Abstract
The capacity to promote cell dedifferentiation is widespread among plant species. We have recently reported that an AP2/ERF transcription factor WOUND INDUCED DEDIFFERENTIATION 1 (WIND1) and its paralogues, WIND2–4, promote cell dedifferentiation in Arabidopsis (Arabidopsis thaliana). Phylogenetic analyses suggest that AtWIND1 orthologs are found in land plants and that the shared peptide motifs between Arabidopsis paralogues are conserved in putative orthologs in dicotyledonous and monocotyledonous plants. In this study we show that AtWIND1 chemically induced rapeseed and tomato, as well as AtWIND1 constitutively expressed tobacco, promote callus formation on phytohormone-free medium. Our results suggest that the WIND1-mediated signaling cascade to promote cell dedifferentiation might be conserved in at least several species of Brassicaceae and Solanaceae.
Keywords: callus induction, cell dedifferentiation, AP2/ERF transcription factor, tissue culture, WIND1
Plant cells have high plasticity for differentiation against various environmental challenges. One of the most striking examples of cellular reprogramming in plants is dedifferentiation of somatic adult cells after wounding, the phenomenon also found in other multicellular organisms.1 Plants often form amorphous mass of dedifferentiated cells at the wound site, and this so-called “callus” not only functions as a coverage but it is also utilized as a source of de novo tissue or organ regeneration.2 We have recently reported that an AP2/ERF transcription factor WOUND INDUCED DEDIFFERENTIATION 1 (WIND1), also called RAP2.4,3,4 and its close homologs WIND2–4 are wound responsive and function as positive regulators of cell dedifferentiation in Arabidopsis (Arabidopsis thaliana).2,5,6 The ectopic overexpression of each of the WIND genes is sufficient to induce callus in Arabidopsis.6 If WIND genes have conserved biological function among plant species is of great interest, especially considering its potential use for the improvement of tissue culture methods. The WIND1 ortholog in an Arabidopsis relative, a salt cress Thellungiella halophila (ThWIND1-L), is reported to be wound-inducible and to have an ability to induce spontaneous callus formation in Arabidopsis.7 However, it is currently unknown how widely molecular entities of WIND genes or signaling pathway regulated by WIND genes is conserved.
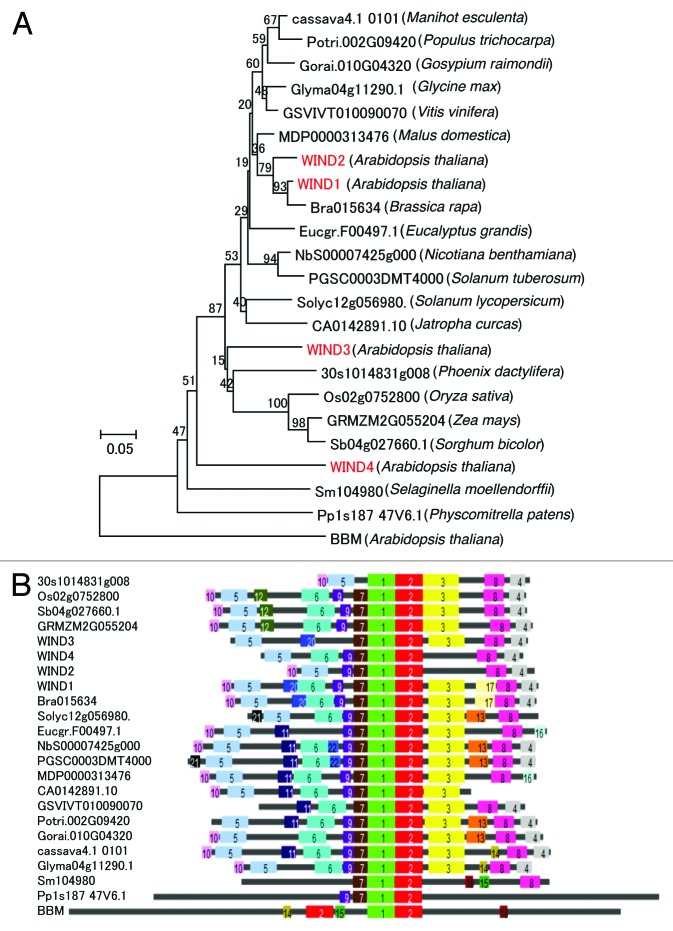
In order to elucidate how widespread WIND transcription factors are, we first performed database searches for 20 plant species: 12 Dicotyledoneae, 4 Monocotyledoneae, 1 Lycopodiophyta, 1 Bryopsida, and 2 Chlorophyceae (Table S1). Putative WIND orthologs were defined by the result of reciprocal BLAST searches (details are described in Materials and Methods), and they were found in all species we examined except Chlamydomonas reinhardtii and Volvox carteri (Fig. 1A; Table 1). Green algae are known to have AP2/ERF transcription family,8 thus WIND subclade likely evolved from AP2/ERF family after the divergence of land plants from green algae. Peptide motif composition is highly conserved in dicotyledons and monocotyledons; however, almost all conserved motifs except the AP2/ERF domain are lacking in the putative orthologs of Selaginella moellendorffii and Physcomitrella patens (Fig. 1B; Table 1). Thus it is plausible that molecular functions of these orthologs in Magnoliophyta are shared with WIND genes in Arabidopsis.
Figure 1. AtWIND1 orthologs are widely conserved in land plants. (A) A phylogenetic tree of putative WIND orthologs in 18 representative plant species. Another Arabidopsis AP2/ERF transcription factor BBM (AT5G17430) was employed as an outgroup. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (500 replicates) is shown next to the branches. Arabidopsis WIND1–4 were highlighted in red. All data set files used in this analysis are shown in Table S1. (B) Overrepresented peptide motifs among putative WIND orthologs. BBM (AT5G17430) was employed as an outgroup. Gene names and their plant sources are provided in Table 1. Each overrepresented peptide motif is automatically numbered and colored by the SALAD program for descriptive purpose.
Table 1. Paralogous genes in A. thaliana and putative orthologs in other plants.
| Gene name | Species | BLAST score | BLAST E-value | |
|---|---|---|---|---|
| AT1G78080 | WIND1 | Arabidopsis thaliana | 680 | 0 |
| AT1G22190 | WIND2 | Arabidopsis thaliana | 278 | 5.00E-74 |
| AT1G36060 | WIND3 | Arabidopsis thaliana | 239 | 3.00E-62 |
| AT5G65130 | WIND4 | Arabidopsis thaliana | 187 | 2.00E-46 |
| Bra015634 | Brassica rapa | 481 | 1.00E-135 | |
| Gorai.010G043200.1 | Gosypium raimondii | 332 | 5.00E-90 | |
| MDP0000313476 | Malus domestica | 327 | 2.00E-88 | |
| cassava4.1_010102m | Manihot esculenta | 326 | 2.00E-88 | |
| Glyma04 g11290.1 | Glycine max | 314 | 8.00E-85 | |
| NbS00007425 g0002.1 | Nicotiana benthamiana | 302 | 4.00E-81 | |
| GSVIVT01009007001 | Vitis vinifera | 293 | 2.00E-78 | |
| Potri.002G094200.1 | Populus trichocarpa | 291 | 2.00E-78 | |
| PGSC0003DMT400024508 | Solanum tuberosum | 282 | 3.00E-75 | |
| Eucgr.F00497.1 | Eucalyptus grandis | 263 | 2.00E-69 | |
| Solyc12 g056980.1.1 | Solanum lycopersicum | 259 | 2.00E-68 | |
| CA0142891.10 | Jatropha curcas | 235 | 5.00E-61 | |
| Os02 g0752800 | Oryza sativa | 232 | 4.00E-60 | |
| GRMZM2G055204_T01 | Zea mays | 224 | 1.00E-57 | |
| Sb04 g027660.1 | Sorghum bicolor | 219 | 4.00E-56 | |
| 30s1014831 g008 | Phoenix dactylifera | 197 | 2.00E-49 | |
| Sm104980 | Selaginella moellendorffii | 135 | 9.00E-31 | |
| Pp1s187_47V6.1 | Physcomitrella patens | 131 | 1.00E-29 |
Next, we asked if cellular and developmental response to WIND1 gain-of-function is common in different species. We chose crop species for this experiment, aiming for future applicative research. We have previously shown that 17β-estradiol-induced AtWIND1 promotes callus formation in Arabidopsis.6 In this study, we extended these observations and found that dexamethasone (DEX)-mediated AtWIND1 induction, namely 35S:AtWIND1-Glucocorticoid Receptor (GR) Arabidopsis treated with DEX, also strongly enhances callus induction on phytohormone-free media (Fig. 2A and B). We introduced the same DEX-mediated AtWIND1 induction system into rapeseed (Brassica napus), which belongs to the same family (Brassicaceae) as Arabidopsis. As shown in Figure 2C-E, the DEX treated-rapeseed plants carrying the 35S:AtWIND1-GR showed defective elongation in hypocotyls. Scanning electron microscopic observation clarified that round-shaped epidermal cells covered the AtWIND1-induced plant hypocotyl (Fig. 2D and E). This phenotype was also observed in AtWIND1-overexpressing Arabidopsis plants,6 suggesting that similar downstream events such as inhibition of normal differentiation is triggered by WIND1 in these 2 species and suppression of differentiation is one of WIND1 functions in these species. Moreover, AtWIND1 overexpressing rapeseed plants exhibited vigorous callus formation on the cotyledons and hypocotyls in the absence of any exogenous phytohormones (Fig. 2F-I). We observed these phenotypes in at least 4 independent transgenic plants, and the induction of callus formation was strictly DEX-dependent in all cases.
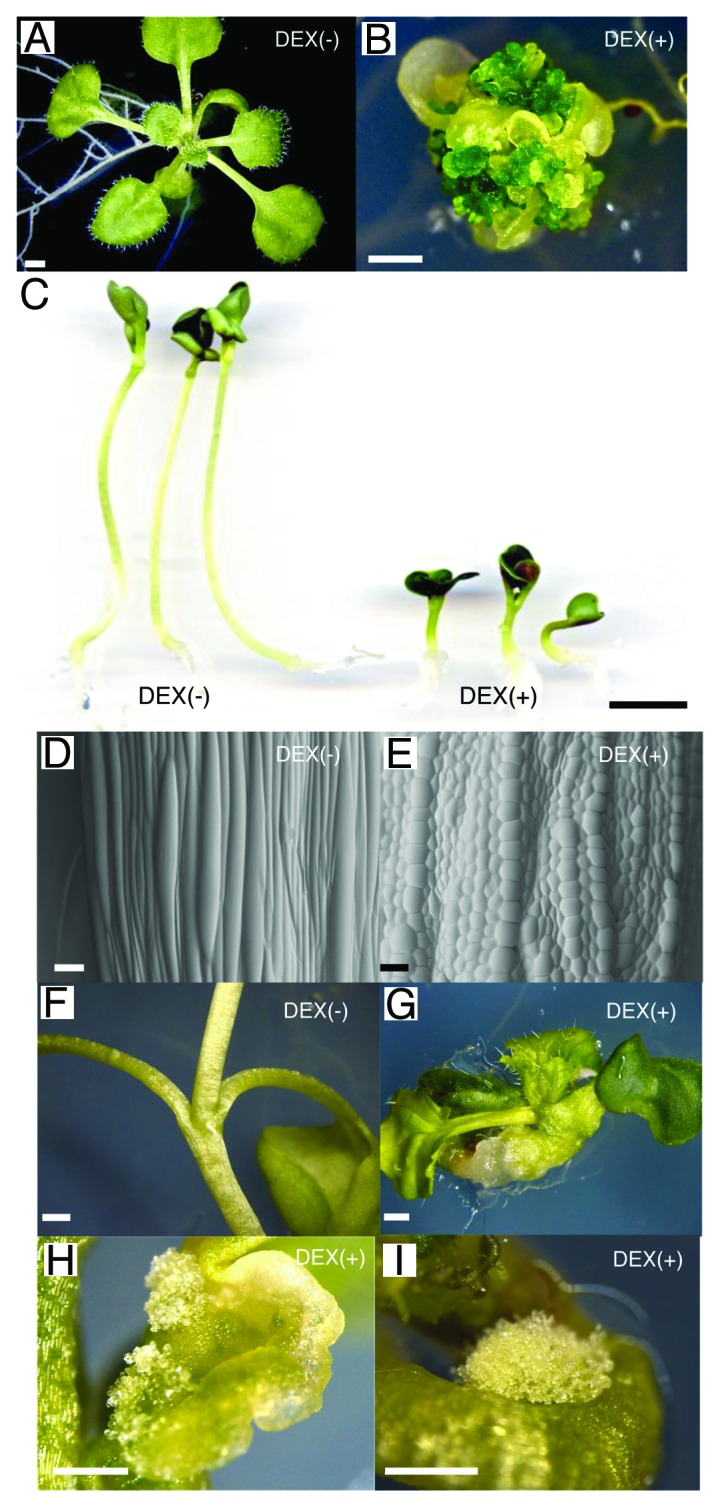
Figure 2. Chemical induction of AtWIND1 is sufficient to promote callus formation in Arabidopsis and rapeseed (Brassica napus). (A) Eighteen-d-old 35S:AtWIND1-Glucocorticoid Receptor (GR) Arabidopsis seedlings germinated on phytohormone-free MS medium without dexamethasone (DEX). (B) Sixty-d-old 35S:AtWIND1-GRArabidopsis seedlings germinated on phytohormone-free MS medium with 10 µM DEX. (C) Seven-d-old 35S:AtWIND1-GR rapeseed seedlings germinated on phytohormone-free MS medium without (-; left) or with (+; right) 10 µM dexamethasone (DEX). (D) and (E) Scanning electron microscopy photographs of rapeseed hypocotyls. Ten-d-old 35S:AtWIND1-GR seedlings germinated on phytohormone-free MS medium without (D) or with (E) 10 µM DEX. (F) to (I) Thirty-d-old 35S:AtWIND1-GR rapeseed seedlings germinated on phytohormone-free MS medium with (G, H, I) or without (F) 10 µM DEX. AtWIND1 causes abnormal swelling hypocotyl (G) and callus formation on cotyledon (H) and hypocotyls (I). Scale bars = 1 mm (A, B, F, G, H, I), 1 cm (C), 100 µm (D, E).
We next introduced the 35S:AtWIND1-GR construct into tomato (Solanum lycopersicum cv Micro-Tom) to see whether AtWIND1 functions in the family of Solanaceae. As shown in Figure 3C and D, 3 independent lines of AtWIND1-GR expressing tomato plants developed callus in the presence of DEX. In addition, vegetative shoots developed on the AtWIND1-induced callus (Fig. 3E). These shoots continued to grow and regenerated roots when excised and placed on the normal (DEX and phytohormone free) MS medium. They even regenerated whole plants (Fig. 3F). Thus, it is evident that tomato cells in the AtWIND1-induced callus is pluripotent. Taken together, our data suggest that the WIND1-dependent cell dedifferentiation pathway is conserved in the rapeseed and tomato and that AtWIND1 is perhaps able to activate innate factors that function in a cell dedifferentiation-signaling cascade, at least in the species we examined. It is also noteworthy that the callus formation in both cases was seen on hypocotyls after about 20 d incubation, while successive shoot regeneration occurred only in the tomato, suggesting certain tissue/species specific factors might contribute to the phenomena.

Figure 3. Chemical induction of AtWIND1 promotes callus formation and shoot regeneration in tomato (Solanum lycopersicum cv Micro-Tom). (A) Twenty-five-d-old 35S:AtWIND1-GR seedlings germinated on phytohormone-free MS medium without (-; left) or with (+; right) 10 µM DEX. (B) and (C) Magnified view of the hypocotyls grown without DEX (B) and with DEX (C). (D) and (E) Sixty-d-old 35S:AtWIND1-GR seedlings grown on the DEX medium (D) and magnified view of the developing callus (E). Arrows indicate ectopic shoots-like structures developing on the callus. (F) Plant regeneration from the 35S:AtWIND1-GR callus. Shoots-like structures that derived from 35S:AtWIND1-GR callus were excised and cultured on the MS medium for 24 d. An arrow marks the regenerating root. Scale bar: 1 cm (A), 1 mm (B, C, D, E, F).
Overexpression of WIND1 in Arabidopsis not only induced callus formation but also enabled us to establish cell culture lines that can be maintained in the absence of exogenous phytohormones.5 Thus, we checked if this is also possible in other species. We introduced the 35S:AtWIND1 construct into Nicotiana tobacum SR1. As a result, 31 transformants out of 36 we isolated (86%) aborted normal development and developed pale green leaves and friable tissue (Fig. 4A-C). Similar to the WIND1 overexpressing Arabidopsis plants, 35S:AtWIND1 tobacco plants that display severe phenotypes developed callus that could be subcultured for at least several years as so-called shooty callus (Fig. 4D).2 These results suggest that WIND1 can be utilized not only for callus induction but also for the establishment of callus cell lines.
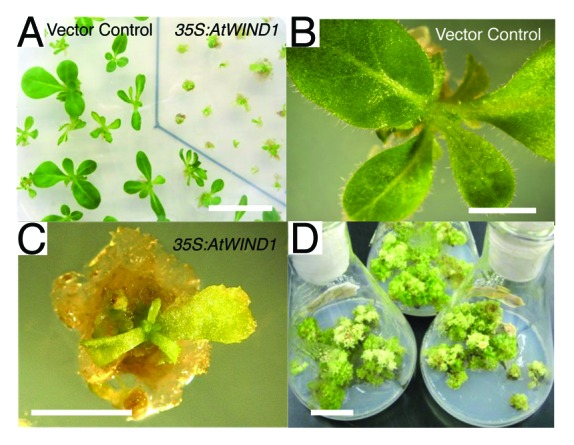
Figure 4. Constitutive expression of AtWIND1 promotes callus formation and can establish callus lines in Nicotiana tabacum (SR1). (A) Three-mo-old 35S:AtWIND1 plants and control plants transformed with an empty vector. (B) and (C) Magnified view of vector control plants (B) and 35S:AtWIND1 plants (C). (D) 35S:AtWIND1 callus can be subcultured and maintained on the MS medium without exogenous phytohormones. Scale bar: 3 cm (A, D), 5 mm (B, C)
Inducing cell fate reprogramming has been attempted for many years in plants to enhance tissue and organ regeneration, and modifying the activity of transcription factors has proved to be one of the most effective strategies. For instance, ectopic overexpression of an AP2/ERF transcription factor BABY BOOM (BBM) from Brassica napus (Bn) induces somatic embryos in both Brassica and Arabidopsis without exogenous plant hormones.9 The BnBBM overexpression system has been also successfully introduced into several crop species.10-12 Our data imply that like BBM, WIND proteins have a potential to improve efficiency of tissue regeneration since the putative orthologs are widely conserved among higher plants, and when Arabidopsis WIND1 is ectopically expressed in different species, it can function as a dedifferentiation promoting factor. Since various organs—for example, shoot, root, and somatic embryo—are regenerated from WIND1 induced-callus in Arabidopsis,2,5,7 the direction of regeneration may not been determined by WIND itself. Thus application of WIND with other organ initiation factors, using them as molecular switches to induce dedifferentiation and redifferentiation, may hold a future promise as one of the strategies for improving tissue culture engineering in plants.
Materials and Methods
Ortholog search and shared peptide motif analysis
The amino acid sequence of AtWIND1 was employed as a TBLASTN query to collect genes with high homology to AtWIND1 from other 20 plant species. The coding sequences of these genes were retrieved from databases for each species listed in Table S1. Each of the top 500 homologous genes from these species was then employed as a BLASTX query against all amino acid sequences in Arabidopsis. When the top-hit gene of this BLASTX search was WIND1, WIND2, WIND3, or WIND4, the query was defined as a WIND ortholog.
For phylogenetic analysis, amino acid sequences of representative putative orthologs in 18 species, WIND1, WIND2, WIND3, WIND4, and BBM were aligned by MAFFT13 and the phylogenetic tree was constructed by Neighbor-Joining method implemented in MEGA5.14
To find peptide motifs shared between the orthologs and paralogues, the same sequences were employed as a query for the interactive SALAD analysis of the SALAD database.15 This analysis found overrepresented short sequences among input multiple sequences based on MEME software.16
Plasmid construction and plants transformation
To generate the 35S:AtWIND1 plasmid that carries resistance to kanamycin, the 35S:AtWIND1 sequence on the p35SG vector, which was used in previous work,5 was cloned into the destination vector pBCKK, which is a derivative of pBCKH,17 using the Gateway LR clonase reaction (Invitrogen). To construct the 35S:AtWIND1-GR plasmid, the coding region of AtWIND15 was cloned into the p35SGRG vector which harbors the GR sequence between the Ω translation enhancer sequence and NOS terminal sequence in the p35SG entry vector.18 The resulting 35S:AtWIND1-GR sequence was then cloned into the pBCKK vector by the LR clonase reaction. Brassica napus cv Westar plants, Solanum lycopersicum cv Micro-Tom, and Nicotiana tobacum SR1 were transformed by the methods previously reported.19-21
Medium conditions
All culture media, except for the tobacco callus culture, contained Murashige and Skoog basal salt mixture (Wako), 1% sucrose and 0.6% Gellan Gum (Wako), and pH was adjusted to 5.8. The tobacco callus culture media contained 3% sucrose, and Gamborg’s B5 vitamin was additionally supplemented. The dexamethasone (Sigma) was dissolved in ethanol to make 10-mM stock solution and added to the medium after autoclaving.
Supplementary Material
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank Chika Ikeda, Mariko Mouri, Yasuko Yatomi, and Youko Ooi for their technical supports. We are also grateful to Dr Miho Ikeda for her advice on tobacco transformation. This work was supported by Grants-in-Aid for Scientific Research on Innovative Areas (Grant No. 22119010) and the Programme for Promotion of Basic and Applied Researches for Innovations in Bio-oriented Industry to Sugimoto K. Iwase A was funded by the RIKEN Special Postdoctoral Researchers Program and a grant from Japan Society for the Promotion of Science (Grant No. 24770053). The tomato resources used in this research were provided by the National BioResource Project (NBRP), Japanese Ministry of Education, Culture, Sports, Science and Technology (MEXT), Japan. The production of transgenic rapeseed and tomato plants was supported by the RIKEN Plant Transformation Network.
References
- 1.Birnbaum KD, Sánchez Alvarado A. Slicing across kingdoms: regeneration in plants and animals. Cell. 2008;132:697–710. doi: 10.1016/j.cell.2008.01.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ikeuchi M, Sugimoto K, Iwase A. Plant callus: mechanisms of induction and repression. Plant Cell. 2013;25:3159–73. doi: 10.1105/tpc.113.116053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Okamuro JK, Caster B, Villarroel R, Van Montagu M, Jofuku KD. The AP2 domain of APETALA2 defines a large new family of DNA binding proteins in Arabidopsis. Proc Natl Acad Sci U S A. 1997;94:7076–81. doi: 10.1073/pnas.94.13.7076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Delessert C, Wilson IW, Van Der Straeten D, Dennis ES, Dolferus R. Spatial and temporal analysis of the local response to wounding in Arabidopsis leaves. Plant Mol Biol. 2004;55:165–81. doi: 10.1007/s11103-004-0112-7. [DOI] [PubMed] [Google Scholar]
- 5.Iwase A, Mitsuda N, Koyama T, Hiratsu K, Kojima M, Arai T, Inoue Y, Seki M, Sakakibara H, Sugimoto K, et al. The AP2/ERF transcription factor WIND1 controls cell dedifferentiation in Arabidopsis. Curr Biol. 2011;21:508–14. doi: 10.1016/j.cub.2011.02.020. [DOI] [PubMed] [Google Scholar]
- 6.Iwase A, Ohme-Takagi M, Sugimoto K. WIND1: a key molecular switch for plant cell dedifferentiation. Plant Signal Behav. 2011;6:1943–5. doi: 10.4161/psb.6.12.18266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhou C, Guo J, Feng Z, Cui X, Zhu J. Molecular characterization of a novel AP2 transcription factor ThWIND1-L from Thellungiella halophila. Plant Cell Tissue Organ Cult. 2012 doi: 10.1007/s11240-012-0163-4. [DOI] [Google Scholar]
- 8.Magnani E, Sjölander K, Hake S. From endonucleases to transcription factors: evolution of the AP2 DNA binding domain in plants. Plant Cell. 2004;16:2265–77. doi: 10.1105/tpc.104.023135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boutilier K, Offringa R, Sharma VK, Kieft H, Ouellet T, Zhang L, Hattori J, Liu CM, van Lammeren AAM, Miki BLA, et al. Ectopic expression of BABY BOOM triggers a conversion from vegetative to embryonic growth. Plant Cell. 2002;14:1737–49. doi: 10.1105/tpc.001941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Srinivasan C, Liu Z, Heidmann I, Supena EDJ, Fukuoka H, Joosen R, Lambalk J, Angenent G, Scorza R, Custers JBM, et al. Heterologous expression of the BABY BOOM AP2/ERF transcription factor enhances the regeneration capacity of tobacco (Nicotiana tabacum L.) Planta. 2007;225:341–51. doi: 10.1007/s00425-006-0358-1. [DOI] [PubMed] [Google Scholar]
- 11.Deng W, Luo K, Li Z, Yang Y. A novel method for induction of plant regeneration via somatic embryogenesis. Plant Sci. 2009;177:43–8. doi: 10.1016/j.plantsci.2009.03.009. [DOI] [Google Scholar]
- 12.Heidmann I, de Lange B, Lambalk J, Angenent GC, Boutilier K. Efficient sweet pepper transformation mediated by the BABY BOOM transcription factor. Plant Cell Rep. 2011;30:1107–15. doi: 10.1007/s00299-011-1018-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Katoh K, Misawa K, Kuma K, Miyata T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002;30:3059–66. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–9. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mihara M, Itoh T, Izawa T. SALAD database: a motif-based database of protein annotations for plant comparative genomics. Nucleic Acids Res. 2010;38:D835–42. doi: 10.1093/nar/gkp831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 2009;37:W202-8. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mitsuda N, Hiratsu K, Todaka D, Nakashima K, Yamaguchi-Shinozaki K, Ohme-Takagi M. Efficient production of male and female sterile plants by expression of a chimeric repressor in Arabidopsis and rice. Plant Biotechnol J. 2006;4:325–32. doi: 10.1111/j.1467-7652.2006.00184.x. [DOI] [PubMed] [Google Scholar]
- 18.Ohta M, Matsui K, Hiratsu K, Shinshi H, Ohme-Takagi M. Repression domains of class II ERF transcriptional repressors share an essential motif for active repression. Plant Cell. 2001;13:1959–68. doi: 10.1105/TPC.010127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kohno-Murase J, Murase M, Ichikawa H, Imamura J. Effects of an antisense napin gene on seed storage compounds in transgenic Brassica napus seeds. Plant Mol Biol. 1994;26:1115–24. doi: 10.1007/BF00040693. [DOI] [PubMed] [Google Scholar]
- 20.Sun H-J, Uchii S, Watanabe S, Ezura H. A highly efficient transformation protocol for Micro-Tom, a model cultivar for tomato functional genomics. Plant Cell Physiol. 2006;47:426–31. doi: 10.1093/pcp/pci251. [DOI] [PubMed] [Google Scholar]
- 21.Horsch RB, Fry JE, Hoffmann NL, Eichholtz D, Rogers SG, Fraley RT. A simple and general method for transferring genes into plants. Science. 1985;227:1229–31. doi: 10.1126/science.227.4691.1229. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.