Abstract
The legume-rhizobia symbioses lead to the formation of a novel adaptive complex organ, termed the root nodule, which arises from cortical cell division and rhizobial infection in the root. Lipochitin oligosaccarides, Nod-Factors (NFs) secreted by rhizobia, are responsible for the onset of nodule development. Here we describe the characterization of Lotus japonicas, Nod factor Signaling Pathway2 (LjNSP2) protein that belongs to the plant GRAS family of transcription factors. Yeast two-hybrid analysis indicates that LjNSP2 alone has a transcription-stimulating ability and for this the SH2(src-homology2)-like domain, vital for function of STAT proteins is required. The ADG4 (the activation domain of GAL4)-LjNSP2 fusion coupled with BDG4 (the DNA binding domain of GAL4)-LjNSP2 increased the expression level, whereas the ADG4-Ljnsp2–1 mutant fusion did not, indicating that LjNSP2 interacts with itself to form a homodimer and this depends on the SH2-like domain. Based on the evidence, we discuss the action of LjNSP2, compared with that of the family of animal-specific STAT transcription factors, which induce developmental programmes in response to external stimuli.
Keywords: Nod factor, NSP2, transcription factor, activator, Lotus japonicus
NFs induce a variety of responses in a host-specific manner, including root hair curling and cortical cell division, during the early steps of nodulation.1-3 NSP2 is a GRAS protein essential for the NF signaling and lies downstream of the calcium spiking.4-7 NSP2 has been shown to form a complex with NPS1 that is associated with the promoters of early nodulin genes in Medicago truncatula.8 On the other hand, L. japonicas nsp2 mutants have been isolated as Ljsym70 and Ljsym35 and characterized,6,7,9 but the function of LjNSP2 remains to be elucidated at protein level.
To test the ability of LjNSP2 to activate transcription and to form dimers, we used the yeast two-hybrid system; LjNSP2 was fused to ADG4 (the activation domain of GAL4) or to BDG4 (the DNA binding domain of GAL4), and their interaction was monitored by measuring the level of expression of the GAL4-lacZ reporter. We also used a DNA fragment derived from Ljnsp2–1 (Ljsym70) mutant that carries substitution mutation of conserved V to E in the SH2 (src-homology2)-like domain, vital for function of STAT protein.7 Even in combination with the unmodified ADG4, BDG4 fused with LjNSP2 increased lacZ expression by 3-fold compared with the unmodified BDG4, while BDG4 fused with Ljnsp2–1 induced no increase of the expression (Fig. 1A). This indicates that LjNSP2 alone has a transcription-stimulating ability and for this the SH2-like region is required. The ADG4-LjNSP2 fusion coupled with BDG4-LjNSP2 increased the expression level a further 50%, whereas the ADG4-Ljnsp2–1 mutant fusion did not (Fig. 1A), indicating that LjNSP2 interacts with itself to form a homodimer and this depends on the SH2-like domain.
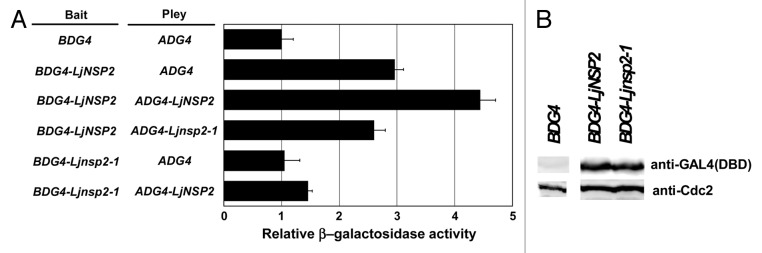
Figure 1. Yeast-two hybrid analyses of LjNSP2 interactions and immunoblotting in yeast cells. (A) Transcription activity of LacZ (β-galactosidase) was measured as the indicator of the interaction between 2 components fused with the prey (ADG4: activation domain of GAL4) and bait (BDG4: DNA binding domain of GAL4). However, an about 3-fold increase of the activity was seen with the combination BDG4-LjNSP2/ADG4 compared with control combination BDG4/ADG4, while there was no increase with BDG4-Ljnsp2–1/ADG4, indicating the presence of a transcription activity of LjNSP2 alone bound to the promoter of LacZ depending the function of the SH2-like domain. A further 50% of increase of the activity was seen in the combination BDG4-LjNSP2/ADG4-LjNSP2, being consistent with the ability of LjNSP2 to form homodimer. This is also dependent on the SH2-like function because this increase was not observed for the combination with ADG4-Ljnsp2–1. (B) Yeast transformants carrying pBD and pAD-LjNSP2, pBD-LjNSP2 and pAD-LjNSP2, pBD-Ljnsp2–1, and pAD-LjNSP2 were grown in YPD medium to late-log phase at 30 °C, cell extracts were prepared and immunoblotted by probing with anti-GAL4 (DBD) or anti-PSTAIRE antibody. The size of the detected proteins is consistent with a predicted pBD-LjNSP2 molecular mass of 71 kD. Cdc28 bands served as the internal control.
To test if the Ljnsp2–1 mutant protein is unstable in yeast cells, we probed yeast extract with an anti-GAL4 DNA binding domain antibody. This showed that there were similar levels of protein of both BDG4-Ljnsp2–1 and BDG4-LjNSP2 (Fig. 1B), indicating that the loss of transcription-stimulation and homodimarisation are not due to instability of the protein.
These results suggest that the SH2-like domain is important for the ability of LjNSP2 to activate transcription and to dimerize, and that dimerization enhances transcription relative to the monomer. These suggested functions of the LjNSP2 SH2-like region for transcription activity and dimerization are equivalent to the role of the SH2 domain in animal STAT proteins.10
An interesting character of LjNSP2 is the requirement of SH2-like domain in LjNSP2 function. The Ljnsp2–1 missense mutation in the SH2-like domain caused a complete lack of nodulation equivalent to that seen with the Ljnsp2–2 null mutant, although both mutants grew normally if supplemented with nitrate. Similarly, missense or deletion mutations in the SH2-like region of the other GRAS proteins, Arabidopsis RGA11 and rice SLR1,12 caused strong mutant phenotypes. Richards et al. pointed out structural homology and conserved amino acids between the plant GRAS proteins and animal STAT proteins especially in the SH2 (-like) domains.13 In animals, its importance in dimerization and nuclear localization has been well characterized.10 Here we have demonstrated that the LjNSP2 SH2-like domain plays an important role in transcription activation as seen in LacZ, and its homo-dimerization, as do animal STAT SH2 domains, along with in nodulation-signaling, verifying Richards’ proposal.13 However, the interaction activity between LjNSP2 proteins was not always high in yeast cells. In the some cases of animal STAT proteins, 1 STAT interacts with another. LjNSP2 also might form the heterodimer with other GRAS proteins related to LjNSP2 as is the case in Medicago.8
STAT proteins can mediate the activation of cell surface receptors in a direct manner after direct phosphorylation by a receptor themselves or by non-receptor tyrosine kinases10 (Levy and Darnell, 2002); in nodulation, the pathway of NF signal transduction seems to be more complicated with the participation of 2 kinds of NF receptors like NFR1, NFR5, with SYMRK another kinase, nucleoporins, ion channels like CASTOR/POLLUX, and then CCaMK.14-16 Furthermore, in animal systems, the function of the STAT SH2 domain strongly depends on the phosphorylation of a conserved Gly-Tyr (GY) motif between the SH2-like domain and C-terminal transactivation domain (Richards et al., 2000). This GY motif is present in LjNSP2 and other GRAS proteins.6,7 Determining whether LjNSP2 is phosphorylated following NF activation of the signaling pathway is an exciting challenge in the future.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
This work was supported by the fund of Promotion of Basic Research Activities for Innovative Biosciences, (BRAIN) and the Special Coordination Funds for Promoting Science and Technology of the Japanese Ministry of Education, Culture, Sports, Science, and Technology.
References
- 1.Fisher RF, Long SR. Rhizobium--plant signal exchange. Nature. 1992;357:655–60. doi: 10.1038/357655a0. [DOI] [PubMed] [Google Scholar]
- 2.Dénarié J, Debellé F, Promé JC. Rhizobium lipo-chitooligosaccharide nodulation factors: signaling molecules mediating recognition and morphogenesis. Annu Rev Biochem. 1996;65:503–35. doi: 10.1146/annurev.bi.65.070196.002443. [DOI] [PubMed] [Google Scholar]
- 3.Schultze M, Kondorosi A. Regulation of symbiotic root nodule development. Annu Rev Genet. 1998;32:33–57. doi: 10.1146/annurev.genet.32.1.33. [DOI] [PubMed] [Google Scholar]
- 4.Oldroyd GE, Long SR. Identification and characterization of nodulation-signaling pathway 2, a gene of Medicago truncatula involved in Nod actor signaling. Plant Physiol. 2003;131:1027–32. doi: 10.1104/pp.102.010710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kaló P, Gleason C, Edwards A, Marsh J, Mitra RM, Hirsch S, Jakab J, Sims S, Long SR, Rogers J, et al. Nodulation signaling in legumes requires NSP2, a member of the GRAS family of transcriptional regulators. Science. 2005;308:1786–9. doi: 10.1126/science.1110951. [DOI] [PubMed] [Google Scholar]
- 6.Heckmann AB, Lombardo F, Miwa H, Perry JA, Bunnewell S, Parniske M, Wang TL, Downie JA. Lotus japonicus nodulation requires two GRAS domain regulators, one of which is functionally conserved in a non-legume. Plant Physiol. 2006;142:1739–50. doi: 10.1104/pp.106.089508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Murakami Y, Miwa H, Imaizumi-Anraku H, Kouchi H, Downie JA, Kawaguchi M, Kawasaki S. Positional cloning identifies Lotus japonicus NSP2, a putative transcription factor of the GRAS family, required for NIN and ENOD40 gene expression in nodule initiation. DNA Res. 2006;13:255–65. doi: 10.1093/dnares/dsl017. [DOI] [PubMed] [Google Scholar]
- 8.Hirsch S, Kim J, Muñoz A, Heckmann AB, Downie JA, Oldroyd GE. GRAS proteins form a DNA binding complex to induce gene expression during nodulation signaling in Medicago truncatula. Plant Cell. 2009;21:545–57. doi: 10.1105/tpc.108.064501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kawaguchi M, Imaizumi-Anraku H, Koiwa H, Niwa S, Ikuta A, Syono K, Akao S. Root, root hair, and symbiotic mutants of the model legume Lotus japonicus. Mol Plant Microbe Interact. 2002;15:17–26. doi: 10.1094/MPMI.2002.15.1.17. [DOI] [PubMed] [Google Scholar]
- 10.Levy DE, Darnell JE., Jr. Stats: transcriptional control and biological impact. Nat Rev Mol Cell Biol. 2002;3:651–62. doi: 10.1038/nrm909. [DOI] [PubMed] [Google Scholar]
- 11.Silverstone AL, Ciampaglio CN, Sun T. The Arabidopsis RGA gene encodes a transcriptional regulator repressing the gibberellin signal transduction pathway. Plant Cell. 1998;10:155–69. doi: 10.1105/tpc.10.2.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Itoh H, Ueguchi-Tanaka M, Sato Y, Ashikari M, Matsuoka M. The gibberellin signaling pathway is regulated by the appearance and disappearance of SLENDER RICE1 in nuclei. Plant Cell. 2002;14:57–70. doi: 10.1105/tpc.010319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Richards DE, Peng J, Harberd NP. Plant GRAS and metazoan STATs: one family? Bioessays. 2000;22:573–7. doi: 10.1002/(SICI)1521-1878(200006)22:6<573::AID-BIES10>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 14.Oldroyd GE, Downie JA. Coordinating nodule morphogenesis with rhizobial infection in legumes. Annu Rev Plant Biol. 2008;59:519–46. doi: 10.1146/annurev.arplant.59.032607.092839. [DOI] [PubMed] [Google Scholar]
- 15.Kouchi H, Imaizumi-Anraku H, Hayashi M, Hakoyama T, Nakagawa T, Umehara Y, Suganuma N, Kawaguchi M. How many peas in a pod? Legume genes responsible for mutualistic symbioses underground. Plant Cell Physiol. 2010;51:1381–97. doi: 10.1093/pcp/pcq107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Madsen LH, Tirichine L, Jurkiewicz A, Sullivan JT, Heckmann AB, Bek AS, Ronson CW, James EK, Stougaard J. The molecular network governing nodule organogenesis and infection in the model legume Lotus japonicus. Nat Commun. 2010;1:10. doi: 10.1038/ncomms1009. [DOI] [PMC free article] [PubMed] [Google Scholar]


