteracted or enhanced activity of the MBW complex further reduces or enhances flavonoid biosynthesis via affecting the expression of LBGs, respectively. This fine-tuning of the flavonoid pathway is therefore achieved by competitive or synergistic protein interactions that disrupt or organize the formation of the MBW complex, allowing plants to respond to numerous developmental and environmental signals during growth and development.3,4,10-12 Intriguingly, 4 closely related R2R3-MYBs (MYB3, MYB4, MYB7, and MYB32) from the R2R3-MYB subgroup 4 may also act as fine-tuners of the flavonoid pathway in a manner similar to MYBL2.6,13 In addition, 3 LATERAL ORGAN BOUNDARY DOMAIN (LBD) proteins (LBD37, LBD38, and LBD39) operate upstream of PAP1 and PAP2 to negatively modulate anthocyanin production.59 Activation of EBGs by TT1 and PAPs as well as the positive feedback regulation of TT8 expression by the MBW complex adds another layer of complexity to the regulatory mechanism of the flavonoid pathway.60-62
Figure 2. A schematic model for the fine-tuning of the flavonoid biosynthetic pathway. MYB11, MYB12, and MYB111 redundantly activate the expression of EBGs (A). TCP3 associates with these 3 R2R3-MYBs to stimulate the transcription of EBGs and thereby enhances flavonol production (B). The ternary R2R3-MYB/bHLH/WD40 (MBW) complex activates the expression of LBGs and thus controls the biosynthesis of anthocyanins and PAs (C). MYBL2 and SPL9 behave as negative fine-tuners of anthocyanin biosynthesis and disrupt the formation of the MBW complex via interacting competitively with bHLHs in the case of MYBL2 (D) or with R2R3-MYBs in the case of SPL9 (E). By contrast, TT1 acts as a positive fine-tuner of the PA biosynthesis in the endothelial cells of the testa (F) and TCP3 functions as a positive fine-tuner of both anthocyanin- and PA-specific MBW complexes (G). The 2 positive fine-tuners TT1 and TCP3 organize the formation of the MBW complex by synergistically associating with R2R3-MYBs. The counteracted or enhanced activity of the MBW complex reduces or heightens the biosynthesis of anthocyanins and PAs via influencing the expression of LBGs, respectively. Thin arrows indicate the transcription of LBGs and EBGs controlled by the ternary MBW complex and MYB11, MYB12, or MYB111, respectively. Hammers denote the inhibitory effects of MYBL2 or SPL9 on the transcription of LBGs. Thick arrows represent the heightened expression of EBGs or LBGs.
The flavonoid biosynthetic pathway has been one of the most intensively investigated metabolic systems in plants. Although numerous transcriptional regulators have been identified and characterized, many gaps still remain in our understanding of how this metabolic pathway is transcriptionally regulated in response to environmental and developmental signals. The availability of genomics data sets for model plant species, including Arabidopsis, rice, mosses, and Medicago truncatula, offers a good opportunity to investigate transcriptional control of flavonoid production from a completely new perspective. Advancing our knowledge of the flavonoid pathway will favor the development of new biotechnological tools for the generation of value-added plants with optimized flavonoid contents.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1.Winkel-Shirley B. Flavonoid biosynthesis. A colorful model for genetics, biochemistry, cell biology, and biotechnology. Plant Physiol. 2001;126:485–93. doi: 10.1104/pp.126.2.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nesi N, Debeaujon I, Jond C, Stewart AJ, Jenkins GI, Caboche M, Lepiniec L. The TRANSPARENT TESTA16 locus encodes the ARABIDOPSIS BSISTER MADS domain protein and is required for proper development and pigmentation of the seed coat. Plant Cell. 2002;14:2463–79. doi: 10.1105/tpc.004127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Appelhagen I, Lu GH, Huep G, Schmelzer E, Weisshaar B, Sagasser M. TRANSPARENT TESTA1 interacts with R2R3-MYB factors and affects early and late steps of flavonoid biosynthesis in the endothelium of Arabidopsis thaliana seeds. Plant J. 2011;67:406–19. doi: 10.1111/j.1365-313X.2011.04603.x. [DOI] [PubMed] [Google Scholar]
- 4.Gou JY, Felippes FF, Liu CJ, Weigel D, Wang JW. Negative regulation of anthocyanin biosynthesis in Arabidopsis by a miR156-targeted SPL transcription factor. Plant Cell. 2011;23:1512–22. doi: 10.1105/tpc.111.084525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mehrtens F, Kranz H, Bednarek P, Weisshaar B. The Arabidopsis transcription factor MYB12 is a flavonol-specific regulator of phenylpropanoid biosynthesis. Plant Physiol. 2005;138:1083–96. doi: 10.1104/pp.104.058032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stracke R, Ishihara H, Huep G, Barsch A, Mehrtens F, Niehaus K, Weisshaar B. Differential regulation of closely related R2R3-MYB transcription factors controls flavonol accumulation in different parts of the Arabidopsis thaliana seedling. Plant J. 2007;50:660–77. doi: 10.1111/j.1365-313X.2007.03078.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stafford HA. Flavonoid evolution: an enzymic approach. Plant Physiol. 1991;96:680–5. doi: 10.1104/pp.96.3.680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ramsay NA, Glover BJ. MYB-bHLH-WD40 protein complex and the evolution of cellular diversity. Trends Plant Sci. 2005;10:63–70. doi: 10.1016/j.tplants.2004.12.011. [DOI] [PubMed] [Google Scholar]
- 9.Ishida T, Hattori S, Sano R, Inoue K, Shirano Y, Hayashi H, Shibata D, Sato S, Kato T, Tabata S, et al. Arabidopsis TRANSPARENT TESTA GLABRA2 is directly regulated by R2R3 MYB transcription factors and is involved in regulation of GLABRA2 transcription in epidermal differentiation. Plant Cell. 2007;19:2531–43. doi: 10.1105/tpc.107.052274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dubos C, Le Gourrierec J, Baudry A, Huep G, Lanet E, Debeaujon I, Routaboul JM, Alboresi A, Weisshaar B, Lepiniec L. MYBL2 is a new regulator of flavonoid biosynthesis in Arabidopsis thaliana. Plant J. 2008;55:940–53. doi: 10.1111/j.1365-313X.2008.03564.x. [DOI] [PubMed] [Google Scholar]
- 11.Matsui K, Umemura Y, Ohme-Takagi M. AtMYBL2, a protein with a single MYB domain, acts as a negative regulator of anthocyanin biosynthesis in Arabidopsis. Plant J. 2008;55:954–67. doi: 10.1111/j.1365-313X.2008.03565.x. [DOI] [PubMed] [Google Scholar]
- 12.Li S, Zachgo S. TCP3 interacts with R2R3-MYB proteins, promotes flavonoid biosynthesis and negatively regulates the auxin response in Arabidopsis thaliana. Plant J. 2013;76:901–13. doi: 10.1111/tpj.12348. [DOI] [PubMed] [Google Scholar]
- 13.Zimmermann IM, Heim MA, Weisshaar B, Uhrig JF. Comprehensive identification of Arabidopsis thaliana MYB transcription factors interacting with R/B-like BHLH proteins. Plant J. 2004;40:22–34. doi: 10.1111/j.1365-313X.2004.02183.x. [DOI] [PubMed] [Google Scholar]
- 14.Dubos C, Stracke R, Grotewold E, Weisshaar B, Martin C, Lepiniec L. MYB transcription factors in Arabidopsis. Trends Plant Sci. 2010;15:573–81. doi: 10.1016/j.tplants.2010.06.005. [DOI] [PubMed] [Google Scholar]
- 15.Feller A, Machemer K, Braun EL, Grotewold E. Evolutionary and comparative analysis of MYB and bHLH plant transcription factors. Plant J. 2011;66:94–116. doi: 10.1111/j.1365-313X.2010.04459.x. [DOI] [PubMed] [Google Scholar]
- 16.Schellmann S, Hülskamp M, Uhrig J. Epidermal pattern formation in the root and shoot of Arabidopsis. Biochem Soc Trans. 2007;35:146–8. doi: 10.1042/BST0350146. [DOI] [PubMed] [Google Scholar]
- 17.Simon M, Lee MM, Lin Y, Gish L, Schiefelbein J. Distinct and overlapping roles of single-repeat MYB genes in root epidermal patterning. Dev Biol. 2007;311:566–78. doi: 10.1016/j.ydbio.2007.09.001. [DOI] [PubMed] [Google Scholar]
- 18.Wang S, Kwak SH, Zeng Q, Ellis BE, Chen XY, Schiefelbein J, Chen JG. TRICHOMELESS1 regulates trichome patterning by suppressing GLABRA1 in Arabidopsis. Development. 2007;134:3873–82. doi: 10.1242/dev.009597. [DOI] [PubMed] [Google Scholar]
- 19.Koes R, Verweij W, Quattrocchio F. Flavonoids: a colorful model for the regulation and evolution of biochemical pathways. Trends Plant Sci. 2005;10:236–42. doi: 10.1016/j.tplants.2005.03.002. [DOI] [PubMed] [Google Scholar]
- 20.Riese M, Höhmann S, Saedler H, Münster T, Huijser P. Comparative analysis of the SBP-box gene families in P. patens and seed plants. Gene. 2007;401:28–37. doi: 10.1016/j.gene.2007.06.018. [DOI] [PubMed] [Google Scholar]
- 21.Xing S, Salinas M, Höhmann S, Berndtgen R, Huijser P. miR156-targeted and nontargeted SBP-box transcription factors act in concert to secure male fertility in Arabidopsis. Plant Cell. 2010;22:3935–50. doi: 10.1105/tpc.110.079343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu G, Park MY, Conway SR, Wang JW, Weigel D, Poethig RS. The sequential action of miR156 and miR172 regulates developmental timing in Arabidopsis. Cell. 2009;138:750–9. doi: 10.1016/j.cell.2009.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yamaguchi A, Wu MF, Yang L, Wu G, Poethig RS, Wagner D. The microRNA-regulated SBP-Box transcription factor SPL3 is a direct upstream activator of LEAFY, FRUITFULL, and APETALA1. Dev Cell. 2009;17:268–78. doi: 10.1016/j.devcel.2009.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yu N, Cai WJ, Wang S, Shan CM, Wang LJ, Chen XY. Temporal control of trichome distribution by microRNA156-targeted SPL genes in Arabidopsis thaliana. Plant Cell. 2010;22:2322–35. doi: 10.1105/tpc.109.072579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dean G, Cao Y, Xiang D, Provart NJ, Ramsay L, Ahad A, White R, Selvaraj G, Datla R, Haughn G. Analysis of gene expression patterns during seed coat development in Arabidopsis. Mol Plant. 2011;4:1074–91. doi: 10.1093/mp/ssr040. [DOI] [PubMed] [Google Scholar]
- 26.Nesi N, Jond C, Debeaujon I, Caboche M, Lepiniec L. The Arabidopsis TT2 gene encodes an R2R3 MYB domain protein that acts as a key determinant for proanthocyanidin accumulation in developing seed. Plant Cell. 2001;13:2099–114. doi: 10.1105/TPC.010098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hichri I, Barrieu F, Bogs J, Kappel C, Delrot S, Lauvergeat V. Recent advances in the transcriptional regulation of the flavonoid biosynthetic pathway. J Exp Bot. 2011;62:2465–83. doi: 10.1093/jxb/erq442. [DOI] [PubMed] [Google Scholar]
- 28.Englbrecht CC, Schoof H, Böhm S. Conservation, diversification and expansion of C2H2 zinc finger proteins in the Arabidopsis thaliana genome. BMC Genomics. 2004;5:39. doi: 10.1186/1471-2164-5-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Crawford BC, Ditta G, Yanofsky MF. The NTT gene is required for transmitting-tract development in carpels of Arabidopsis thaliana. Curr Biol. 2007;17:1101–8. doi: 10.1016/j.cub.2007.05.079. [DOI] [PubMed] [Google Scholar]
- 30.Petricka JJ, Clay NK, Nelson TM. Vein patterning screens and the defectively organized tributaries mutants in Arabidopsis thaliana. Plant J. 2008;56:251–63. doi: 10.1111/j.1365-313X.2008.03595.x. [DOI] [PubMed] [Google Scholar]
- 31.Sagasser M, Lu GH, Hahlbrock K, Weisshaar B. A. thaliana TRANSPARENT TESTA 1 is involved in seed coat development and defines the WIP subfamily of plant zinc finger proteins. Genes Dev. 2002;16:138–49. doi: 10.1101/gad.212702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Martín-Trillo M, Cubas P. TCP genes: a family snapshot ten years later. Trends Plant Sci. 2010;15:31–9. doi: 10.1016/j.tplants.2009.11.003. [DOI] [PubMed] [Google Scholar]
- 33.Kosugi S, Ohashi Y. DNA binding and dimerization specificity and potential targets for the TCP protein family. Plant J. 2002;30:337–48. doi: 10.1046/j.1365-313X.2002.01294.x. [DOI] [PubMed] [Google Scholar]
- 34.Luo D, Carpenter R, Vincent C, Copsey L, Coen E. Origin of floral asymmetry in Antirrhinum. Nature. 1996;383:794–9. doi: 10.1038/383794a0. [DOI] [PubMed] [Google Scholar]
- 35.Aguilar-Martínez JA, Poza-Carrión C, Cubas P. Arabidopsis BRANCHED1 acts as an integrator of branching signals within axillary buds. Plant Cell. 2007;19:458–72. doi: 10.1105/tpc.106.048934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Niwa M, Daimon Y, Kurotani K, Higo A, Pruneda-Paz JL, Breton G, Mitsuda N, Kay SA, Ohme-Takagi M, Endo M, et al. BRANCHED1 interacts with FLOWERING LOCUS T to repress the floral transition of the axillary meristems in Arabidopsis. Plant Cell. 2013;25:1228–42. doi: 10.1105/tpc.112.109090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kieffer M, Master V, Waites R, Davies B. TCP14 and TCP15 affect internode length and leaf shape in Arabidopsis. Plant J. 2011;68:147–58. doi: 10.1111/j.1365-313X.2011.04674.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nath U, Crawford BC, Carpenter R, Coen E. Genetic control of surface curvature. Science. 2003;299:1404–7. doi: 10.1126/science.1079354. [DOI] [PubMed] [Google Scholar]
- 39.Nag A, King S, Jack T. miR319a targeting of TCP4 is critical for petal growth and development in Arabidopsis. Proc Natl Acad Sci U S A. 2009;106:22534–9. doi: 10.1073/pnas.0908718106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Palatnik JF, Allen E, Wu X, Schommer C, Schwab R, Carrington JC, Weigel D. Control of leaf morphogenesis by microRNAs. Nature. 2003;425:257–63. doi: 10.1038/nature01958. [DOI] [PubMed] [Google Scholar]
- 41.Koyama T, Furutani M, Tasaka M, Ohme-Takagi M. TCP transcription factors control the morphology of shoot lateral organs via negative regulation of the expression of boundary-specific genes in Arabidopsis. Plant Cell. 2007;19:473–84. doi: 10.1105/tpc.106.044792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sarvepalli K, Nath U. Hyper-activation of the TCP4 transcription factor in Arabidopsis thaliana accelerates multiple aspects of plant maturation. Plant J. 2011;67:595–607. doi: 10.1111/j.1365-313X.2011.04616.x. [DOI] [PubMed] [Google Scholar]
- 43.Schommer C, Palatnik JF, Aggarwal P, Chételat A, Cubas P, Farmer EE, Nath U, Weigel D. Control of jasmonate biosynthesis and senescence by miR319 targets. PLoS Biol. 2008;6:e230. doi: 10.1371/journal.pbio.0060230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sugio A, Kingdom HN, MacLean AM, Grieve VM, Hogenhout SA. Phytoplasma protein effector SAP11 enhances insect vector reproduction by manipulating plant development and defense hormone biosynthesis. Proc Natl Acad Sci U S A. 2011;108:E1254–63. doi: 10.1073/pnas.1105664108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Guo Z, Fujioka S, Blancaflor EB, Miao S, Gou X, Li J. TCP1 modulates brassinosteroid biosynthesis by regulating the expression of the key biosynthetic gene DWARF4 in Arabidopsis thaliana. Plant Cell. 2010;22:1161–73. doi: 10.1105/tpc.109.069203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Koyama T, Sato F, Ohme-Takagi M. A role of TCP1 in the longitudinal elongation of leaves in Arabidopsis. Biosci Biotechnol Biochem. 2010;74:2145–7. doi: 10.1271/bbb.100442. [DOI] [PubMed] [Google Scholar]
- 47.Gonzalez A, Zhao M, Leavitt JM, Lloyd AM. Regulation of the anthocyanin biosynthetic pathway by the TTG1/bHLH/Myb transcriptional complex in Arabidopsis seedlings. Plant J. 2008;53:814–27. doi: 10.1111/j.1365-313X.2007.03373.x. [DOI] [PubMed] [Google Scholar]
- 48.Dharmasiri N, Dharmasiri S, Estelle M. The F-box protein TIR1 is an auxin receptor. Nature. 2005;435:441–5. doi: 10.1038/nature03543. [DOI] [PubMed] [Google Scholar]
- 49.Yin XJ, Volk S, Ljung K, Mehlmer N, Dolezal K, Ditengou F, Hanano S, Davis SJ, Schmelzer E, Sandberg G, et al. Ubiquitin lysine 63 chain forming ligases regulate apical dominance in Arabidopsis. Plant Cell. 2007;19:1898–911. doi: 10.1105/tpc.107.052035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Leyser HM, Lincoln CA, Timpte C, Lammer D, Turner J, Estelle M. Arabidopsis auxin-resistance gene AXR1 encodes a protein related to ubiquitin-activating enzyme E1. Nature. 1993;364:161–4. doi: 10.1038/364161a0. [DOI] [PubMed] [Google Scholar]
- 51.Kuhn BM, Geisler M, Bigler L, Ringli C. Flavonols accumulate asymmetrically and affect auxin transport in Arabidopsis. Plant Physiol. 2011;156:585–95. doi: 10.1104/pp.111.175976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Murphy A, Peer WA, Taiz L. Regulation of auxin transport by aminopeptidases and endogenous flavonoids. Planta. 2000;211:315–24. doi: 10.1007/s004250000300. [DOI] [PubMed] [Google Scholar]
- 53.Peer WA, Bandyopadhyay A, Blakeslee JJ, Makam SN, Chen RJ, Masson PH, Murphy AS. Variation in expression and protein localization of the PIN family of auxin efflux facilitator proteins in flavonoid mutants with altered auxin transport in Arabidopsis thaliana. Plant Cell. 2004;16:1898–911. doi: 10.1105/tpc.021501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Petrásek J, Friml J. Auxin transport routes in plant development. Development. 2009;136:2675–88. doi: 10.1242/dev.030353. [DOI] [PubMed] [Google Scholar]
- 55.Santelia D, Henrichs S, Vincenzetti V, Sauer M, Bigler L, Klein M, Bailly A, Lee Y, Friml J, Geisler M, et al. Flavonoids redirect PIN-mediated polar auxin fluxes during root gravitropic responses. J Biol Chem. 2008;283:31218–26. doi: 10.1074/jbc.M710122200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Buer CS, Djordjevic MA. Architectural phenotypes in the transparent testa mutants of Arabidopsis thaliana. J Exp Bot. 2009;60:751–63. doi: 10.1093/jxb/ern323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Peer WA, Brown DE, Tague BW, Muday GK, Taiz L, Murphy AS. Flavonoid accumulation patterns of transparent testa mutants of arabidopsis. Plant Physiol. 2001;126:536–48. doi: 10.1104/pp.126.2.536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shirley BW, Kubasek WL, Storz G, Bruggemann E, Koornneef M, Ausubel FM, Goodman HM. Analysis of Arabidopsis mutants deficient in flavonoid biosynthesis. Plant J. 1995;8:659–71. doi: 10.1046/j.1365-313X.1995.08050659.x. [DOI] [PubMed] [Google Scholar]
- 59.Rubin G, Tohge T, Matsuda F, Saito K, Scheible WR. Members of the LBD family of transcription factors repress anthocyanin synthesis and affect additional nitrogen responses in Arabidopsis. Plant Cell. 2009;21:3567–84. doi: 10.1105/tpc.109.067041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Baudry A, Caboche M, Lepiniec L. TT8 controls its own expression in a feedback regulation involving TTG1 and homologous MYB and bHLH factors, allowing a strong and cell-specific accumulation of flavonoids in Arabidopsis thaliana. Plant J. 2006;46:768–79. doi: 10.1111/j.1365-313X.2006.02733.x. [DOI] [PubMed] [Google Scholar]
- 61.Borevitz JO, Xia Y, Blount J, Dixon RA, Lamb C. Activation tagging identifies a conserved MYB regulator of phenylpropanoid biosynthesis. Plant Cell. 2000;12:2383–94. doi: 10.1105/tpc.12.12.2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tohge T, Nishiyama Y, Hirai MY, Yano M, Nakajima J, Awazuhara M, Inoue E, Takahashi H, Goodenowe DB, Kitayama M, et al. Functional genomics by integrated analysis of metabolome and transcriptome of Arabidopsis plants over-expressing an MYB transcription factor. Plant J. 2005;42:218–35. doi: 10.1111/j.1365-313X.2005.02371.x. [DOI] [PubMed] [Google Scholar]