Abstract
Stress perception and communication play important roles in the adaptation of plants to changing environmental conditions. Plant roots are the first organs to detect changes in the soil water potential induced by salt stress. In the presence of salinity stress, root-to-shoot communication occurs to adjust the growth of the whole plant. So far, the phytohormone abscisic acid (ABA), hydraulic signals and reactive oxygen species (ROS) have been proposed to mediate this communication under salt stress. Recently, we identified the rice transcription factor SALT-RESPONSIVE ERF1 (SERF1), which regulates a ROS-dependent transcriptional cascade in roots required for salinity tolerance. Upon salt stress, SERF1 knockout mutant plants show an increased leaf temperature as compared with wild type. As this occurs within the first 20 min of salt stress, we here evaluated the involvement of SERF1 in the perception of salt stress in the shoot. By metabolic profiling and expression analysis we show that the action of SERF1 in signal communication to the shoot is independent from ABA, but does affect the accumulation of ROS-related metabolites and transcripts under short-term salt stress.
Keywords: reactive oxygen species, transcription factor, metabolism, root-to-shoot, salt stress, Oryza sativa
Introduction
Saline soils represent a major abiotic stress that adversely affects plant growth and agricultural production. Since rice is a significant food crop that delivers 20% of the daily calory intake for human consumption, it is important to realize that it represents one of the most salt-sensitive cereals.1 The understanding of the molecular basis of salt stress tolerance in rice is therefore of great importance for future breeding programs.2 Key aspects within this process are signal perception and signal transduction as they represent the first components of the plant's adaptive response to stress.3
The role of reactive oxygen species (ROS) during salinity tolerance has long been associated with the adverse effect they can have on cell viability.4 However, ROS, especially hydrogen peroxide (H2O2), play a pivotal and central regulatory role in plants, as they can actively regulate their production and degradation.5 Because ROS production and scavenging occur continuously, alterations in the balance between both processes would result in a rapid change in cellular redox status creating a stress signal.6 Within minutes of salt stress, rice roots produce a so-called ROS burst.7 Recently, it has been established that plasma membrane-located NADPH oxidases are not only required for the ROS burst but are essential to establish salinity tolerance.8,9 In addition, the initial peak of ROS production can trigger a cascade of cell-to-cell communication involving the formation of a ROS wave that propagates throughout different tissues to carry the signal over a long distance.10,11
We recently demonstrated that the DEHYDRATION-RESPONSE ELEMENT-BINDING (DREB) transcription factor (TF) SALT-RESPONSIVE ERF1 (SERF1) functions as a bottleneck in the conversion of the salt-induced ROS burst into a transcriptional response in rice roots.12SERF1 is root-specifically induced by both H2O2 and salt stress. Despite the fact that SERF1 knockout plants (serf1) show an increase in H2O2 levels upon salt treatment, the induction of ROS-responsive genes, including those encoding mitogen-activated protein kinases (MAPKs) and TFs, is severely attenuated. SERF1 directly controls the expression of 2 H2O2-inducible MAPK cascade genes, namely MAPK5, known to enhance salt tolerance when overexpressed, and MAP3K6.13,14 Moreover, MAPK5 specifically phosphorylates SERF1 at Ser-105 in vitro, enabling amplification of the salt stress signal.12 Furthermore, SERF1 directly regulates the expression of the TF genes DEHYDRATION-RESPONSIVE ELEMENT-BINDING 2A (DREB2A) and ZINC-FINGER PROTEIN 179 (ZFP179), which both are positive regulators of salt stress tolerance.15,16 Similar to the disruption of the ROS signal in the NADPH oxidase mutant rbohf in Arabidopsis thaliana,9 an early accumulation of sodium ions (Na+) in the shoot is observed for serf1 upon salt stress.12 Thus, ROS-dependent signaling is essential to establish salinity tolerance in plants.
So far, only little is known about the communication between the root and the shoot during salt stress. Abscisic acid (ABA) has been thought of having a fundamental role in root-to-shoot signaling during drought stress.17 However, such a prominent role for ABA in long-distance communication during salt stress and water shortage could not be confirmed.18,19 As the shoot response to decreased water supply is not affected by the ability to synthesize ABA in the root, but depends on ABA synthesized in the shoot, it indicates that a specific long-distance signal precedes ABA signaling during water shortage.19 One possible form of communication between the root and the shoot is through hydraulic signals, although the exact mechanism behind such a signal is still elusive. In addition, ROS-dependent salt stress signaling is essential for controlling root-to-shoot Na+ delivery.9,12 In Arabidopsis, root-perceived stress signals that are mediated by ROS are rapidly transferred to the shoot.10
Interestingly, serf1 leaves display a significantly higher temperature than the wild type already after 20 min of salt stress, which is likely caused by reduced transpiration rates and stomatal closure.12 At the metabolic level, osmoprotectants accumulate during long-term salt stress.20 Recently, a distinction has been made between metabolites that respond in either an ABA-dependent or ABA-independent manner upon drought stress.21 Still, relatively little is known regarding the initial response of metabolites during salt stress, although the expected fluctuations in metabolite pools during short-term stress might be rather small.
In this study, for a better understanding of the role of SERF1 in protecting the shoot from salt stress, we determined its effect on metabolism during short-term salt stress. In addition, we examined the consequence of loss of SERF1 on the transcriptional response of the shoot to salt stress. Next to salinity stress, we assessed the induction of salt-responsive genes in the shoot upon ROS, ABA and mannitol treatment of the root. Our analysis indicates that ABA has a minor role during salt stress-induced root-to-shoot communication, while ROS signals appear to dominate this process.
Results
Metabolic responses in roots after short-term salt stress
We previously showed that serf1 plants have an impaired salt stress tolerance resulting in early accumulation of Na+ in leaves and roots.12 As the Na+/K+ ratio during the first 24 h of salt stress is not affected in serf1, it is likely that the increased leaf temperature observed in serf1 after 20 min of salt stress is caused by an osmotic imbalance. To assess the consequence of the loss of SERF1 on early metabolic events during salt stress, we performed GC-MS-based metabolite profiling. In total, more than 80 compounds were identified including amino acids, organic acids and sugars. Only those metabolites that were significantly affected by either 3 h of salt stress or the genotype are shown in Table 1.
Table 1. Relative metabolite content in roots of 4-wk-old wild type and serf1 under control conditions and after 3 h of salt stress.
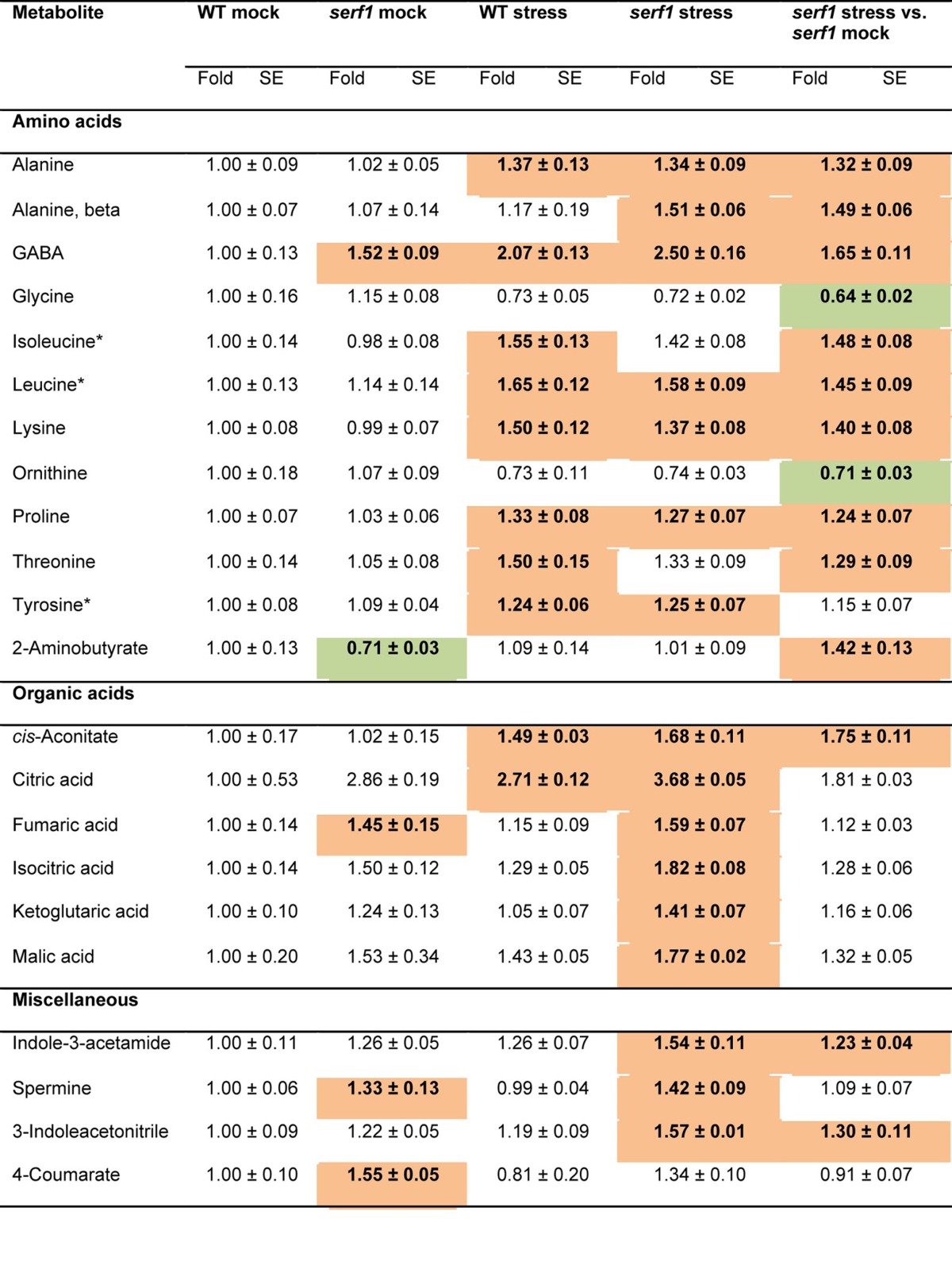
Data for serf1 (mock and stress) and the wild type under salt stress (100 mM NaCl) are normalized to the mean levels of wild type under control conditions. In addition, the metabolite content of serf1 upon stress is shown, normalized to the mean levels of serf1 mock. Values are presented as means ± SE of 6 biological determinations. Bold indicates values that were determined by Student's t test to be significantly different (P≤ 0.05) from the control. Orange and green shading indicate significant up- and downregulation, respectively, as compared with controls. Asterisks indicate ABA-dependent metabolites.21
In roots of wild-type and serf1 plants, salt stress triggered a significant accumulation of the free amino acid content of Ala, γ-aminobutyrate (GABA), Ile, Leu, Lys, Pro, Thr and Tyr (Table 1). Previously, we found that after 24 h of salt stress, the levels of Ala, GABA, Lys and Thr increased in roots,22 which overlaps with their response after 3 h of salt stress. Furthermore, we observed a specific increase in 2-aminobutyrate (2-AB) and β-Ala levels in salt-stressed roots of the mutant after 3 h of treatment. Recently, 2-AB has been linked to ophthalmate biosynthesis in plants,23 and it is an oxidative stress biomarker in mammals.24 In addition, a decrease in Gly and ornithine was observed in both the mutant and the wild type, albeit this was only significant for the mutant. Under control conditions, GABA levels were elevated in serf1, while those of 2-AB were reduced as compared with wild-type roots.
The tricarboxylic acid (TCA) cycle intermediates showed a strong response upon salt treatment, as previously reported in barley (Table 1).25 Upon salt stress, both wild-type and serf1 roots displayed a significant increase in the levels of citrate and cis-aconitate. In addition, the levels of isocitrate, α-ketoglutarate and malate were significantly elevated in serf1 roots under salt stress as compared with the wild type, but not when compared with serf1 control roots. Under control conditions, the level of fumarate was increased in serf1 roots as compared with the wild type. In line with this, after 24 h of salt stress a significant increase of nearly all TCA cycle intermediates in roots of rice was reported.22 Apparently, first citrate and cis-aconitate accumulate during salt stress, of which citrate constitutes the entry point into the TCA cycle.
The 15 sugars detected by GC-MS did not show any difference in abundance upon 3 h of salt stress in roots of the wild type or serf1. In contrast, 24 h of salt stress causes a reduction in the level of several sugars,22 indicating that salt-related changes in sugar levels in the root are relatively slow.
Of the 11 miscellaneous compounds determined, spermine and 4-coumarate were elevated in serf1 roots under control conditions as compared with the wild type. Although spermine levels have been shown to increase during abiotic stress, it does not confer tolerance but is required during stress recovery.26 Furthermore, salt-stressed serf1 roots displayed significantly higher levels of indole-3-acetamide and 3-indoleacetonitrile, which are intermediates of auxin biosynthesis, than the stressed wild type.27
Metabolic responses in leaves after short-term salt stress
Although salt stress is perceived by the root, an inhibition of shoot growth occurs within hours in salt-sensitive plants.28 The observed rapid increase in leaf temperature in stressed serf1 plants as compared with the wild type indicates a rapid decline in leaf transpiration due to the closure of stomata.12,29 One outcome of the decreased stomatal conductance is a reduced uptake of carbon dioxide which leads to reduced photosynthesis.30
In leaves, the level of β-Ala was significantly lowered upon salt stress in both the wild type and serf1 (Table 2). In addition, Ala, Asp, Gln and 2-AB levels were significantly reduced in the wild type upon salt stress, but showed only a slight decrease in serf1 leaves, which might be due to the lower accumulation of these compounds already under control conditions. Furthermore, His, Trp and Val levels were decreased in stressed serf1 leaves. On the other hand, Gln levels were lowered in serf1 leaves under control conditions, while GABA levels significantly dropped during salt stress, indicating a differential metabolic response to salinity stress in leaves. Although elevated GABA levels are commonly linked to stress tolerance, an increased GABA level does not provide salt stress tolerance by itself. Notably, the breakdown of GABA is required to provide tolerance toward ionic stress in Arabidopsis.31 The lower GABA level in serf1 leaves might reflect a more rapid turn-over of this metabolite during salt stress.
Table 2. Relative metabolite content in leaves of 4-wk-old wild type and serf1 under control conditions and after 3 h of salt stress.
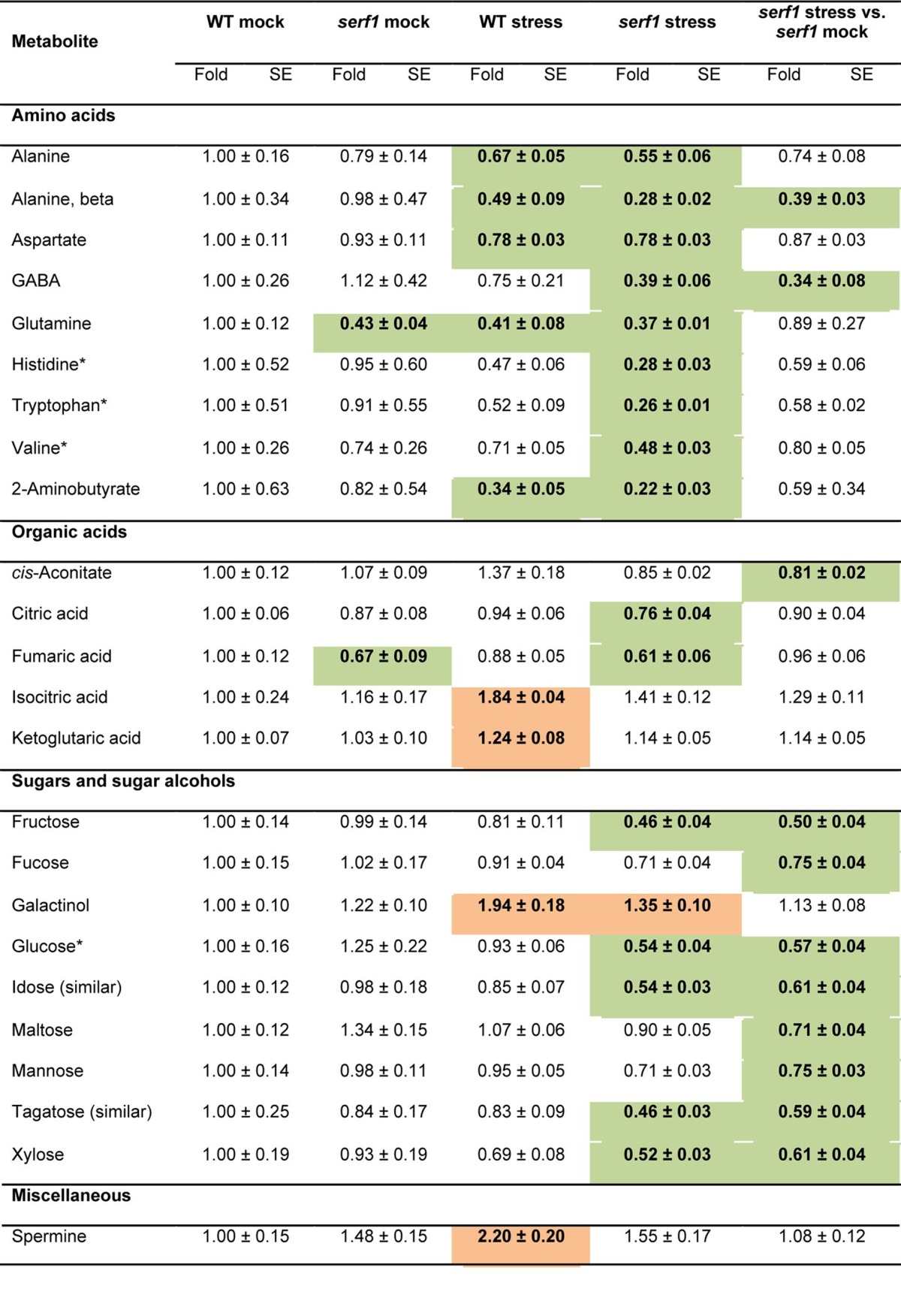
Data for serf1 (mock and stress) and the wild type under salt stress (100 mM NaCl) are normalized to the mean levels of wild type under control conditions. In addition, the metabolite content of serf1 upon stress is shown, normalized to the mean levels of serf1 mock. Values are presented as means ± SE of 6 biological determinations. Bold indicates values that were determined by Student's t test to be significantly different (P≤ 0.05) from the control. Orange and green shading indicate significant up- and downregulation, respectively, as compared with controls. Asterisks indicate ABA-dependent metabolites.21
For the TCA cycle intermediates a significant upregulation of isocitrate and α-ketoglutarate was observed in wild-type leaves upon salt stress but not in serf1. In contrast, cis-aconitate showed significantly decreased levels in serf1 leaves. Furthermore, serf1 leaves had lower levels of fumarate under control conditions. For several plant species it has been demonstrated that fumarate is linked to nitrogen assimilation and osmotic adjustment,32 indicating a potential difference for these processes in serf1.
Under non-stress conditions, no difference in the levels of sugars in roots and leaves was observed between serf1 and wild type. Intriguingly, the sugar levels in serf1 leaves already significantly declined upon 3 h of salt stress (Table 2), while in the wild type even after 24 h of salt stress only a minor effect on sugar levels was observed.22 This indicates a specific effect of SERF1 on maintaining sugar homeostasis under salt stress. Sugars involved in major carbohydrate metabolism like fructose, glucose and maltose showed a significantly lower level in serf1 leaves but not in the wild type. Furthermore, reduced levels of fucose, mannose and xylose were observed in serf1 leaves. On the other hand, in the wild type, a strong accumulation of the ROS scavenger galactinol was detected,33 which did not occur in serf1. The strong reduction of sugars in serf1 leaves, indicating reduced carbon dioxide uptake and photosynthesis, correlates well with the rapid closure of stomata upon salt stress as determined by infrared thermography.12
Among the miscellaneous compounds, an increase in the level of spermine was observed within 3 h of salt stress for the wild type but not serf1. We found only a moderate effect of salt stress on leaf metabolism after 24 h,22 suggesting that the initial changes in metabolism in the wild type observed after 3 h of treatment might be in part restored thereafter.
Loss of SERF1 affects the transcriptional response to salt stress in leaves
To determine whether SERF1 affects the transcriptional response in leaves upon salt stress we tested the expression of 70 previously identified early salt stress marker genes in the wild type and the mutant.12 Of note, SERF1 transcript levels in leaves are not affected upon salt stress, suggesting that the observed changes might be due to the absence of the SERF1-dependent transcriptional cascade in roots.12 In addition to the response of these genes to salt stress, we determined the effect of short-term exposure (30 min and 3 h) of roots to H2O2, mannitol (causing osmotic stress) or ABA on their expression in leaves. We reasoned that these compounds and/or an osmotic signal might be involved in the root-to-shoot communication during salt stress.
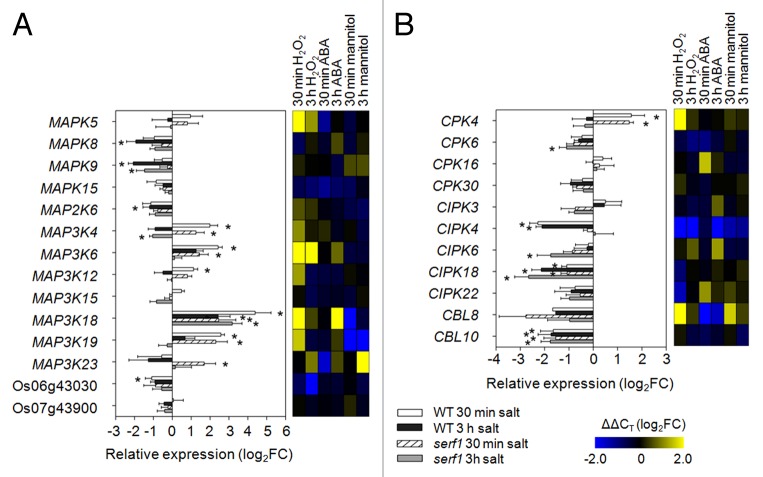
Interestingly, during the initial response to salt stress we found that most of the selected marker genes were downregulated in leaves, which is in contrast to their response in roots.12 Among the MAPK superfamily genes, 9 responded to salt stress in wild-type leaves and 6 in serf1 leaves after 30 min and 3 h of treatment. Notably, the direct SERF1 target genes MAPK5 and MAP3K6 were not affected in their transcriptional response in the mutant background, indicating that SERF1 specifically regulates their expression in roots.12 Furthermore, MAP3K4, MAP3K18 and MAP3K19 showed a similar response in wild-type and serf1 leaves (Fig. 1A). Of note, application of H2O2 to roots induced the expression of MAPK5, MAP3K4, MAP3K6, MAP3K18, and MAP3K19 in leaves, which also responded to salt stress in serf1 (Fig. 1A). This observation suggests that SERF1 is not essential for the propagation of a ROS signal to the shoot. On the other hand, MAP3K23 was specifically upregulated in serf1 leaves. In addition to being mannitol-responsive, it was shown before to be induced upon drought stress.14
Figure 1. Loss of SERF1 alters the transcriptional response of signaling components in leaves. Expression levels of (A) MAPK signaling genes and (B) calcium signaling-related genes were measured in leaves of wild-type and serf1 plants subjected to salt stress for 30 min or 3 h (100 mM NaCl). ACTIN (Os03g50885) was used as reference gene. Data represent means ± SE from 3 biological replicates. An asterisk (*) indicates a significant difference between treated and control plants (P ≤ 0.05; the Student t test). In the heat map the gene expression under H2O2 (5 mM), ABA (5 µM) or mannitol (100 mM) treatment is shown in log2FC relative to the control situation with n = 3. Os06g43030 and Os07g43900 encode protein kinases. WT; wild type; FC, fold change.
Propagation of stress signals from the root involves both ROS and calcium signals.11 Here, we tested the expression of genes encoding calcium-dependent protein kinases (CPKs), calcineurin B-like proteins (CBLs) and CBL-interacting protein kinases (CIPKs) (Fig. 1B). CPK4 was significantly induced by salt stress in both, serf1 and wild type, while CPK6 showed a significantly lower expression in serf1 leaves. On the other hand, CIPK18 was significantly downregulated in the wild type and serf1, while the expression of CIPK4 only decreased in wild-type leaves, which overlapped with the response of this gene to H2O2 treatment. In addition, CBL10 expression was decreased upon salt stress in both the mutant and the wild type. Thus, in contrast to roots,12 most genes encoding signaling components still respond to salt stress in the leaves of serf1.
Long-term salt stress results in the accumulation of toxic Na+ levels in the shoot, which impairs plant growth and viability.30 The transport of Na+ and K+ is performed by different sets of ion channels and transporters. Upon salt stress, a downregulation of genes encoding K+ channels was observed in wild-type and serf1 leaves, including KAT1 and Os06 g14030 (SKOR) (Fig. 2A). Glutamate-like receptor (GLR) genes displayed a similar response in wild-type and serf1, with the exception of GLR2.8, which was downregulated in serf1 (Fig. 2A). The high-affinity K+ transporters (HKT) are mainly involved in the transport of Na+ via the plasma membrane.34 During the initial response to salinity stress, we noticed a downregulation of HKT1 expression in both wild-type and serf1 leaves (Fig. 2B). The downregulation of HKT1 overlapped with the initial response of this gene to the treatment of roots with H2O2. In addition, downregulation of HKT6, HKT7 and HKT8 was observed in serf1, but not the wild type. These downregulated genes were not affected by the treatment with mannitol and ABA, but were mildly induced by H2O2, which indicates an altered stress perception in serf1. The HAK family represents another group of transporters involved in the transport of K+.35HAK2, HAK18 and HAK24 were downregulated in leaves of both serf1 and the wild type. However, HAK4 and HAK13 were differentially expressed in serf1 as compared with the wild type.
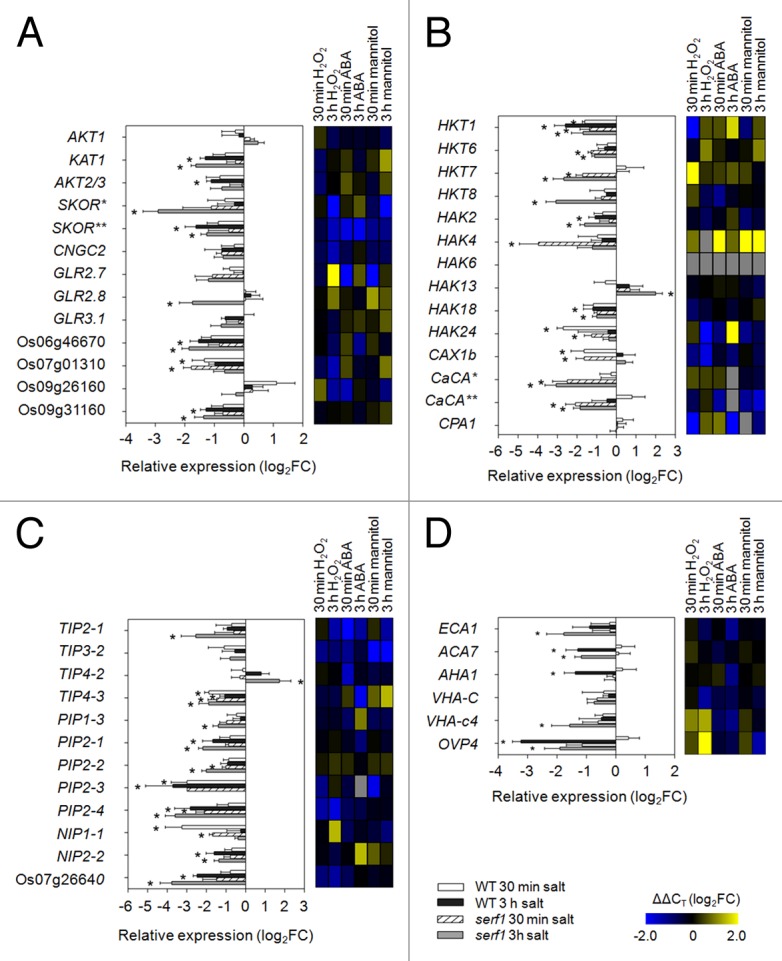
Figure 2. SERF1 influences the transcriptional response of genes relevant for ionic and osmotic homeostasis in leaves. Expression levels of genes encoding (A) ion channels, (B) transporters, (C) aquaporins and (D) ATPases were measured in leaves of wild-type and serf1 plants subjected to salt stress for 30 min or 3 h (100 mM NaCl). ACTIN (Os03g50885) was used as reference gene. Data represent means ± SE from 3 biological replicates. An asterisk (*) indicates a significant difference between treated and control plants (P ≤ 0.05; the Student t test). In the heat map the gene expression under H2O2 (5 mM), ABA (5 µM) or mannitol (100 mM) treatment is shown in log2FC relative to the control situation with n = 3. The potassium channels SKOR* and SKOR** are encoded by Os04g36740 and Os06g14030, respectively. Os06g46670, Os07g01310, Os09g26160 and Os09g31160 encode GLRs. CaCA* and CaCA** are encoded by Os11g01580 and Os12g42910, respectively. Os07g26640 encodes an aquaporin. FC, fold change; WT, wild type; GLR, glutamate-like receptor; CaCA, calcium cation antiporter.
Wild-type and serf1 leaves strongly responded to salt stress with respect to genes encoding aquaporins (Fig. 2C). Seven leaf-expressed aquaporin genes had a reduced expression upon salt stress in wild-type leaves, whereas 10 genes responded in serf1. TIP2–1, TIP4–2, PIP1–3 and PIP2-2 were differentially expressed in the leaves of the mutant. In contrast to roots, in serf1 leaves more proton- and calcium-transporting ATPase genes responded than in the wild type (Fig. 2D). ACA7 and OVP4 were downregulated in both, wild-type and serf1 leaves. AHA1 was only downregulated in wild-type plants, while the expression for the ER- and vacuole-associated ATPase genes (ECA1 and VHA-c4, respectively) was significantly decreased in serf1 upon salt stress.
As SERF1 is a direct transcriptional regulator of several TF genes in roots,12 we tested the expression of genes coding for known salt tolerance-promoting TFs (Fig. 3). For most of them, H2O2 treatment of roots resulted in their expressional induction in leaves, even stronger than observed under salt treatment. Among the NAC TFs, SNAC1 was induced in both wild type and serf1, while for SNAC2 a partially impaired response was observed in serf1 leaves. Furthermore, all thr3ee tested ZFP genes were induced in the wild type within 30 min of salt stress, while in serf1 leaves only ZFP182 responded. In addition, DREB2A and bZIP23 expression was attenuated in serf1 leaves, while the response of AP37 in serf1 was comparable to that of the stressed wild type. Besides this, the drought/salt stress marker genes LEA and LEA3 exhibited a rapid induction in wild-type leaves but an attenuated or no response in the mutant (Fig. 3).
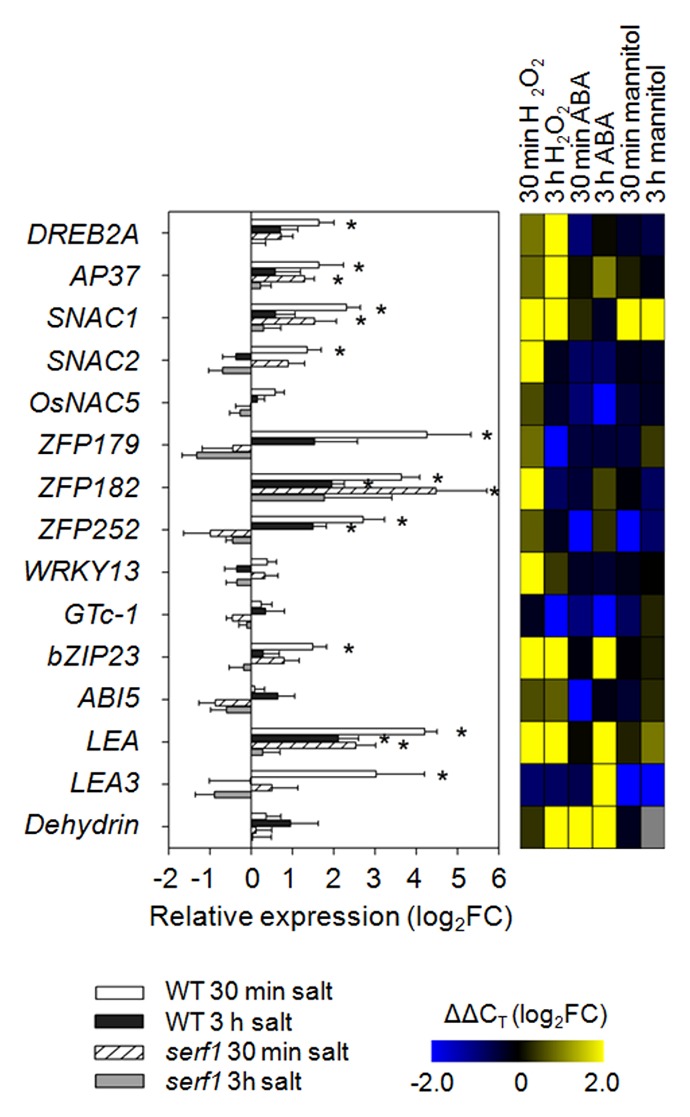
Figure 3. Effect of SERF1 on the expressional response of transcription factor genes in leaves. Expression levels of transcription factor genes and salt stress markers were measured in leaves of wild-type and serf1 plants subjected to salt stress for 30 min or 3 h (100 mM NaCl). ACTIN (Os03g50885) was used as reference gene. Data represent means ± SE from 3 biological replicates. An asterisk (*) indicates a significant difference between treated and control plants (P ≤ 0.05; the Student t test). In the heat map the gene expression under H2O2 (5 mM), ABA (5 µM) or mannitol (100 mM) treatment is shown in log2FC relative to the control situation with n = 3. WT; wild type; FC, fold change.
Discussion
We previously showed that SERF1 controls the initial transcriptional response of the root toward salt stress in a ROS-dependent manner.12 Here, we reveal that the impaired salt stress tolerance in serf1 is characterized by a rapid decrease in carbohydrate (sugar) content of the leaves during the early phase of salt stress. As the roots of serf1 mutant plants exhibit a ROS burst upon salt stress, we expected that root-to-shoot communication during salt stress should still be intact.11,12 However, the increase in leaf temperature of serf1 plants within 20 min of salt stress indicates that the leaves of the mutant perceive salt stress or execute the response to salt stress in a different manner than those of the wild type.12 The metabolite and expression profiles of roots and shoots upon salt stress display several features that suggest an altered root-to-shoot signal involving an ABA-independent component, potentially ROS. Furthermore, the rapid decline in sugars might be symptomatic to the salt sensitivity of serf1, as sugars not only act in energy metabolism but also function as osmoprotectants.36
The rapid increase in leaf temperature observed in serf1 mutant plants under salt stress is most likely the consequence of stomatal closure.12 As ABA is the major phytohormone that controls stomatal closure during abiotic stress,37 ABA might also be related to a different perception of salt stress by serf1 leaves. The effect of ABA on the metabolic profile during abiotic stress is well documented, allowing distinction of ABA-dependent and -independent metabolic changes.21 The roots of salt-stressed serf1 and wild-type plants both showed an increase in the levels of Ile, Leu and Tyr, which are ABA-dependently regulated metabolites. On the other hand, metabolites that are regulated ABA-independently, like citrate and Gly, were affected in their response in serf1 (Table 1). Also in leaves, mainly ABA-independent metabolites were affected by the loss of SERF1, including GABA, citrate, galactinol and xylose (Table 2). These data fit with the observation that the growth response of serf1 toward ABA treatment is similar to that of the wild type.38 Furthermore, in roots, mainly the response of H2O2-responsive genes is impaired in serf1, but not of those controlled by ABA.12 Thus, the differential perception of salt stress in leaves seems to occur independent from ABA. In this respect it is important to note that ABA-induced stomatal closure requires the formation of ROS.37 Therefore, the rapid increase in leaf temperature might primarily be the consequence of altered ROS signaling. However, SERF1 does not affect the ROS burst itself and presumably also not the cell-to-cell propagation of the ROS signal.11,12
Since SERF1 is not induced in leaf tissues during short-term exposure to salt or H2O2,12 we expected that any observed difference in the transcriptional response in leaves is the consequence of an altered root-to-shoot signal. In roots, SERF1 is required for the expression of MAPK cascade genes. However, in leaves of both the wild type and serf1 most MAPK genes responded to salt stress, including MAP3K6 and MAPK5, which are directly activated by SERF1 in the root (Fig. 1A).12 Interestingly, application of H2O2 to roots induced the expression of MAPK5, MAP3K4, MAP3K6, MAP3K12, MAP3K18 and MAP3K19 in leaves. As in serf1 leaves all these genes responded similarly to the wild type under salt stress (Fig. 1A), it supports the idea that SERF1 is not required for the cell-to-cell propagation of the ROS signal from the root to the shoot. Still, as discussed below, several ROS-responsive genes were affected in serf1. Moreover, ABA treatment of roots hardly affected the expression of salt-responsive genes in leaves, supporting the notion that ABA does not act as a long-distance signal during salt stress.19 In addition, previous studies suggested the existence of a hydraulic signal that is required for the root-to-shoot communication during water deficit.19 In this sense, it is interesting to note that only in serf1 leaves the osmotic stress-related MAP3K23 gene was induced (Fig. 1A).14 In agreement with this is the finding that among the salt-responsive genes tested in roots, only aquaporin genes are more responsive in serf1 than the wild type.12
Although ionic stress is not expected to occur within the initial phase of salt stress,30 we found that several of the H2O2-inducible HKT genes (HKT6, HKT7, and HKT8) are specifically downregulated in serf1 leaves (Fig. 2). HKT transporters can function as high-affinity Na+ transporters in rice. To prevent Na+ influx during salt stress, HKT1 (OsHKT2;1) expression is downregulated,39 which we also observed here for the wild type and serf1. HKT6 (OsHKT1;3) has been shown to be permeable for Na+ but not K+, however, its biological role is not clear yet.40 The additional downregulation of HKT8 (SKC1) in serf1 is rather surprising as it provides salinity tolerance by maintaining K+ homeostasis in the shoot.41 The differential expression of the HKT genes in serf1 leaves suggests an early adaptation of ion homeostasis.
Similar to MAPK encoding genes, most of the tested TF genes were found to be upregulated in leaves upon treatment of roots with H2O2. Again, in comparison to roots, the majority of the tested TF genes was still induced in the leaves of serf1 under salt stress (Fig. 3). However, 2 ROS-responsive zinc-finger TF genes, ZFP179 and ZFP252, were not induced in serf1 leaves. Previously, we showed that ZFP179 is a direct target of SERF1.12 Moreover, ZFP179 is required to provide oxidative stress tolerance in leaves, which is also impaired in serf1.12,16 In addition, the ZFP179-regulated gene LEA3 is not responding in serf1 leaves (Fig. 3). Although the leaf perceives a ROS signal from the root, as indicated by the differential expression of the majority of the ROS-responsive genes, the mechanism that controls the expression of ZFP179 might be distinctly different.
Here, we analyzed the contribution of the SERF1 signaling cascade to root-to-shoot communication during salt stress in rice. Our data indicate that SERF1-dependent signaling occurs independent of ABA. Furthermore, the perception of salt stress at the shoot still occurs and appears to cause a partially stronger osmotic stress signal. The transcriptional data indicate that in roots SERF1 can propagate a ROS signal to communicate with the shoot, however, the perception of the signal is altered as less ROS-related genes responded in serf1 leaves. Whereas SERF1 is essential for the expression of ROS-responsive genes during salt stress in the root, its effect on the transcriptional response of the shoot is less prominent.
Materials and Methods
Plant material and growth conditions
Homozygous SERF1 knockout plants (serf1) were grown as described previously.12 Treatment of hydroponically-grown transgenic lines with salt (100 mM NaCl), ABA (5 µM), mannitol (100 mM) or H2O2 (5 mM) was performed as described.12
Metabolic profiling analysis
Rice tissue (roots and leaves) were harvested after 3 h of salt stress (100 mM NaCl) or mock treatment, and metabolites were extracted from 6 replicates. Extraction and derivatization of metabolites from tissues for gas chromatography - mass spectrometry (GC-MS) analysis were performed as reported.42 GC-MS data were obtained using an Agilent 7683 series auto-sampler (Agilent Technologies GmbH, Waldbronn, Germany), coupled to an Agilent 6890 gas chromatograph - Leco Pegasus 2 time-of-flight mass spectrometer (LECO, St. Joseph, MI, USA). Identical chromatogram acquisition parameters were used as those previously described.43 Chromatograms were exported from Leco ChromaTOF software (version 3.25) to R software (http://www.r-project.org/). Peak detection, retention time (RT) alignment and library matching were obtained using the TargetSearch package from bioconductor.44 Metabolites were quantified by the peak intensity of a selective mass. Metabolite intensities were normalized by the fresh weight, followed by the internal standard (13C sorbitol) and log2 transformation.
RNA extraction and qRT-PCR
RNA isolation and cDNA synthesis for expression profiling on root and leaf tissues were performed as described previously.45 Three biological replicates were used for each experiment. Oligonucleotide sequences for expression profiling of early salt-responsive marker genes and TF genes with a known role in salt tolerance can be found in Schmidt et al.12
Acknowledgments
This work was in part conducted within the ERA-NET Plant Genomics project TRIESTER - Trilateral Initiative for Enhancing Salt Tolerance in Rice for which funding was provided by the Federal Ministry of Education and Research (BMBF) in Germany (grant no. 0313993A). Romy Schmidt thanks the FAZIT Stiftung for providing a PhD fellowship. We thank Aenne Eckardt and Gudrun Wolte for technical assistance.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1.Negrão S, Courtois B, Ahmadi N, Abreu I, Saibo N, Oliveira MM. Recent updates on salinity stress in rice: from physiological to molecular responses. Crit Rev Plant Sci. 2011;30:329–77. doi: 10.1080/07352689.2011.587725. [DOI] [Google Scholar]
- 2.Kovach MJ, McCouch SR. Leveraging natural diversity: back through the bottleneck. Curr Opin Plant Biol. 2008;11:193–200. doi: 10.1016/j.pbi.2007.12.006. [DOI] [PubMed] [Google Scholar]
- 3.Negrão S, Almadanim MC, Pires IS, Abreu IA, Maroco J, Courtois B, Gregorio GB, McNally KL, Oliveira MM. New allelic variants found in key rice salt-tolerance genes: an association study. Plant Biotechnol J. 2013;11:87–100. doi: 10.1111/pbi.12010. [DOI] [PubMed] [Google Scholar]
- 4.Dionisio-Sese ML, Tobita S. Antioxidant responses of rice seedlings to salinity stress. Plant Sci. 1998;135:1–9. doi: 10.1016/S0168-9452(98)00025-9. [DOI] [Google Scholar]
- 5.Schippers JHM, Nguyen HM, Lu D, Schmidt R, Mueller-Roeber B. ROS homeostasis during development: an evolutionary conserved strategy. Cell Mol Life Sci. 2012;69:3245–57. doi: 10.1007/s00018-012-1092-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Foyer CH, Noctor G. Redox homeostasis and antioxidant signaling: a metabolic interface between stress perception and physiological responses. Plant Cell. 2005;17:1866–75. doi: 10.1105/tpc.105.033589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hong CY, Chao YY, Yang MY, Cheng SY, Cho SC, Kao CH. NaCl-induced expression of glutathione reductase in roots of rice (Oryza sativa L.) seedlings is mediated through hydrogen peroxide but not abscisic acid. Plant Soil. 2009;320:103–15. doi: 10.1007/s11104-008-9874-z. [DOI] [Google Scholar]
- 8.Ma L, Zhang H, Sun L, Jiao Y, Zhang G, Miao C, Hao F. NADPH oxidase AtrbohD and AtrbohF function in ROS-dependent regulation of Na⁺/K⁺homeostasis in Arabidopsis under salt stress. J Exp Bot. 2012;63:305–17. doi: 10.1093/jxb/err280. [DOI] [PubMed] [Google Scholar]
- 9.Jiang C, Belfield EJ, Mithani A, Visscher A, Ragoussis J, Mott R, Smith JA, Harberd NP. ROS-mediated vascular homeostatic control of root-to-shoot soil Na delivery in Arabidopsis. EMBO J. 2012;31:4359–70. doi: 10.1038/emboj.2012.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Miller G, Schlauch K, Tam R, Cortes D, Torres MA, Shulaev V, Dangl JL, Mittler R. The plant NADPH oxidase RBOHD mediates rapid systemic signaling in response to diverse stimuli. Sci Signal. 2009;2:ra45. doi: 10.1126/scisignal.2000448. [DOI] [PubMed] [Google Scholar]
- 11.Mittler R, Vanderauwera S, Suzuki N, Miller G, Tognetti VB, Vandepoele K, Gollery M, Shulaev V, Van Breusegem F. ROS signaling: the new wave? Trends Plant Sci. 2011;16:300–9. doi: 10.1016/j.tplants.2011.03.007. [DOI] [PubMed] [Google Scholar]
- 12.Schmidt R, Mieulet D, Hubberten HM, Obata T, Hoefgen R, Fernie AR, Fisahn J, San Segundo B, Guiderdoni E, Schippers JHM, et al. Salt-responsive ERF1 regulates reactive oxygen species-dependent signaling during the initial response to salt stress in rice. Plant Cell. 2013;25:2115–31. doi: 10.1105/tpc.113.113068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xiong L, Yang Y. Disease resistance and abiotic stress tolerance in rice are inversely modulated by an abscisic acid-inducible mitogen-activated protein kinase. Plant Cell. 2003;15:745–59. doi: 10.1105/tpc.008714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jung KH, Cao P, Seo YS, Dardick C, Ronald PC. The Rice Kinase Phylogenomics Database: a guide for systematic analysis of the rice kinase super-family. Trends Plant Sci. 2010;15:595–9. doi: 10.1016/j.tplants.2010.08.004. [DOI] [PubMed] [Google Scholar]
- 15.Matsukura S, Mizoi J, Yoshida T, Todaka D, Ito Y, Maruyama K, Shinozaki K, Yamaguchi-Shinozaki K. Comprehensive analysis of rice DREB2-type genes that encode transcription factors involved in the expression of abiotic stress-responsive genes. Mol Genet Genomics. 2010;283:185–96. doi: 10.1007/s00438-009-0506-y. [DOI] [PubMed] [Google Scholar]
- 16.Sun SJ, Guo SQ, Yang X, Bao YM, Tang HJ, Sun H, Huang J, Zhang HS. Functional analysis of a novel Cys2/His2-type zinc finger protein involved in salt tolerance in rice. J Exp Bot. 2010;61:2807–18. doi: 10.1093/jxb/erq120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sauter A, Davies WJ, Hartung W. The long-distance abscisic acid signal in the droughted plant: the fate of the hormone on its way from root to shoot. J Exp Bot. 2001;52:1991–7. doi: 10.1093/jexbot/52.363.1991. [DOI] [PubMed] [Google Scholar]
- 18.Fricke W, Akhiyarova G, Veselov D, Kudoyarova G. Rapid and tissue-specific changes in ABA and in growth rate in response to salinity in barley leaves. J Exp Bot. 2004;55:1115–23. doi: 10.1093/jxb/erh117. [DOI] [PubMed] [Google Scholar]
- 19.Christmann A, Weiler EW, Steudle E, Grill E. A hydraulic signal in root-to-shoot signalling of water shortage. Plant J. 2007;52:167–74. doi: 10.1111/j.1365-313X.2007.03234.x. [DOI] [PubMed] [Google Scholar]
- 20.Sanchez DH, Siahpoosh MR, Roessner U, Udvardi M, Kopka J. Plant metabolomics reveals conserved and divergent metabolic responses to salinity. Physiol Plant. 2008;132:209–19. doi: 10.1111/j.1399-3054.2007.00993.x. [DOI] [PubMed] [Google Scholar]
- 21.Urano K, Maruyama K, Ogata Y, Morishita Y, Takeda M, Sakurai N, Suzuki H, Saito K, Shibata D, Kobayashi M, et al. Characterization of the ABA-regulated global responses to dehydration in Arabidopsis by metabolomics. Plant J. 2009;57:1065–78. doi: 10.1111/j.1365-313X.2008.03748.x. [DOI] [PubMed] [Google Scholar]
- 22.Schmidt R, Schippers JHM, Mieulet D, Obata T, Fernie AR, Guiderdoni E, Mueller-Roeber B. MULTIPASS, a rice R2R3-type MYB transcription factor, regulates adaptive growth by integrating multiple hormonal pathways. Plant J. 2013;76:258–73. doi: 10.1111/tpj.12286. [DOI] [PubMed] [Google Scholar]
- 23.Oliver MJ, Guo L, Alexander DC, Ryals JA, Wone BW, Cushman JC. A sister group contrast using untargeted global metabolomic analysis delineates the biochemical regulation underlying desiccation tolerance in Sporobolus stapfianus. Plant Cell. 2011;23:1231–48. doi: 10.1105/tpc.110.082800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Soga T, Baran R, Suematsu M, Ueno Y, Ikeda S, Sakurakawa T, Kakazu Y, Ishikawa T, Robert M, Nishioka T, et al. Differential metabolomics reveals ophthalmic acid as an oxidative stress biomarker indicating hepatic glutathione consumption. J Biol Chem. 2006;281:16768–76. doi: 10.1074/jbc.M601876200. [DOI] [PubMed] [Google Scholar]
- 25.Widodo, Patterson JH, Newbigin E, Tester M, Bacic A, Roessner U. Metabolic responses to salt stress of barley (Hordeum vulgare L.) cultivars, Sahara and Clipper, which differ in salinity tolerance. J Exp Bot. 2009;60:4089–103. doi: 10.1093/jxb/erp243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Peremarti A, Bassie L, Christou P, Capell T. Spermine facilitates recovery from drought but does not confer drought tolerance in transgenic rice plants expressing Datura stramonium S-adenosylmethionine decarboxylase. Plant Mol Biol. 2009;70:253–64. doi: 10.1007/s11103-009-9470-5. [DOI] [PubMed] [Google Scholar]
- 27.Mano Y, Nemoto K. The pathway of auxin biosynthesis in plants. J Exp Bot. 2012;63:2853–72. doi: 10.1093/jxb/ers091. [DOI] [PubMed] [Google Scholar]
- 28.Munns R. Comparative physiology of salt and water stress. Plant Cell Environ. 2002;25:239–50. doi: 10.1046/j.0016-8025.2001.00808.x. [DOI] [PubMed] [Google Scholar]
- 29.Sirault XR, James RA, Furbank RT. A new screening method for osmotic component of salinity tolerance in cereals using infrared thermography. Funct Plant Biol. 2009;36:970–7. doi: 10.1071/FP09182. [DOI] [PubMed] [Google Scholar]
- 30.Munns R, Tester M. Mechanisms of salinity tolerance. Annu Rev Plant Biol. 2008;59:651–81. doi: 10.1146/annurev.arplant.59.032607.092911. [DOI] [PubMed] [Google Scholar]
- 31.Renault H, Roussel V, El Amrani A, Arzel M, Renault D, Bouchereau A, Deleu C. The Arabidopsis pop2-1 mutant reveals the involvement of GABA transaminase in salt stress tolerance. BMC Plant Biol. 2010;10:20. doi: 10.1186/1471-2229-10-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Araújo WL, Nunes-Nesi A, Fernie AR. Fumarate: Multiple functions of a simple metabolite. Phytochemistry. 2011;72:838–43. doi: 10.1016/j.phytochem.2011.02.028. [DOI] [PubMed] [Google Scholar]
- 33.Nishizawa A, Yabuta Y, Yoshida E, Maruta T, Yoshimura K, Shigeoka S. Arabidopsis heat shock transcription factor A2 as a key regulator in response to several types of environmental stress. Plant J. 2006;48:535–47. doi: 10.1111/j.1365-313X.2006.02889.x. [DOI] [PubMed] [Google Scholar]
- 34.Sassi A, Mieulet D, Khan I, Moreau B, Gaillard I, Sentenac H, Véry AA. The rice monovalent cation transporter OsHKT2;4: revisited ionic selectivity. Plant Physiol. 2012;160:498–510. doi: 10.1104/pp.112.194936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gupta M, Qiu X, Wang L, Xie W, Zhang C, Xiong L, Lian X, Zhang Q. KT/HAK/KUP potassium transporters gene family and their whole-life cycle expression profile in rice (Oryza sativa) Mol Genet Genomics. 2008;280:437–52. doi: 10.1007/s00438-008-0377-7. [DOI] [PubMed] [Google Scholar]
- 36.Garcia AB, Engler J, Iyer S, Gerats T, Van Montagu M, Caplan AB. Effects of Osmoprotectants upon NaCl Stress in Rice. Plant Physiol. 1997;115:159–69. doi: 10.1104/pp.115.1.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Desikan R, Cheung MK, Bright J, Henson D, Hancock JT, Neill SJ. ABA, hydrogen peroxide and nitric oxide signalling in stomatal guard cells. J Exp Bot. 2004;55:205–12. doi: 10.1093/jxb/erh033. [DOI] [PubMed] [Google Scholar]
- 38.Schmidt R, Schippers JHM, Mieulet D, Watanabe M, Hoefgen R, Guiderdoni E, Mueller-Roeber B. SALT-RESPONSIVE ERF1 is a negative regulator of grain filling and gibberellin-mediated seedling establishment in rice. Mol Plant. 2014;7:404–21. doi: 10.1093/mp/sst131. [DOI] [PubMed] [Google Scholar]
- 39.Horie T, Costa A, Kim TH, Han MJ, Horie R, Leung HY, Miyao A, Hirochika H, An G, Schroeder JI. Rice OsHKT2;1 transporter mediates large Na+ influx component into K+-starved roots for growth. EMBO J. 2007;26:3003–14. doi: 10.1038/sj.emboj.7601732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jabnoune M, Espeout S, Mieulet D, Fizames C, Verdeil JL, Conéjéro G, Rodríguez-Navarro A, Sentenac H, Guiderdoni E, Abdelly C, et al. Diversity in expression patterns and functional properties in the rice HKT transporter family. Plant Physiol. 2009;150:1955–71. doi: 10.1104/pp.109.138008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ren ZH, Gao JP, Li LG, Cai XL, Huang W, Chao DY, Zhu MZ, Wang ZY, Luan S, Lin HX. A rice quantitative trait locus for salt tolerance encodes a sodium transporter. Nat Genet. 2005;37:1141–6. doi: 10.1038/ng1643. [DOI] [PubMed] [Google Scholar]
- 42.Lisec J, Schauer N, Kopka J, Willmitzer L, Fernie AR. Gas chromatography mass spectrometry-based metabolite profiling in plants. Nat Protoc. 2006;1:387–96. doi: 10.1038/nprot.2006.59. [DOI] [PubMed] [Google Scholar]
- 43.Weckwerth W, Wenzel K, Fiehn O. Process for the integrated extraction, identification and quantification of metabolites, proteins and RNA to reveal their co-regulation in biochemical networks. Proteomics. 2004;4:78–83. doi: 10.1002/pmic.200200500. [DOI] [PubMed] [Google Scholar]
- 44.Cuadros-Inostroza A, Caldana C, Redestig H, Kusano M, Lisec J, Peña-Cortés H, Willmitzer L, Hannah MA. TargetSearch--a Bioconductor package for the efficient preprocessing of GC-MS metabolite profiling data. BMC Bioinformatics. 2009;10:428. doi: 10.1186/1471-2105-10-428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schmidt R, Schippers JHM, Welker A, Mieulet D, Guiderdoni E, Mueller-Roeber B. Transcription factor OsHsfC1b regulates salt tolerance and development in Oryza sativa ssp. japonica. AoB Plants. 2012;2012:pls011. doi: 10.1093/aobpla/pls011. [DOI] [PMC free article] [PubMed] [Google Scholar]