Abstract
Phakopsora pachyrhizi, a fungus that causes rust disease on soybean, has potential to impart significant yield loss and disrupt food security and animal feed production. Rpp1 is a soybean gene that confers immunity to soybean rust, and it is important to understand how it regulates the soybean defense system and to use this knowledge to protect commercial crops. It was previously discovered that some soybean proteins resembling transcription factors accumulate in the nucleus of Rpp1 soybeans. To determine if they contribute to immunity, Bean pod mottle virus was used to attenuate or silence the expression of their genes. Rpp1 plants subjected to virus-induced gene silencing exhibited reduced amounts of RNA for 5 of the tested genes, and the plants developed rust-like symptoms after subsequent inoculation with fungal spores. Symptoms were associated with the accumulation of rust fungal RNA and protein. Silenced plants also had reduced amounts of RNA for the soybean Myb84 transcription factor and soybean isoflavone O-methyltransferase, both of which are important to phenylpropanoid biosynthesis and lignin formation, crucial components of rust resistance. These results help resolve some of the genes that contribute to Rpp1-mediated immunity and improve upon the knowledge of the soybean defense system. It is possible that these genes could be manipulated to enhance rust resistance in otherwise susceptible soybean cultivars.
Keywords: soybean rust, transcription factor, disease resistance, gene silencing, Rpp1
Introduction
Soybean rust is caused by an obligate, biotrophic, basidiomycete fungus, Phakopsora pachyrhizi.1 The disease is currently controlled with fungicides. Farmers would rather grow resistant soybeans, but the few known resistance (R) genes to soybean rust are not deployed in commercial cultivars. Worse still, soybean rust isolates have the potential to overcome these genes.2 Not to be deterred by a seemingly Sisyphean problem, scientists hoping to improve resistance and curtail fungicide application have concentrated their research efforts over the last decade to identify the genetic positions of the few known R-genes, supporting the goal of determining how the genes function and how the fungus overcomes them.3,4,5,6,7,8
During soybean invasion, there are multiple points where the rust fungus makes contact with leaf cells where it could be recognized by the plant immune system.9,10,11,12 One opportunity for detection is at the fungal haustorium/plant cell interface where the fungus acquires its nutrients from the plant.13 While not yet proven for soybean rust, it is known for other rust fungi that rusts secrete effector proteins from their haustoria to facilitate pathogenicity. This includes disabling the plant immune system, stabilizing the host cell, and inducing the plant cell to produce nutrients for the fungus.14,15,16,17,18,19
If a pathogen effector protein is detected or recognized by a plant R protein, a plant defense response called effector-triggered immunity (ETI) ensues.20 The phenotypic hallmark of ETI is hypersensitive resistance, which often leads to programmed death of the plant cells at the site of infection and cessation of pathogen accumulation and spread. In Arabidopsis thaliana responding to bacterial pathogen effector proteins, the biochemistry associated with ETI includes Mitogen-Activated Protein (MAP) kinase signaling, reactive oxidative species generation, gene expression and protein alterations, salicylic acid and ethylene hormonal signaling, fortification of cell walls, and production of antimicrobial chemicals, peptides, and enzymes.21 In resistant soybeans with R-genes Resistance to P. pachyrhizi (Rpp) 2 and 3, the expressions of genes controlling similar biochemical processes are induced during rust infection.22,23 In soybeans with R-genes overcome by rusts, susceptibility may be predicated on evasive effector proteins.
The dominant Rpp1 rust resistance gene was introduced in the 1980s into soybean cultivar Williams 82 (W82) by a cross with soybean accession PI200492.24 Progeny were backcrossed to W82 5 times and selected for W82 agronomic traits and Rpp1-mediated immunity. The W82/Rpp1 isogenic line, herein called Rpp1, was analyzed for genomic sequence polymorphisms and was indistinguishable from the parent W82 except for the 0.4 cM Rpp1 locus.5 These data imply that W82 harbors all genes required for rust immunity except those within the Rpp1 locus. It was with Rpp1 that a proteomic screen was performed to identify transcription factors (TFs) and other proteins inherent to W82 that might exhibit increased accumulation in the nucleus during Rpp1-mediated immunity.25 The hypothesis was that the activated Rpp1 protein led to the migration of proteins to the nucleus or led to a change in accumulation of proteins in the nucleus that then influenced defense-related gene expression. The screen resolved 111 proteins, many of which had sequence homology to TFs and other nuclear proteins found in other plants, or which had predicted nuclear localization signals. There was little overlap between these proteins and the genes with altered expressions in rust-infected Rpp2 plants,22 and this implied that the proteins may be important to Rpp1-mediated immunity.
Scientists are now exploiting a process known as virus-induced gene silencing (VIGS) to determine whether soybean genes are important to disease resistance.26,27 VIGS involves challenging soybean plants with recombinant Bean pod mottle virus (BPMV) expressing a small fragment of a soybean gene thought to be involved in disease resistance against another pathogen. VIGS occurs prior to test pathogen inoculation and if successful, induces the endogenous RNAi-mediated defense system in soybean and lowers the gene expression of the candidate gene. A concomitant reduction in resistance after challenge with the pathogen implicates the silenced gene in defense. Pandey et al. (2011) used VIGS to confirm that the soybean orthologs of A. thaliana genes Enhanced Disease Susceptibility 1 (GmEDS1), Phytoalexin Deficient 4 (GmPAD4), and Nonexpressor of Pathogenesis-Related genes 1 (GmNPR1), regulators of salicylic acid signaling crucial for defense against biotrophic fungal and bacterial pathogens, function as part of Rpp2-mediated resistance to soybean rust.28 In addition, they showed that the genes for the enzymatic precursors for lignin and antimicrobial compound biosynthesis, Phenylalanine-Ammonia Lyase 1 (GmPAL1), Isoflavone O-Methyltransferase (GmO-MT), and Cytochrome P450 family 83 E12 (Cyp83E12) and 5 TFs that regulate the expression of these genes, also are part of the Rpp2-mediated resistance system.28 VIGS also has been used to identify the rust-resistance gene Rpp4 and other resistance genes for other pathogens of soybean.6,29,30,31 Because VIGS is well-suited for dissecting the soybean defense system, we used it to investigate several of the genes of the proteins linked to Rpp1-mediated immunity.25 The results imply that some of the genes are important to Rpp1-mediated defense and add to our much-needed knowledge of the regulation of the soybean immune system.
Results
Candidate gene isolation
Genes for 10 of the proteins with increased accumulation in rust-challenged Rpp1 plants were selected for further characterization (Table 1).25 Their sequences were initially determined from the W82 genome gene models.32 Six of the genes, dubbed C, I, K, J, L, and N, are among a set of soybean TFs predicted from the genome.33 These genes are not classified as WRKY TFs and are not similar to those known to be involved in Rpp2-mediated defense.28 The other 4 candidates were selected for different reasons. Gene A is similar to an A. thaliana candidate lignin biosynthesis gene induced by an Oomycetes pathogen effector.34 Genes E, M, and O are also similar to A. thaliana genes, but have unresolved functions. cDNAs for each of these genes were cloned from W82 RNA. Clones with sequences that perfectly matched the gene models were obtained for all genes except for A, which had a single, silent mutation.
Table 1. Soybean genes targeted for BPMV-induced gene silencing, their features and phenotypes when silenced in Rpp1 plants.
| Soybean gene | Alias | Gene domains | A. thaliana homolog (BLASTP e-value) |
Potential function | Rust symptoms on VIGS-Rpp1 | P. pachyrhizi (RNA or protein) in VIGS-Rpp1 | Reduced gene expression in VIGS-Rpp1 |
|---|---|---|---|---|---|---|---|
| Glyma01 g31830 | A | Dirigent superfamily | AT1G55210 (9e-53) |
Lignin biosynthesis | Yes | Yes | Yes |
| Glyma08 g07830 | E | COG4260 | AT5g64160 (2e-71) |
Unknown | Yes | Yes | Yes |
| Glyma08 g23720 | C | GATA zinc-finger | AT5G15840 (3e-188) |
CONSTANS | No | No | Not tested |
| Glyma12 g30600 | I | None identified | AT5G03740 (1e-23) |
TF | Yes | Yes | Yes |
| Glyma13 g17840 | K | None identified | AT1G09520 (4e-6) |
TF | No | No | No |
| Glyma13 g44430 | J | U1 like zinc finger | AT2G32600 (4e-128) |
TF | Yes | Yes | Yes |
| Glyma14 g11400 | L | PHD superfamily | AT5G20510 (8e-107) |
TF | No | No | Not tested |
| Glyma16 g32940 | M | DUF296 superfamily | AT5G62260 (7e-56) |
Unknown | Yes | Yes | Yes |
| Glyma17 g04670 | N | None identified | AT1G09520 (3e-10) |
TF | No | No | No |
| Glyma18 g01150 | O | CTD superfamily | AT1G24310 (9e-166) |
Unknown | No | No | No |
Virus induced gene silencing
Small, 240–300 bp DNA fragments of coding sequence proximal to the stop codon of the soybean genes were amplified and cloned into a plasmid encoding RNA2 of BPMV. Since the goal was to achieve silencing specificity to the target gene, fragments were chosen to distinguish the target gene from paralogs and homologs. These RNA2 plasmid constructs were co-bombarded with BPMV RNA1M plasmid to soybean seedlings, and the virus was propagated. The recombinant BPMV silencing constructs exhibited typical mottle symptoms on W82 and Rpp1 leaves similarly to the control BPMV 1037 that expresses the green fluorescent protein (Fig. 1).
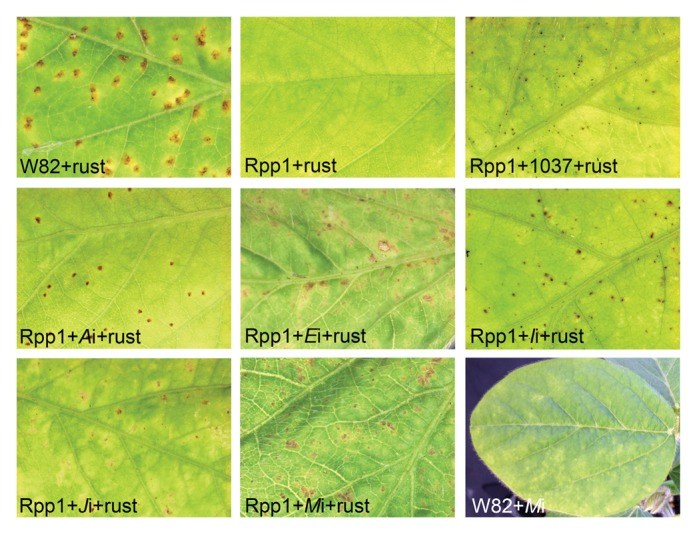
Figure 1. Symptoms on soybeans challenged with soybean rust and BPMV. The W82 variety is susceptible to soybean rust and exhibits lesions and rust pustules by 2 wk after challenge. The Rpp1 isoline is immune to soybean rust and is usually symptomless, but exhibits small sites of necrosis when preinfected with BPMV 1037. Rpp1 challenged with soybean rust exhibits a range of lesion sizes when preinfected with the various BPMV constructs expressing fragments intended to induce silencing of the denoted genes. W82 infected with BPMV-Mi 8 d after inoculation (no rust) exhibit typical viral mottle symptoms on upper leaves (bottom right panel).
Eight to 12 Rpp1 plants were infected with each BPMV construct, alongside 2 W82 and 2 Rpp1 plants infected with 1037. For each experiment, there were also 2 W82 and 2 Rpp1 plants not inoculated with virus. The plants were then challenged with soybean rust isolate Louisiana 4–1 and were monitored for 2 to 3 wk. Louisiana 4–1 produces an immune phenotype on Rpp1 and a susceptible phenotype on W82 (Fig. 1).25,35 Immunity is associated with few visible symptoms and is different than the resistance phenotype controlled by genes Rpp2–4, which produce larger, hypersensitive local lesions. The immune phenotype also can be differentiated from the symptomatic necrotic lesions and rust pustules that are signs of infection in susceptible W82 (Fig. 1). All W82 plants developed rust pustules within 2 wk after inoculation, whereas all Rpp1 plants exhibited no signs of rust infection, except when preinfected with 1037 when small, speckled sites of necrosis and cell death were observed (Fig. 1). We interpreted this as BPMV rendering visible the otherwise imperceptible hypersensitivity in rust-inoculated Rpp1 plants. We therefore sought to identify a compromised-immunity phenotype that was between the susceptible phenotype of a rust-infected W82 and the speckled-immune phenotype for a rust-challenged Rpp1 plant infected with 1037. We observed that rust-challenged plants infected with BPMV-Ai, Ei, Ii, Ji, or Mi exhibited necrotic lesions and rust-like symptoms, albeit in absence of signs of rust sporulation. We attributed this phenotype to compromised Rpp1-mediated immunity (Fig. 1).
The most severe compromised-immunity phenotypes appeared on Rpp1 plants infected with BPMV-Ji and Mi, whereas the least severe phenotype appeared on plants with BPMV-Ii. Not all of the plants tested with each construct exhibited compromised-immunity phenotypes, which implied that there might be different degrees of gene silencing that occurred in each plant. Rpp1 plants inoculated with rust and infected with BPMV-Ci, Ki, Li, Ni, and Oi had phenotypes more similar to rust-challenged Rpp1 plants infected with 1037. Rust challenges were performed at least 5 times over the course of 2 years. The compromised-immunity phenotypes observed on the Rpp1 plants infected with silencing constructs BPMV-Ai, Ei, Ii, Ji, and Mi were consistently reproducible.
Pathogen detection in compromised Rpp1 plants
RNA was extracted from soybean trifoliate leaves from challenges 1 and 2, and quantitative reverse transcriptase-polymerase chain reaction (RT-PCR) was performed to determine if the compromised-immunity phenotypes were associated with increased amounts of pathogen accumulation. The detected amounts of soybean rust tubulin RNA were normalized to the amounts of RNA from a soybean gene constitutively expressed during rust infection.28,36 P. pachyrhizi α-tubulin RNA was detected in positive control W82 plants inoculated with rust, but was not detected in plants not inoculated with rust (Fig. 2). These results confirmed that the assay was specific to soybean rust α-tubulin and did not inadvertently detect soybean RNA. By contrast to W82 plants, only small, but statistically significant amounts of P. pachyrhizi α-tubulin RNA could be detected in Rpp1 plants infected with 1037 and challenged with rust. This confirmed that the plants had been sprayed with rust spores, but that the fungus did not spread as a result of Rpp1-mediated immunity. By comparison to these positive and negative controls, P. pachyrhizi α-tubulin RNA was reliably detected in rust-challenged Rpp1 plants preinfected with BPMV-Ai, Ei, Ii, Ji, and Mi (Fig. 2). Consistent with the varying degrees of the compromised-immunity phenotype observed between plants tested with any one silencing construct, silenced Rpp1 plants also accumulated varied amounts of P. pachyrhizi α-tubulin RNA. This observation could be related to differing degrees of gene silencing or virus titer. It was not possible to distinguish these 2 possibilities by quantitative RT-PCR in 30-d old trifoliate leaves undergoing senescence and deteriorating from disease.
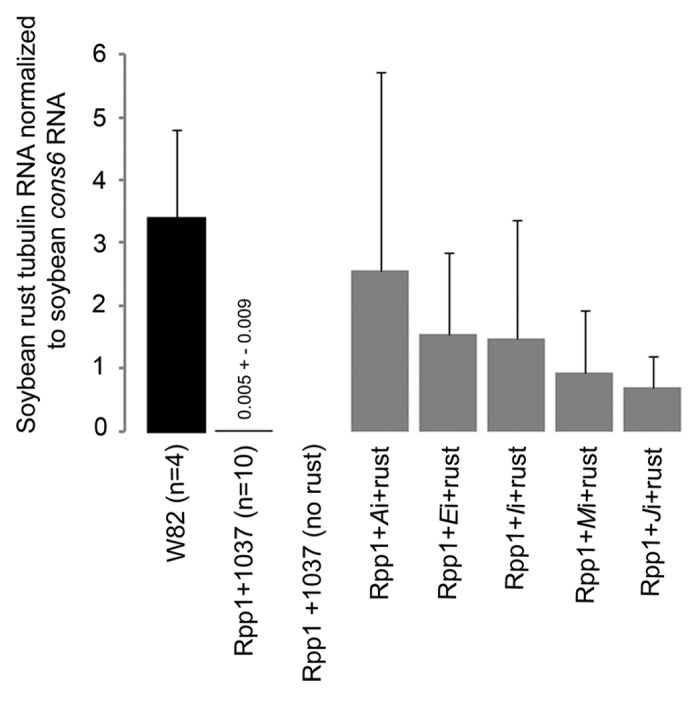
Figure 2. Relative levels of soybean rust tubulin RNA detected in soybean variety W82 and isoline Rpp1 challenged with rust and BPMV. The amount of rust tubulin was normalized to the amount of soybean cons6. Rpp1 plants were preinfected with BPMV 1037 (control) or with the denoted BPMV silencing constructs. Data for silenced plants from replicate rust challenges 1 and 2 are shown. Rust tubulin was not detected in Rpp1 plants not challenged with rust.
To independently confirm that P. pachyrhizi accumulated in Rpp1 plants showing a compromised-immunity phenotype, protein was extracted from the trifoliate leaves from rust challenges 4 and 5 and tested in western blots against an antibody specific to an extracellular P. pachyrhizi protein, PHEP 369, expressed in spores and germlings.37 The PHEP 369 protein was detected in rust-challenged Rpp1 plants infected with BPMV-Ai, Ei, Ii, Ji, and Mi, but the protein was not readily found in rust-challenged 1037-infected Rpp1 controls (Fig. 3). Basing our decision upon the total amount of protein analyzed, we estimated that the amount of rust protein detected in Rpp1 plants exhibiting compromised-immunity was about 100 times less than that found in susceptible W82 plants. As with the quantitative RT-PCR assays, different amounts of P. pachyrhizi protein accumulated between Rpp1 plants infected with the same silencing construct and challenged with rust.
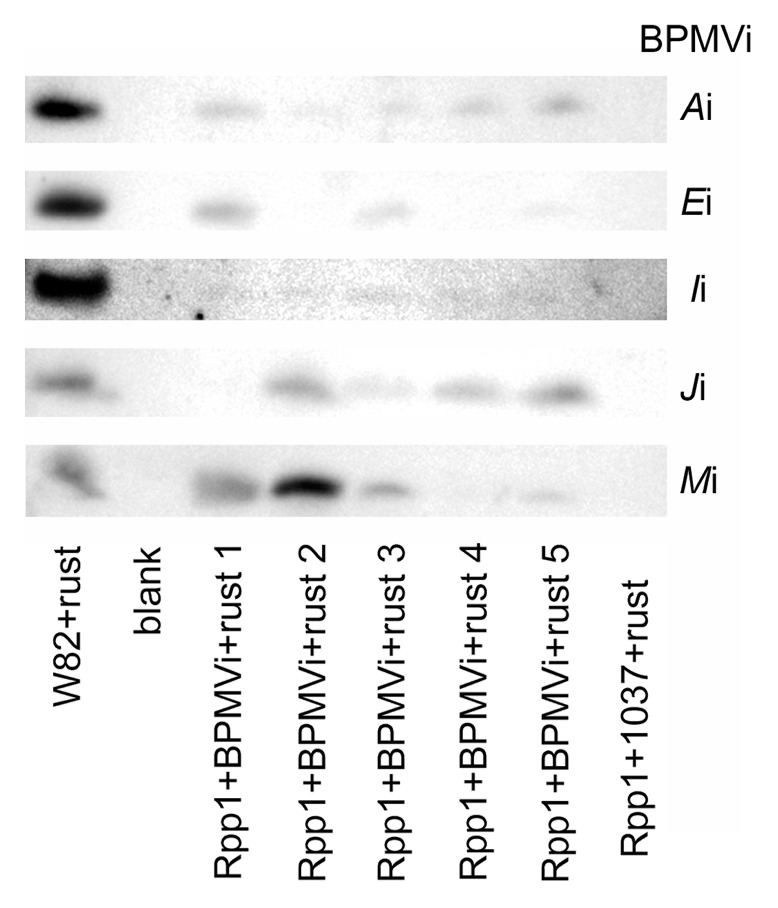
Figure 3. Soybean rust protein PHEP 369 in soybean variety W82 and isoline Rpp1 challenged with rust and BPMV. Rpp1 plants were preinfected with BPMV 1037 (control) or with the denoted BPMV silencing constructs. PHEP 369 was detected with anti-PHEP 369 antibody. One hundred times less total protein from the W82+rust sample was evaluated. Data are from rust challenge 5. Five independent replicates with silencing constructs are shown.
Effects of VIGS on gene expression
We sought to confirm that the compromised-immunity phenotypes were attributed to VIGS. This proved difficult to accomplish in plants challenged with both rust and BPMV, so Rpp1 plants were inoculated with each BPMV silencing construct and were examined 7 d later without any subsequent rust challenge. Using the same low-cycle RT-PCR method from Pandey et al. (2011) to verify VIGS, we observed reduced accumulation of A, E, I, J, and M in Rpp1 plants infected with the respective VIGS constructs (Fig. 4). These results are consistent with the compromised-immunity phenotype associated with each construct. Not all Rpp1 plants infected with any single BPMV construct, however, exhibited the same degree of reduced expression. This observation is consistent with the variation in the accumulation of P. pachyrhizi RNA and protein in plants infected with the same silencing construct.
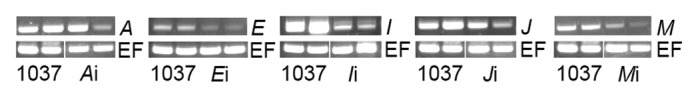
Figure 4. RT-PCR evaluation of the expression of genes targeted by BPMV gene silencing. Two Rpp1 plants inoculated with BPMV 1037 (control) and 2 plants inoculated with the denoted silencing construct are shown. The samples were amplified with primers to the respective gene and elongation factor 1b (EF; control). Reactions were terminated after 27 cycles.
To learn more about how immunity was compromised, we examined after VIGS the expression of marker genes with known roles in soybean rust defense. We examined GmEDS1, GmPAD4, and GmNPR1, which are regulators of salicylic acid-mediated defense signals in soybean,38 and we examined a soybean ortholog of Ethylene Insensitive 2 (EIN2), the central regulator of ethylene signaling that also functions in pathogen defense in A. thaliana.39,40,41 We examined GmPAL1 and GmO-MT, which catalyze reactions for phenylpropanoid biosynthesis and lignin formation, which are crucial to soybean rust defense.22,28,42 We also examined the expression of 2 TF genes, Myb domain protein 84 (GmMyb84), which is linked to GmPAL1 expression, and WRKY DNA-binding protein 36 (GmWRKY36), which is linked to the expression of GmO-MT.28 Low-cycle RT-PCR was performed on Rpp1 plant leaves inoculated with each BPMV silencing construct using primers specific to the marker genes. In all silenced soybeans, but not the 1037 inoculated controls, there were reduced amounts of expression of GmMyb84 and GmO-MT (Fig. 5). Reduced expression of GmO-MT was least pronounced in BPMV-Ei infected plants, which implies that E plays a less prominent regulatory role for GmO-MT. Reduced expression of GmMyb84 was greatest in BPMV-Ji infected plants, which implies that J plays a more prominent regulatory role for GmMyb84. These results imply that phenylpropanoid biosynthesis and lignin formation are necessary for Rpp1-mediated immunity as they are for Rpp2-mediated resistance.
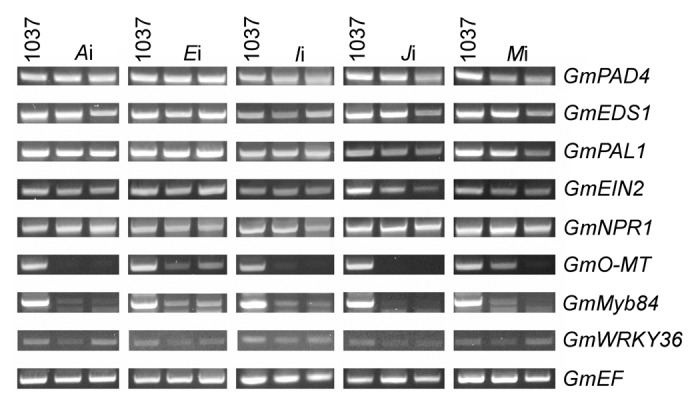
Figure 5. RT-PCR evaluation of the expression of non-target genes after BPMV gene silencing. One Rpp1 plant inoculated with BPMV 1037 (control) and 2 plants inoculated with the denoted silencing construct are shown. Amplification of soybean elongation factor 1b (GmEF) serves as a control. Reactions were terminated after 28 cycles.
Other genes of the soybean defense system may have been affected by VIGS as well (Fig. 5). VIGS with BPMV-Ji may have also lowered GmEDS1, GmPAD4, and GmEIN2 expression, while VIGS with BPMV-Mi may have led to reduced expression of GmEDS1, GmPAD4, and GmPAL1. These results suggest that the salicylic acid and ethylene hormonal signaling pathways may be regulated by Rpp1. These results, however, are partially inconsistent with those reported by Pandey et al. (2011) because neither GmNPR1 nor GmWRKY36, which are transcriptionally downstream of GmPAD4 and GmEDS1 in Rpp2-mediated resistance,28 appeared to be affected in Rpp1 plants. Since these data are at the limits of the dynamic range of measurement for low-cycle RT-PCR, it will be worthwhile to apply more sensitive, complementary assays that can resolve whether GmPAD4, GmEDS1, and GmEIN2 are controlled by J and M and further delineate on a broader scale the genetic networks in which A, E, I, J, and M reside in Rpp1 plants.
Discussion
Our knowledge of the immune system of soybean is limited compared with that for the model plant A. thaliana, but it has been growing in recent years. We now know that Rpp1, Rpp2, and Rpp3 regulate the expression and accumulation of hundreds of genes and proteins within 24 h of soybean rust inoculation.22,23,25 Induced genes include GmEDS1, GmPAD4, and GmNPR1, which regulate the salicylic acid defense pathway, leading to increased phenylpropanoid metabolism through enzymes like GmO-MT.28 These findings are important because they reveal the genes and defense pathways transcriptionally downstream of Rpp1–3, but they are also important because they demonstrate that there are commonalities between the A. thaliana and soybean defense systems. This indicates that plants use conserved mechanisms to combat biotrophic pathogens. And although there are no known rusts that infect A. thaliana, the conservation of defense mechanism means that basic knowledge gained in the A. thaliana model system against pathogens other than rusts may help us understand why soybeans are so susceptible to soybean rust. At the same time, the comparison may allow us to identify the specific soybean defenses best suited for fighting soybean rust.
It is now clear that rust infection leads to the accumulation of proteins in the soybean nucleus.25 These proteins include many apparent TFs which may control the expressions of other genes necessary to regulate the myriad branches of the defense system sufficient to impede rust infection.25 It is not clear, however, how R-proteins like Rpp1, Rpp2, and Rpp3 act upon the nucleus to modulate gene expression, but there is ample evidence in soybeans and in A. thaliana to presume that R-proteins do so directly by acting as TFs themselves or indirectly through signal transduction to other TFs. Under such circumstances, the R-proteins and TFs would need to migrate to the nucleus or they would need to be activated/deactivated in the nucleus after pathogen infection. Evidence supporting such models includes the migration of R-proteins N, RPS4, RRS1, and MLA10 to the nucleus after pathogen recognition,43,44,45,46 the accumulation of EDS1 and NPR1 in the nucleus and the interaction between PAD4 and EDS1 there during defense responses,47,48,49 the nuclear localization of EIN2 and its activation of TFs during defense response,39,40,41,50 and the release of WRKY33 from MPK4 in the nucleus.51
Assuming that these models explain why some proteins accumulate in the nucleus during Rpp1-mediated immunity,25 we sought to validate the roles of nuclear proteins in disease resistance by using VIGS to reduce their expressions and by testing plants for compromised immunity. When the expressions of 5 candidates A, E, I, J, and M were reduced in Rpp1 plants and when these normally immune plants were challenged with rust, the plants exhibited rust-like symptoms and accumulated greater amounts of soybean rust RNA and protein. These results imply that Rpp1 acts upon A, E, I, J, and M to confer immunity to soybean rust. It remains unknown whether the proteins for A, E, I, J, and M indeed act in the nucleus, serve as TFs, or physically interact with Rpp1.
Although reducing the expression of A, E, I, J, and M by VIGS compromised Rpp1-mediated immunity, we never observed the formation of uredial rust pustules required for the successful completion of the rust asexual life cycle. This implies that the rust defenses controlled by Rpp1 were only partially deactivated or that other defense genes were sufficient to suppress uredia formation. We noticed that Rpp1 plants accumulated the greatest amount of soybean rust protein PHEP 369 when the expressions of GmPAD4 and GmEDS1, 2 regulators of salicylic acid signaling, and GmEIN2, a regulator of ethylene signaling, appeared to be reduced. Thus, a rust fungus may need to disrupt several defense pathways to overcome Rpp1-mediated immunity. Likewise, Rpp1-mediated immunity may comprise both redundant and distinct genetic pathways, the sum of which contributes to the limitation of fungal spread.
The results presented here suggest that A, E, I, J, and M are linked to the expression of GmMyb84 and GmO-MT, which may influence the accumulation of phenylpropanoids necessary for lignification and phytoalexin production.28,42 We remark that gene A is similar to an A. thaliana candidate lignin biosynthesis gene that is induced by an Oomycetes effector (Table 1).34 Hence, the regulation of genes for phenylpropanoid metabolism may be essential to Rpp1 immunity, as it is for Rpp2 and Rpp3 resistance to soybean rust and Phaseolus vulgaris Ur-4 resistance to common bean rust.22,23,52 Thus, the genetic control of phenylpropanoid metabolism should be considered when developing soybean varieties with improved resistance or tolerance to soybean rust.
The results from this study on the Rpp1-mediated defense response can be compared with the results reported by Pandey et al. (2011) on the Rpp2-mediated defense response.28 Pandey et al. revealed that VIGS of GmMyb84 reduced the expression of GmPAL, but not GmO-MT, whereas VIGS of GmWRKY36 reduced the expression of GmO-MT, but not GmPAL.28 We, however, show in all silenced plants that a reduction of GmMyb84 expression correlated with reduced expression of GmO-MT, but not GmPAL. We also observed no reduced expression for GmWRKY36 when the expression of GmO-MT was reduced. In light of these possible contradictions, it is important to recognize that these genetic relationships were only evaluated by testing a few marker genes in each study and that the regulation of these genes could be more complex. There may be sophistication to the regulation of Rpp1-mediated immunity that is not yet resolved. We will continue to investigate the transcriptional and proteomic bases of rust pathogenicity and disease resistance accordingly.25,37,52,53,54,55
Materials and Methods
Plants
Glycine max cv Williams 82 (W82) and a Williams 82/Rpp1 inbred isoline (Rpp1) were studied.24,32
DNA cloning
Soybean gene models were obtained from the W82 genome assembly v. 1.0 (www.phytozome.net).32 Genes A, C, E, I, J, K, L, M, N, and O are listed in Table 1, and the DNA oligonucleotide primers used to clone and sequence them are provided in Table 2. cDNAs were amplified from 100 ng W82 RNA using a gene specific 3′ primer and the SuperScript III One-Step RT-PCR kit (Life Technologies, #12574–018). cDNAs were size selected and gel purified using QIAquick (Qiagen, #28704), inserted into pCR2.1 (Life Technologies, #K450001SC), propagated in Escherichia coli, and sequenced using the Sanger method by Genewiz (Germantown, MD).
Table 2. Sequences of oligonucleotides.
| Gene | Name | Purpose | Oligo name | Oligo Sequence |
|---|---|---|---|---|
| Glyma01 g31830.1 | A | gene cloning | Glyma01 g31830.1 F | ATGCCAACCC TCAACCATGT |
| Glyma01 g31830.2 | A | gene cloning | Glyma01 g31830.1 R | TCAATAATGC ATAACATAAA C |
| Glyma08 g07830.1 | E | gene cloning | Glyma08 g07830.1 F | ATGAGCTTGT CTCTTATTCA |
| Glyma08 g07830.1 | E | gene cloning | Glyma08 g07830.1 R | CTAATCAAAC TGTTGCCTTA |
| Glyma08 g23720.1 | C | gene cloning | Glyma08 g23720.1 F | ATGGATGGTA TTCATGGGG |
| Glyma08 g23720.1 | C | gene cloning | Glyma08 g23720.1 R | TCATGATGAA TCATCAGCTT |
| Glyma12 g30600.1 | I | gene cloning | Glyma12 g30600.1 F | ATGGAGTTTT GGGGTGCCG |
| Glyma12 g30600.1 | I | gene cloning | Glyma12 g30600.1 R | TCACTGACCA CCATGCTTTG |
| Glyma13 g17840.1 | K | gene cloning | Glyma13 g17840.1 F | ATGAACTCAC GGCAATCCTC |
| Glyma13 g17840.1 | K | gene cloning | Glyma13 g17840.1 R | TTACGCGCCA CGCCTCCG |
| Glyma13 g44430.1 | J | gene cloning | Glyma13 g44430 up F | CTAAACTAAA AATGGATCGA GA |
| Glyma13 g44430.1 | J | gene cloning | Glyma13 g44430.1 R | CTACTGTCCC ATGTTAGG |
| Glyma14 g11400.1 | L | gene cloning | Glyma14 g11400.1 F | ATGGAGGCAG GTTACAATC |
| Glyma14 g11400.1 | L | gene cloning | Glyma14 g11400.1 R | TCAAGGTCGA GCTCGCTT |
| Glyma16 g32940.1 | M | gene cloning | Glyma16 g32940.1 F | ATGGAGGAAA GAGAGATTTT |
| Glyma16 g32940.1 | M | gene cloning | Glyma16 g32940 down R | AGGGAGCATG TTTAGCAT |
| Glyma17 g04670.1 | N | gene cloning | Glyma17 g04670.1 F | ATGAACTCAC GGCAAGC |
| Glyma17 g04670.1 | N | gene cloning | Glyma17 g04670.1 R | TTACGCGCCA CGCCTTC |
| Glyma18 g01150.1 | O | gene cloning | Glyma18 g01150.1 F | ATGTTTGGCT CCGCTCAACC |
| Glyma18 g01150.1 | O | gene cloning | Glyma18 g01150.1 R | TCAGATTAAA TTCCCATTTT |
| Glyma01 g31830.1 | A | VIGS | A4 = 1g31830 F291 | AAAGGGATCC CACGTGTTGA GCTCCAAGCT TGTG |
| Glyma01 g31830.2 | A | VIGS | A4 = 1g31830 R291 | TTGGGTACCT ATAATGCATA ACATAAAC |
| Glyma08 g23720.1 | C | VIGS | C2 = 8g23720 F243 | AAAGGGATCC CACGTGCAGC AGCAAGATAT TGTT |
| Glyma08 g23720.1 | C | VIGS | C2 = 8g23720 R243 | TTGGGTACCT TGAATCATCA GCTTCTCC |
| Glyma08 g07830.1 | E | VIGS | E1 = 8g07830 264F | AAAGGGATCC CACGTGACTG CAACAAATCC TAAT |
| Glyma08 g07830.1 | E | VIGS | E1 = 8g07830 264R | TTGGGTACCT ATCAAACTGT TGCCTTAG |
| Glyma13 g17840.1 | K | VIGS | K1 = 13 g17840 267F | AAAGGGATCC CACGTGCCTA CTTGTCTCGA TTCT |
| Glyma13 g17840.1 | K | VIGS | K1 = 13 g17840 267R | TTGGGTACCT GTTCTGTTCC TCCTCGTC |
| Glyma14 g11400.1 | L | VIGS | L1 = 14 g11400 300F | AAAGGGATCC CACGTGTCTA AGTCAAACTC TAAG |
| Glyma14 g11400.1 | L | VIGS | L1 = 14 g11400 300R | TTGGGTACCT AGGTCGAGCT CGCTTGTT |
| Glyma17 g04670.1 | N | VIGS | N1 = 17 g04670 297F | AAAGGGATCC CACGTGCTCG ATTCCAACTT CACC |
| Glyma17 g04670.1 | N | VIGS | N1 = 17 g04670 297R | TTGGGTACCT GCCACGCCTT CGGCCAGC |
| Glyma18 g01150.1 | O | VIGS | O4 = 18 g01150 279F | AAAGGGATCC CACGTGAAAT TAGCCACAAT AACT |
| Glyma18 g01150.1 | O | VIGS | O4 = 18 g01150 279R | TTGGGTACCT TTCAGTTCTT GCTGTGTC |
| Glyma12 g30600.1 | I | VIGS | I1 = 12 g30600 300F | AAAGGGATCC CACGTGAAGA AGGCAGATCT AGGA |
| Glyma12 g30600.1 | I | VIGS | I1 = 12 g30600 300R | TTGGGTACCT CTGACCACCA TGCTTTGC |
| Glyma13 g44430.1 | J | VIGS | J+1 = 13 g44430 258F | AAAGGGATCC CACGTGCCAG AGGCGAACAA ACCA |
| Glyma13 g44430.1 | J | VIGS | J+1 = 13 g44430 258R | TTGGGTACCT AGATGGCATT CCACCAGA |
| Glyma16 g32940.1 | M | gene synthesis | M+1FP1 255 | CAGCAGAAAC CGAAGAAGCC AAGGGTGGAG CATATAATAT CAATGGCTGC CCCTATGCAT GTCAACCCTA CTTCAGCTGC |
| Glyma16 g32940.1 | M | gene synthesis | M+1 RP2 255 | TGAAAAGCAGCTGGTGTCATGATTGGCTTTACTCCACCAAGACCAATTCTTATTTCTTCAGCAGCTGAAGTAGGGTTGAC |
| Glyma16 g32940.1 | M | gene synthesis | M+1 FP3 255 | ATGACACCAG CTGCTTTTCA AGTGGACCAC ATTTTTGGCA ATGGCCAAAG CTCTGGGAAC TCAGCTTCTG ATGATTCAGC |
| Glyma16 g32940.1 | M | gene synthesis | M+1 FP4 255 | GCATGCAACT CCAGCATCTG CATGGCTGGG GTTGGACTCA TTTTCAGGGA AAGAGGCTGA ATCATCAGAA GCTGA |
| Glyma16 g32940.1 | M | VIGS | M+1 = 16 g32940 255F | AAAGGGATCC CACGTGCAGC AGAAACCGAA GAAG |
| Glyma16 g32940.1 | M | VIGS | M+1 = 16 g32940 255R | TTGGGTACCT GCATGCAACT CCAGCATC |
Recombinant BPMV RNA2
DNA primers were designed to amplify 240–300 base pair fragments proximal to the stop codon in each cDNA clone. The primers contained unique KpnI and BamHI restriction endonuclease sites and were designed to maintain the translation of the BPMV RNA2 polyprotein (Table 2). Fragments for Ai, Ci, Ei, Ii, Ji, Ki, Li, Ni, and Oi were inserted between the KpnI and BamHI restriction sites of pIAD35, which encodes the second genomic segment of BPMV.26 A fragment for Mi was synthesized from overlapping oligonucleotides [0.5 µl of each 80-mer (100 µM) was reacted in amplification buffer with 2.5 µl 2 mM deoxyribonucleotide triphosphates and 1 unit Taq polymerase for 5 PCR cycles. One µl of that reaction was reacted with 0.5 µl of each ampoligo (100 µM) under the same conditions for 35 PCR cycles] and inserted into the same sites. Clones were sequenced by Genewiz.
Pathogen inoculation
pBPMV-R1M (encoding a genome segment 1 of a BPMV mutant that helps produce more visible mottle symptoms) and engineered pBPMV-R2 RNAi silencing constructs were delivered by biolistics using a PDS-1000/He Gene Gun (Bio-Rad) to the primary leaves of 10-d-old Rpp1 and W82 seedlings.26,27 pIA1037 (1037), expressing the green fluorescent protein from BPMV genome segment 2, was used as a control. Bombarded plants developed symptoms on the first trifoliate leaf between 10–14 d later. Leaves were collected from symptomatic plants, desiccated, and used as inocula for subsequent experiments. Virus-inoculated W82 and Rpp1 plants were taken into the USDA-ARS BSL-3 plant pathogen containment greenhouse facility at Ft Detrick, MD, for rust inoculation and processing under the appropriate USDA-APHIS permit.56 Expanded trifoliate leaves showing viral symptoms were sprayed with a water suspension of uredospores of P. pachyrhizi clonal isolate Louisiana 4–1 adjusted to produce the highest possible density of pustules per surface area on W82. Sprayed plants were placed in an 18 °C dew chamber for 24 h and then moved into the greenhouse. Plants were monitored for rust symptoms for 2–3 wk. Five replicate experiments were performed.
RT-PCR
RNA was purified from inoculated leaves 2 wk after inoculation with rust spores. Fifty ng DNase-treated RNA were tested with the QuantiFast Probe RT-PCR Plus kit (Qiagen, #204482) in an Mx3000P machine (Stratagene) according to the manufacturer’s instructions. Primers and 6-FAM 5′ end-labeled probes were designed to specifically amplify the P. pachyrhizi α-tubulin gene and G. max cons6, a gene demonstrated to be constitutively expressed in soybean leaves after rust infection.28,36 All reactions were performed in triplicate. A standard curve consisting of serial 1:5 dilutions was prepared with total RNA concentrations of 50, 10, 2, 0.4, and 0.08 ng. RNA amounts of test genes were interpolated from standard curves with a correlation coefficient of 95% or greater. RNA amounts for the P. pachyrhizi α-tubulin gene were normalized to those for cons6. To evaluate relative levels of gene silencing and marker gene expression, RNA was isolated from leaves 7 d after inoculation with BPMV silencing constructs but not challenged with soybean rust. Ten ng of RNA were amplified for 27–28 cycles using the cDNA cloning primers or primers for marker genes28 and the SuperScript III One-Step RT-PCR kit. The expression of soybean gene EF1b was studied as a reference control.28 The reaction products were separated on 1.0% agarose gels stained with ethidium bromide.
Western blots
Soybean rust protein PHEP 369 was detected as previously described.37 One trifoliate leaf was pulverized in liquid nitrogen, ground in phosphate buffered saline, 3 mM DTT and 0.2% sodium dodecyl sulfate, heated at 95 °C for 5 min, and centrifuged at 14000 x g for 5 min. The protein concentration of the supernatant was determined by bicinchoninic assay (Thermo Scientific, #23225). 200 µg of protein for all samples except the positive control W82 infected with rust (100 times less was used) were separated on NuPAGE 4–12% Bis-Tris gels (Life Technologies, #NP0355BOX). Proteins were transferred to a nitrocellulose membrane, reacted with anti–rPHEP 369 polyclonal antibody at 1:1000 dilution, reacted with horseradish peroxidase conjugated goat anti-rabbit antibody at 1:25000 dilution and detected with Super Signal West Pico chemiluminescent substrate (Thermo Scientific, #34078).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank SA Whitham at Iowa State University for the BPMV vectors and Wendy Maddox at USDA-ARS, Ft Detrick, for assistance in the containment greenhouse. This research was funded entirely by USDA-ARS.
Glossary
Abbreviations:
- R
resistance
- ETI
effector-triggered immunity
- MAP kinase
mitogen-activated protein kinase
- Rpp
Resistance to Phakopsora pachyrhizi
- W82
Glycine max cv. Williams 82
- Rpp1
Glycine max cv. Williams 82/Rpp1 isoline
- TF
transcription factor
- VIGS
virus-induced gene silencing
- BPMV
Bean pod mottle virus
- EDS1
Enhanced Disease Susceptibility 1
- PAD4
Phytoalexin Deficient 4
- NPR1
Nonexpressor of Pathogenesis-Related genes 1
- PAL1
Phenylalanine Lyase 1
- O-MT
Isoflavone O-Methyltransferase
- Cyp83E12
Cytochrome P450 family 83 E12
- RT-PCR
reverse transcriptase-polymerase chain reaction
- EIN2
Ethylene Insensitive 2
- Myb84
Myb domain protein 84
- WRKY36
WRKY DNA-binding protein 36
References
- 1.Goellner K, Loehrer M, Langenbach C, Conrath U, Koch E, Schaffrath U. Phakopsora pachyrhizi, the causal agent of Asian soybean rust. Mol Plant Pathol. 2010;11:169–77. doi: 10.1111/j.1364-3703.2009.00589.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bonde MR, Nester SE, Austin CN, Stone CL, Frederick RD, Hartman GL, et al. Evaluation of Virulence of Phakopsora pachyrhizi and P. meibomiae Isolates. Plant Dis. 2006;90:708–16. doi: 10.1094/PD-90-0708. [DOI] [PubMed] [Google Scholar]
- 3.Chakraborty N, Curley J, Frederick RD, Hyten DL, Nelson RT, Hartman GL, et al. Mapping and confirmation of a new allele at Rpp1 from soybean PI 594538A conferring RB lesion-type resistance to soybean rust. Crop Sci. 2009;49:783–90. doi: 10.2135/cropsci2008.06.0335. [DOI] [Google Scholar]
- 4.Garcia A, Calvo ES, de Souza Kiihl RA, Harada A, Hiromoto DM, Vieira LG. Molecular mapping of soybean rust (Phakopsora pachyrhizi) resistance genes: discovery of a novel locus and alleles. Theor Appl Genet. 2008;117:545–53. doi: 10.1007/s00122-008-0798-z. [DOI] [PubMed] [Google Scholar]
- 5.Hyten DL, Hartman RL, Nelson RD, Frederick RD, Concibido JM, Narvel JM, et al. Map location of the Rpp1 locus that confers resistance to soybean rust in soybean. Crop Sci. 2007;47:837–8. doi: 10.2135/cropsci2006.07.0484. [DOI] [Google Scholar]
- 6.Meyer JD, Silva DC, Yang C, Pedley KF, Zhang C, van de Mortel M, Hill JH, Shoemaker RC, Abdelnoor RV, Whitham SA, et al. Identification and analyses of candidate genes for rpp4-mediated resistance to Asian soybean rust in soybean. Plant Physiol. 2009;150:295–307. doi: 10.1104/pp.108.134551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li S, Smith JR, Ray JD, Frederick RD. Identification of a new soybean rust resistance gene in PI 567102B. Theor Appl Genet. 2012;125:133–42. doi: 10.1007/s00122-012-1821-y. [DOI] [PubMed] [Google Scholar]
- 8.Kendrick MD, Harris DK, Ha BK, Hyten DL, Cregan PB, Frederick RD, Boerma HR, Pedley KF. Identification of a second Asian soybean rust resistance gene in Hyuuga soybean. Phytopathology. 2011;101:535–43. doi: 10.1094/PHYTO-09-10-0257. [DOI] [PubMed] [Google Scholar]
- 9.Bonde MR, Melching JS, Bromfield KR. Histology of the suscept-pathogen relationship between Glycine max and Phakopsora pachyrhizi, the cause of soybean rust. Phytopathology. 1976;66:1290–4. doi: 10.1094/Phyto-66-1290. [DOI] [Google Scholar]
- 10.Marchetti MA, Melching JS, Bromfield KR. The effects of temperature and dew period on germination and infection by uredospores of Phakopsora pachyrhizi. Phytopathology. 1976;•••:66. [Google Scholar]
- 11.Marchetti MA, Uecker FA, Bromfield KR. Uredial development of Phakopsora pachyrhizi in soybeans. Phytopathology. 1975;65:822–3. doi: 10.1094/Phyto-65-822. [DOI] [Google Scholar]
- 12.McLean RJ. Histological studies of resistance to soybean rust, Phakopsora pachyrhizi Syd. Aust J Agric Res. 1979;30:77–84. doi: 10.1071/AR9790077. [DOI] [Google Scholar]
- 13.Voegele RT, Struck C, Hahn M, Mendgen K. The role of haustoria in sugar supply during infection of broad bean by the rust fungus Uromyces fabae. Proc Natl Acad Sci U S A. 2001;98:8133–8. doi: 10.1073/pnas.131186798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Catanzariti AM, Dodds PN, Lawrence GJ, Ayliffe MA, Ellis JG. Haustorially expressed secreted proteins from flax rust are highly enriched for avirulence elicitors. Plant Cell. 2006;18:243–56. doi: 10.1105/tpc.105.035980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dodds PN, Lawrence GJ, Catanzariti AM, Teh T, Wang CI, Ayliffe MA, Kobe B, Ellis JG. Direct protein interaction underlies gene-for-gene specificity and coevolution of the flax resistance genes and flax rust avirulence genes. Proc Natl Acad Sci U S A. 2006;103:8888–93. doi: 10.1073/pnas.0602577103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dodds PN, Lawrence GJ, Catanzariti AM, Ayliffe MA, Ellis JG. The Melampsora lini AvrL567 avirulence genes are expressed in haustoria and their products are recognized inside plant cells. Plant Cell. 2004;16:755–68. doi: 10.1105/tpc.020040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kemen E, Kemen A, Ehlers A, Voegele R, Mendgen K. A novel structural effector from rust fungi is capable of fibril formation. Plant J. 2013;75:767–80. doi: 10.1111/tpj.12237. [DOI] [PubMed] [Google Scholar]
- 18.Rafiqi M, Gan PH, Ravensdale M, Lawrence GJ, Ellis JG, Jones DA, Hardham AR, Dodds PN. Internalization of flax rust avirulence proteins into flax and tobacco cells can occur in the absence of the pathogen. Plant Cell. 2010;22:2017–32. doi: 10.1105/tpc.109.072983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Link TI, Lang P, Scheffler BE, Duke MV, Graham MA, Cooper B, Tucker ML, van de Mortel M, Voegele RT, Mendgen K, et al. The haustorial transcriptomes of Uromyces appendiculatus and Phakopsora pachyrhizi and their candidate effector families. Mol Plant Pathol. 2013 doi: 10.1111/mpp.12099. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jones JD, Dangl JL. The plant immune system. Nature. 2006;444:323–9. doi: 10.1038/nature05286. [DOI] [PubMed] [Google Scholar]
- 21.Spoel SH, Dong X. How do plants achieve immunity? Defence without specialized immune cells. Nat Rev Immunol. 2012;12:89–100. doi: 10.1038/nri3141. [DOI] [PubMed] [Google Scholar]
- 22.van de Mortel M, Recknor JC, Graham MA, Nettleton D, Dittman JD, Nelson RT, Godoy CV, Abdelnoor RV, Almeida AM, Baum TJ, et al. Distinct biphasic mRNA changes in response to Asian soybean rust infection. Mol Plant Microbe Interact. 2007;20:887–99. doi: 10.1094/MPMI-20-8-0887. [DOI] [PubMed] [Google Scholar]
- 23.Schneider KT, van de Mortel M, Bancroft TJ, Braun E, Nettleton D, Nelson RT, Frederick RD, Baum TJ, Graham MA, Whitham SA. Biphasic gene expression changes elicited by Phakopsora pachyrhizi in soybean correlate with fungal penetration and haustoria formation. Plant Physiol. 2011;157:355–71. doi: 10.1104/pp.111.181149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bernard RL, Nelson RL, Cremeens CR. USDA soybean genetics collection: isoline collection. Soybean Genetics Newsl. 1991;18:27–57. [Google Scholar]
- 25.Cooper B, Campbell KB, Feng J, Garrett WM, Frederick R. Nuclear proteomic changes linked to soybean rust resistance. Mol Biosyst. 2011;7:773–83. doi: 10.1039/c0mb00171f. [DOI] [PubMed] [Google Scholar]
- 26.Zhang C, Bradshaw JD, Whitham SA, Hill JH. The development of an efficient multipurpose bean pod mottle virus viral vector set for foreign gene expression and RNA silencing. Plant Physiol. 2010;153:52–65. doi: 10.1104/pp.109.151639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang C, Yang C, Whitham SA, Hill JH. Development and use of an efficient DNA-based viral gene silencing vector for soybean. Mol Plant Microbe Interact. 2009;22:123–31. doi: 10.1094/MPMI-22-2-0123. [DOI] [PubMed] [Google Scholar]
- 28.Pandey AK, Yang C, Zhang C, Graham MA, Horstman HD, Lee Y, Zabotina OA, Hill JH, Pedley KF, Whitham SA. Functional analysis of the Asian soybean rust resistance pathway mediated by Rpp2. Mol Plant Microbe Interact. 2011;24:194–206. doi: 10.1094/MPMI-08-10-0187. [DOI] [PubMed] [Google Scholar]
- 29.Liu S, Kandoth PK, Warren SD, Yeckel G, Heinz R, Alden J, Yang C, Jamai A, El-Mellouki T, Juvale PS, et al. A soybean cyst nematode resistance gene points to a new mechanism of plant resistance to pathogens. Nature. 2012;492:256–60. doi: 10.1038/nature11651. [DOI] [PubMed] [Google Scholar]
- 30.Liu JZ, Horstman HD, Braun E, Graham MA, Zhang C, Navarre D, Qiu WL, Lee Y, Nettleton D, Hill JH, et al. Soybean homologs of MPK4 negatively regulate defense responses and positively regulate growth and development. Plant Physiol. 2011;157:1363–78. doi: 10.1104/pp.111.185686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang C, Grosic S, Whitham SA, Hill JH. The requirement of multiple defense genes in soybean Rsv1-mediated extreme resistance to soybean mosaic virus. Mol Plant Microbe Interact. 2012;25:1307–13. doi: 10.1094/MPMI-02-12-0046-R. [DOI] [PubMed] [Google Scholar]
- 32.Schmutz J, Cannon SB, Schlueter J, Ma J, Mitros T, Nelson W, Hyten DL, Song Q, Thelen JJ, Cheng J, et al. Genome sequence of the palaeopolyploid soybean. Nature. 2010;463:178–83. doi: 10.1038/nature08670. [DOI] [PubMed] [Google Scholar]
- 33.Wang Z, Libault M, Joshi T, Valliyodan B, Nguyen HT, Xu D, Stacey G, Cheng J. SoyDB: a knowledge database of soybean transcription factors. BMC Plant Biol. 2010;10:14. doi: 10.1186/1471-2229-10-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Qutob D, Kemmerling B, Brunner F, Küfner I, Engelhardt S, Gust AA, Luberacki B, Seitz HU, Stahl D, Rauhut T, et al. Phytotoxicity and innate immune responses induced by Nep1-like proteins. Plant Cell. 2006;18:3721–44. doi: 10.1105/tpc.106.044180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pham TA, Miles MR, Frederick RD, Hill CB, Hartman GL. Differential responses of resistant soybean entries to isolates of Phakopsora pachyrhizi. Plant Dis. 2009;93:224–8. doi: 10.1094/PDIS-93-3-0224. [DOI] [PubMed] [Google Scholar]
- 36.Libault M, Thibivilliers S, Bilgin DD, Radwan O, Benitez M, Clough SJ, et al. Identification of four soybean reference genes for gene expression normalization. The Plant Genome. 2008;1:44–54. doi: 10.3835/plantgenome2008.02.0091. [DOI] [Google Scholar]
- 37.Luster DG, McMahon MB, Edwards HH, Boerma BL, Lewis Ivey ML, Miller SA, Dorrance AE. Novel Phakopsora pachyrhizi extracellular proteins are ideal targets for immunological diagnostic assays. Appl Environ Microbiol. 2012;78:3890–5. doi: 10.1128/AEM.07079-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Glazebrook J. Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu Rev Phytopathol. 2005;43:205–27. doi: 10.1146/annurev.phyto.43.040204.135923. [DOI] [PubMed] [Google Scholar]
- 39.Tintor N, Ross A, Kanehara K, Yamada K, Fan L, Kemmerling B, Nürnberger T, Tsuda K, Saijo Y. Layered pattern receptor signaling via ethylene and endogenous elicitor peptides during Arabidopsis immunity to bacterial infection. Proc Natl Acad Sci U S A. 2013;110:6211–6. doi: 10.1073/pnas.1216780110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu Z, Wu Y, Yang F, Zhang Y, Chen S, Xie Q, Tian X, Zhou JM. BIK1 interacts with PEPRs to mediate ethylene-induced immunity. Proc Natl Acad Sci U S A. 2013;110:6205–10. doi: 10.1073/pnas.1215543110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Boutrot F, Segonzac C, Chang KN, Qiao H, Ecker JR, Zipfel C, Rathjen JP. Direct transcriptional control of the Arabidopsis immune receptor FLS2 by the ethylene-dependent transcription factors EIN3 and EIL1. Proc Natl Acad Sci U S A. 2010;107:14502–7. doi: 10.1073/pnas.1003347107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lygin AV, Li S, Vittal R, Widholm JM, Hartman GL, Lozovaya VV. The importance of phenolic metabolism to limit the growth of Phakopsora pachyrhizi. Phytopathology. 2009;99:1412–20. doi: 10.1094/PHYTO-99-12-1412. [DOI] [PubMed] [Google Scholar]
- 43.Burch-Smith TM, Schiff M, Caplan JL, Tsao J, Czymmek K, Dinesh-Kumar SP. A novel role for the TIR domain in association with pathogen-derived elicitors. PLoS Biol. 2007;5:e68. doi: 10.1371/journal.pbio.0050068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wirthmueller L, Zhang Y, Jones JD, Parker JE. Nuclear accumulation of the Arabidopsis immune receptor RPS4 is necessary for triggering EDS1-dependent defense. Curr Biol. 2007;17:2023–9. doi: 10.1016/j.cub.2007.10.042. [DOI] [PubMed] [Google Scholar]
- 45.Deslandes L, Olivier J, Peeters N, Feng DX, Khounlotham M, Boucher C, Somssich I, Genin S, Marco Y. Physical interaction between RRS1-R, a protein conferring resistance to bacterial wilt, and PopP2, a type III effector targeted to the plant nucleus. Proc Natl Acad Sci U S A. 2003;100:8024–9. doi: 10.1073/pnas.1230660100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shen QH, Saijo Y, Mauch S, Biskup C, Bieri S, Keller B, Seki H, Ulker B, Somssich IE, Schulze-Lefert P. Nuclear activity of MLA immune receptors links isolate-specific and basal disease-resistance responses. Science. 2007;315:1098–103. doi: 10.1126/science.1136372. [DOI] [PubMed] [Google Scholar]
- 47.García AV, Blanvillain-Baufumé S, Huibers RP, Wiermer M, Li G, Gobbato E, Rietz S, Parker JE. Balanced nuclear and cytoplasmic activities of EDS1 are required for a complete plant innate immune response. PLoS Pathog. 2010;6:e1000970. doi: 10.1371/journal.ppat.1000970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kinkema M, Fan W, Dong X. Nuclear localization of NPR1 is required for activation of PR gene expression. Plant Cell. 2000;12:2339–50. doi: 10.1105/tpc.12.12.2339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Feys BJ, Moisan LJ, Newman MA, Parker JE. Direct interaction between the Arabidopsis disease resistance signaling proteins, EDS1 and PAD4. EMBO J. 2001;20:5400–11. doi: 10.1093/emboj/20.19.5400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ju C, Yoon GM, Shemansky JM, Lin DY, Ying ZI, Chang J, Garrett WM, Kessenbrock M, Groth G, Tucker ML, et al. CTR1 phosphorylates the central regulator EIN2 to control ethylene hormone signaling from the ER membrane to the nucleus in Arabidopsis. Proc Natl Acad Sci U S A. 2012;109:19486–91. doi: 10.1073/pnas.1214848109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Qiu JL, Fiil BK, Petersen K, Nielsen HB, Botanga CJ, Thorgrimsen S, Palma K, Suarez-Rodriguez MC, Sandbech-Clausen S, Lichota J, et al. Arabidopsis MAP kinase 4 regulates gene expression through transcription factor release in the nucleus. EMBO J. 2008;27:2214–21. doi: 10.1038/emboj.2008.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lee J, Feng J, Campbell KB, Scheffler BE, Garrett WM, Thibivilliers S, Stacey G, Naiman DQ, Tucker ML, Pastor-Corrales MA, et al. Quantitative proteomic analysis of bean plants infected by a virulent and avirulent obligate rust fungus. Mol Cell Proteomics. 2009;8:19–31. doi: 10.1074/mcp.M800156-MCP200. [DOI] [PubMed] [Google Scholar]
- 53.Cooper B, Garrett WM, Campbell KB. Shotgun identification of proteins from uredospores of the bean rust Uromyces appendiculatus. Proteomics. 2006;6:2477–84. doi: 10.1002/pmic.200500630. [DOI] [PubMed] [Google Scholar]
- 54.Cooper B, Neelam A, Campbell KB, Lee J, Liu G, Garrett WM, Scheffler B, Tucker ML. Protein accumulation in the germinating Uromyces appendiculatus uredospore. Mol Plant Microbe Interact. 2007;20:857–66. doi: 10.1094/MPMI-20-7-0857. [DOI] [PubMed] [Google Scholar]
- 55.Luster DG, McMahon MB, Carter ML, Fortis LL, Nuñez A. Proteomic analysis of germinating urediniospores of Phakopsora pachyrhizi, causal agent of Asian soybean rust. Proteomics. 2010;10:3549–57. doi: 10.1002/pmic.200900469. [DOI] [PubMed] [Google Scholar]
- 56.Melching JS, Bromfield KR, Kingsolver CH. The plant pathogen containment facility at Frederick, Maryland. Plant Dis. 1983;67:717–22. doi: 10.1094/PD-67-717. [DOI] [Google Scholar]


