Abstract
RNA editing mediated by adenosine deaminases acting on RNA (ADARs) converts adenosine (A) to inosine (I) residues in dsRNA templates. While ADAR-1-mediated editing was essentially described for RNA viruses, the present work addresses the issue for two δ-retroviruses, human T-cell leukemia virus type 2 and simian T-cell leukemia virus type 3 (HTLV-2 and STLV-3). We examined whether ADAR-1 could edit HTLV-2 and STLV-3 virus genomes in cell culture and in vivo. Using a highly sensitive PCR-based method, referred to as 3DI-PCR, we showed that ADAR-1 could hypermutate adenosine residues in HTLV-2. STLV-3 hypermutation was obtained without using 3DI-PCR, suggesting a higher mutation frequency for this virus. Detailed analysis of the dinucleotide editing context showed preferences for 5′ ArA and 5′ UrA. In conclusion, the present observations demonstrate that ADAR-1 massively edits HTLV-2 and STLV-3 retroviruses in vitro, but probably remains a rare phenomenon in vivo.
RNA editing mediated by adenosine deaminases acting on RNA (ADARs) has been shown to be one of the most prevalent post-transcriptional RNA modification mechanisms in higher eukaryotes (Bass, 2002; Samuel, 2001). These enzymes convert adenosine (A) to inosine (I) residues in dsRNA templates. Inosine is essentially recognized by the translational machinery as guanine (G), leading to proteins that are frequently non-functional (Li et al., 1991). Three ADAR genes are known. They are specific for dsRNA. While ADAR-1 and ADAR-2 are expressed in many tissues, ADAR-3 is only expressed in the nervous system (Bass, 1997, 2002; Chen et al., 2000; Melcher et al., 1996). ADAR-1 gene consists of 17 exons across a 30 kb sequence (George & Samuel, 1999; Wang et al., 1995). ADAR-1 transcription is initiated from multiple promoters, one being inducible by type I and II interferons (IFNs), while the others are constitutively active (George & Samuel, 1999; Liu et al., 1997). Interestingly, of the ADAR-1 gene transcripts i.e. ADAR-1L and -1S, only the former can be induced by IFN-α/β and γ, underlining its role in antiviral responses. ADAR-1 editing was initially described in the context of subacute sclerosing panencephalitis, a rare chronic degenerative disease that occurs several years after measles virus infection (Cattaneo et al., 1987, 1988; Patterson et al., 2001; Wong et al., 1989, 1991). ADAR-1 editing was originally confined to negative-stranded viruses such as measles virus, vesicular stomatitis virus (O’Hara et al., 1984), human parainfluenza virus (Murphy et al., 1991), lymphocytic choriomeningitis virus (Grande-Pérez et al., 2002), respiratory syncytial virus (Martínez et al., 1997; Rueda et al., 1994), influenza virus (Suspène et al., 2011; Tenoever et al., 2007) and Rift Valley fever virus (Suspène et al., 2008). Recently, measles and influenza virus genomes derived from inactivated seasonal influenza and live-attenuated measles vaccines were also shown to be edited by ADAR-1 (Suspène et al., 2011).
ADAR editing is not restricted to negative-stranded viruses since the hepatitis C virus (Taylor et al., 2005) genome was also found to be edited. Among retroviruses, A→G editing was first described for Rous-associated virus RAV-1 (Hajjar & Linial, 1995), avian leukosis virus (Felder et al., 1994) and more recently for human immunodeficiency virus-1 (HIV-1) (Doria et al., 2009; Phuphuakrat et al., 2008) although at a low frequency. The δ-retrovirus group includes four human T-cell leukemia viruses (HTLV-1−4), and their simian T-cell leukemia virus counterparts (STLV-1, -2 and -3) (Mahieux & Gessain, 2011; Slattery et al., 1999). STLV-1 is widely distributed in Asian and African non-human primates with STLV-3 being only found in African non-human primates (Mahieux & Gessain, 2011).
HTLVs- or STLVs-edited genomes have not been described so far. Since, these viruses mostly replicate through clonal expansion of the infected cell, they may be less prone to genetic editing (Wattel et al., 1995). We have recently developed a PCR-based method, referred to as 3DI-PCR, that allows selective amplification of ADAR-edited RNAs (Suspène et al., 2008). Here, we show that HTLV-2 and STLV-3 RNAs can be efficiently and massively edited by ADAR-1.
In the first series of experiments, 293T cells were transfected (PolyFect; Qiagen) for 48 h with 2 µg plasmid encoding the full-length HTLV-2 genome (pH 6neo) (Chen et al., 1983), in the presence or absence of 0.5 µg ADAR-1 expression plasmid. Total RNA was recovered using Trizol and cDNA was synthesized by using random primers. A fragment of the HTLV-2 pX gene (nt 6660–6937) was amplified by using the previously described 3DI-PCR technique (Suspène et al., 2008). pH 2Sout 5′-CATAACCAGTATTCCCTTATCAACCC-3′ and pH 2Rout 5′-TTCTGCAGGAGCGTGAGGAGCGGGAGC-3′ primers were used for the first PCR round, while pH 2Rin 5′-GCTATAATAGACCTGCTAGCTTCTGC-3′ and pH 2Sin 5′-CGGCGCAGAAAGGAGCGCCTGCGG-3′ primers were used for the second PCR round. 3DI-PCR products corresponding to extracts obtained only from cells that have been transfected with both HTLV-2 and the ADAR-1 plasmid were recovered at a PCR denaturation temperature as low as 64.8 °C. They were then cloned and sequenced.
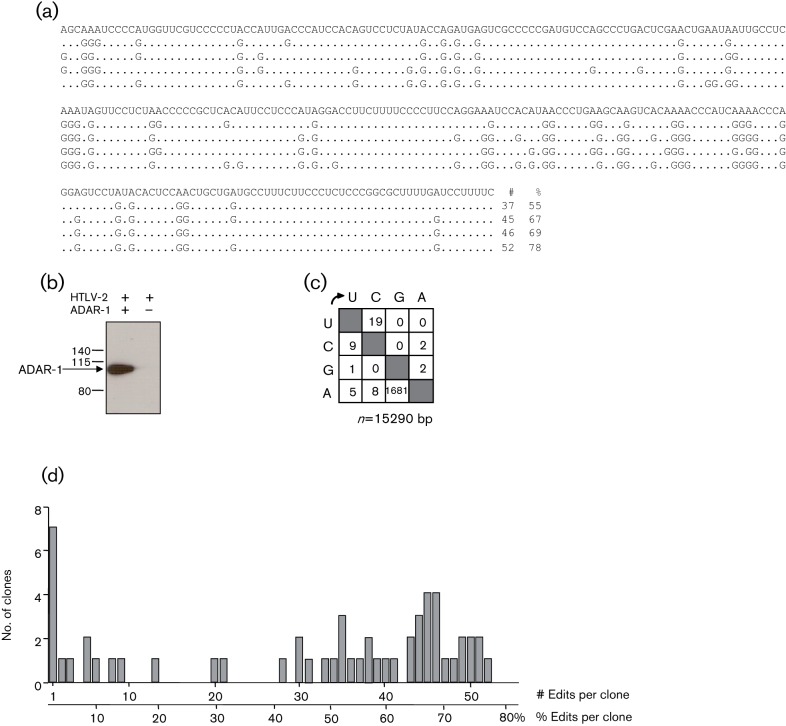
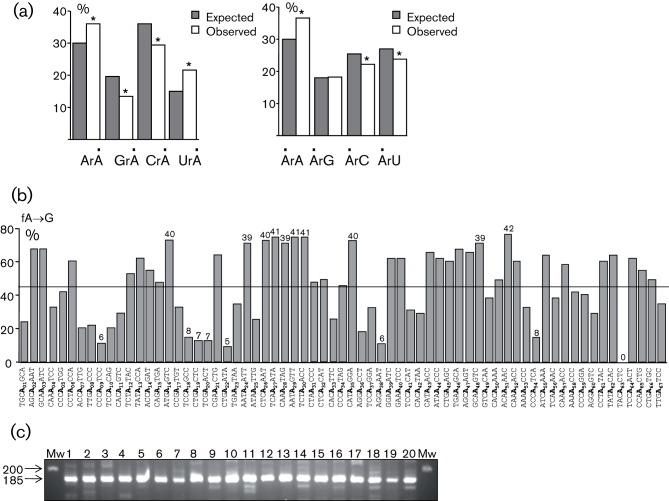
In the first series of analyses, 55 extensive and monotonously A→G-edited HTLV-2 pX sequences were recovered (Fig. 1a). As a control, a Western blot analysis was performed in cells transfected or not with the ADAR-1 expression plasmid (Liu et al., 1997) (Fig. 1b). ADAR-1 enzyme was able to extensively deaminate HTLV-2 RNA (Fig. 1a, c). Of note, hyperedited sequences could not be recovered in the absence of exogenously expressed ADAR-1 (data not shown). The A→G editing frequency distribution per clone shows some lightly edited genome with 1–5 mutations (~20 %) and a majority of highly mutated genomes (i.e. >20 mutations ~73 % of all A nucleotides, Fig. 1d). The mean editing frequency was ~46 % (range 1.5–78 %) (Fig. 1d). The dinucleotide context associated with adenosine editing showed a clear preference for 5′ArA and 5′ UrA and an aversion for 5′ GrA and 5′ CrA (Fig. 2a, left panel), which is in agreement with the literature (Lehmann & Bass, 2000; Suspène et al., 2008, 2011). In contrast, we could not detect any obvious 3′ context (Fig. 2a, right panel).
Fig. 1.
HTLV-2 genome is susceptible to ADAR-1 editing. (a) Selection of ADAR-1 hypermutated HTLV-2 sequences. The HTLV-2 sequence is given with respect to the retroviral plus strand. Only differences among hypermutated sequences are shown. The number of substitutions per sequence is indicated to the right. # and % indicates the number of A→G transitions and the percentage of A targets edited to G, respectively. (b) 293T cells were transfected or not with an ADAR-1 expression plasmid. Twenty-four hours after transfection, cell extracts were processed for Western blot analysis using an anti-ADAR-1 (ab88574; Abcam) antibody. (c) Mutation matrices of HTLV-2 hyperedited sequences. The 19 U→C transitions are distributed among several sequences. No U→C hyperedited sequence was found. (d) Frequency distribution of A→G editing per clone [# and % as in (a)].
Fig. 2.
HTLV-2 sequence context among sites of ADAR-1 deamination. (a) Dinucleotide analysis in 5′ (left) and 3′ (right) of HTLV-2-edited genomes. Dots indicate the edited base. χ2 analysis indicates dinucleotide frequencies that significantly deviate from the expected values (P<0.05, *). (b) Deamination frequencies across the HTLV-2 target sequence. Base-specific deamination frequencies were calculated among a collection of 55 sequences from all the reactions and are given as a function of local sequence context. The horizontal bar indicates the mean site deamination frequency, assuming no effect of the sequence context. Values on the top of each histogram represent the number of deaminated A among the 55 ADAR-1 HTLV-2-edited sequences. (c) Amplification of ADAR-1 cDNA obtained from 20 HTLV-2-infected individuals. The expected size of the amplified product is 185 bp. Mw, Molecular mass marker.
With a large number of clones sequenced, we defined site-specific editing frequency (Fig. 2b). Among the sixty-seven potential adenosine targets that are present in the HTLV-2 pX RNA template, we observed that some positions were highly refractory to mutations (see for example A09, A22 and A38). On the other hand, others were strongly deaminated. As an example, A51 was edited in 42 out of 55 clones sequenced (i.e. ~76 %). One residue (A63) was totally refractory to ADAR-1 editing.
In order to demonstrate a possible effect of ADAR-1 editing in vivo, peripheral blood mononuclear cells were obtained from 20 HTLV-2-infected individuals and immediately frozen (Douceron et al., 2012). Total RNA was extracted and cDNA was subsequently obtained using random primers. Given the fact that tax is usually expressed in less than 50 % of all HTLV-infected individuals, we amplified an env region by PCR and 3DI-PCR. However, hyperedited sequences could not be recovered upon cloning and sequencing of the 20 different 3DI-PCR products, suggesting that ADAR-1 editing is a rare event in vivo in HTLV-2-infected individuals (data not shown). As a control, ADAR-1 expression was detected by RT-PCR among the 20 HTLV-2-infected individuals, demonstrating that all ex vivo samples contained detectable levels of the ADAR-1 transcript (Fig. 2c).
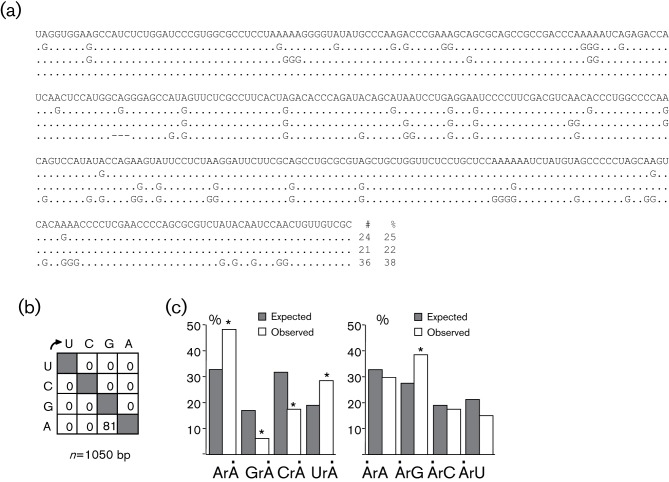
To determine whether ADAR-1 editing could occur for another δ-retrovirus, 293T cells were transfected (Polyfect; Qiagen) with an STLV-3 (PPAF-3) infectious molecular clone (Calattini et al., 2006; Chevalier et al., 2007) without overexpressing ADAR-1. Forty-eight hours post-transfection, total cellular RNA was recovered and cDNA was synthesized with random primers as described above. A fragment of the PPAF-3 pX mRNA was amplified by a nested PCR procedure. PCR products obtained at a 95 °C denaturation temperature were cloned and sequenced. Surprisingly, out of five clones sequenced, we recovered three ADAR-edited sequences (Fig. 3a). These sequences uniquely displayed A→G transitions (Fig. 3b), with a ~29 % mean adenosine substitution frequency per clone (range ~22–38 %). Once again and similar to the HTLV-2 results (Fig. 2), a significant preference for 5′ ArA and 5′ UrA contexts was observed (Fig. 3c). As ADAR-1 is constitutively expressed in 293T (Wang & Samuel, 2009), the majority of STLV-3-edited sequences can probably be ascribed to the ADAR-1 deaminase.
Fig. 3.
STLV-3 genome is susceptible to ADAR-1 editing. (a) The sequence is given with respect to the retroviral plus strand. Only differences among hypermutated sequences are shown. The number of substitutions per sequence is indicated to the right. # and % indicate the number of A→G transitions and the percentage of A→G edited, respectively. (b) Mutation matrices of STLV-3 hyperedited sequences. (c) Dinucleotide analysis in 5′ (left) and 3′ (right) of STLV-3-edited genomes. Dots indicate the edited base. χ2 analysis indicates dinucleotide frequencies that significantly deviate from the expected values (P<0.05, *).
The present study shows that HTLV-2 and STLV-3, two primate retroviruses, can be massively edited by ADAR-1 in cell culture. For HTLV-2, the selective and sensitive 3DI-PCR method was necessary to recover ADAR-1-edited sequences. By contrast ADAR-1-edited STLV-3 sequences were recovered after conventional nested-PCR. Since 293T cells were used for both experiments and since HTLV-2 and STLV-3 sequences were cloned in the same backbone SV2neo vector, this differential sensitivity to ADAR-1 is likely to be related to the viral genome. HTLV-2 and STLV-3 have different genetic structure at the 3′ end of their genome, although none of their gene products are known to be IFN antagonists. Another variable could be the degree of secondary structure in the target sequence. Indeed, ADAR-1 editing occurs by flipping out the adenosine in a dsRNA structure. Local structural differences might therefore explain the differences between HTLV-2 and STLV-3 results. If ADAR-1 was packaged more efficiently into STLV-3 capsids, the viral genome would probably be more efficiently edited. In any case, the susceptibility of STLV-3 to restriction by ADAR-1 is striking. The present data do not exclude editing of viral mRNAs in the cytoplasm as opposed to editing of genomic RNA within the virion.
The contrasts between the ADAR-1 and APOBEC3G editing enzymes are remarkable. Indeed, both are induced by IFN-α and target HTLV-1 or HIV-1 retroviruses. While these two retroviruses infect the same CD4+ T-lymphocytes, ADAR-1 massively edits HTLV-2 sequences in vitro, albeit at low frequency. While recent work has shown that HIV-1 can be edited by ADAR-1, very few A→G mutations could be detected (Doria et al., 2009; Phuphuakrat et al., 2008). Of note, we also failed to detect massive editing of HIV-1 TAR or env RNA with our 3DI-PCR approach (data not shown). In contrast, in the absence of Vif, HIV-1 cDNA is massively edited by APOBEC3G and 3DPCR(Suspène et al., 2005) is not needed to recover these sequences. By contrast, HTLV-1 cDNA is susceptible to APOBEC3G editing, but sensitive 3DPCR is necessary to recover edited sequences.
In conclusion, the present observation demonstrates that ADAR-1 massively edits HTLV-2 and STLV-3 retroviruses in vitro, but probably remains a rare phenomenon in vivo.
Acknowledgements
This work was supported by funds from the Institut Pasteur the CNRS and the DGA (Direction Générale des Armées). The Molecular Retrovirology Unit is ‘Equipe labellisée LIGUE 2010’. R. M. is supported by Ecole Normale Supérieure de Lyon, by INSERM and through funding from the Programme interdisciplinaire CNRS Maladies infectieuses émergentes and from NIH (grant AI072495-01). We thank Edward L. Murphy and Estelle Douceron for HTLV-2 samples and Antoine Gessain and Jocelyn Turpin for discussion and helpful comments.
References
- Bass B. L. (1997). RNA editing and hypermutation by adenosine deamination. Trends Biochem Sci 22, 157–162 10.1016/S0968-0004(97)01035-9 [DOI] [PubMed] [Google Scholar]
- Bass B. L. (2002). RNA editing by adenosine deaminases that act on RNA. Annu Rev Biochem 71, 817–846 10.1146/annurev.biochem.71.110601.135501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calattini S., Chevalier S. A., Duprez R., Afonso P., Froment A., Gessain A., Mahieux R. (2006). Human T-cell lymphotropic virus type 3: complete nucleotide sequence and characterization of the human tax3 protein. J Virol 80, 9876–9888 10.1128/JVI.00799-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cattaneo R., Rebmann G., Schmid A., Baczko K., ter Meulen V., Billeter M. A. (1987). Altered transcription of a defective measles virus genome derived from a diseased human brain. EMBO J 6, 681–688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cattaneo R., Schmid A., Eschle D., Baczko K., ter Meulen V., Billeter M. A. (1988). Biased hypermutation and other genetic changes in defective measles viruses in human brain infections. Cell 55, 255–265 10.1016/0092-8674(88)90048-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen I. S., Quan S. G., Golde D. W. (1983). Human T-cell leukemia virus type II transforms normal human lymphocytes. Proc Natl Acad Sci U S A 80, 7006–7009 10.1073/pnas.80.22.7006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C. X., Cho D. S., Wang Q., Lai F., Carter K. C., Nishikura K. (2000). A third member of the RNA-specific adenosine deaminase gene family, ADAR3, contains both single- and double-stranded RNA binding domains. RNA 6, 755–767 10.1017/S1355838200000170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevalier S. A., Walic M., Calattini S., Mallet A., Prévost M. C., Gessain A., Mahieux R. (2007). Construction and characterization of a full-length infectious simian T-cell lymphotropic virus type 3 molecular clone. J Virol 81, 6276–6285 10.1128/JVI.02538-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doria M., Neri F., Gallo A., Farace M. G., Michienzi A. (2009). Editing of HIV-1 RNA by the double-stranded RNA deaminase ADAR1 stimulates viral infection. Nucleic Acids Res 37, 5848–5858 10.1093/nar/gkp604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douceron E., Kaidarova Z., Miyazato P., Matsuoka M., Murphy E. L., Mahieux R. (2012). HTLV-2 APH-2 expression is correlated with proviral load but APH-2 does not promote lymphocytosis. J Infect Dis 205, 82–86 10.1093/infdis/jir708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felder M. P., Laugier D., Yatsula B., Dezélée P., Calothy G., Marx M. (1994). Functional and biological properties of an avian variant long terminal repeat containing multiple A to G conversions in the U3 sequence. J Virol 68, 4759–4767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- George C. X., Samuel C. E. (1999). Human RNA-specific adenosine deaminase ADAR1 transcripts possess alternative exon 1 structures that initiate from different promoters, one constitutively active and the other interferon inducible. Proc Natl Acad Sci U S A 96, 4621–4626 10.1073/pnas.96.8.4621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grande-Pérez A., Sierra S., Castro M. G., Domingo E., Lowenstein P. R. (2002). Molecular indetermination in the transition to error catastrophe: systematic elimination of lymphocytic choriomeningitis virus through mutagenesis does not correlate linearly with large increases in mutant spectrum complexity. Proc Natl Acad Sci U S A 99, 12938–12943 10.1073/pnas.182426999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajjar A. M., Linial M. L. (1995). Modification of retroviral RNA by double-stranded RNA adenosine deaminase. J Virol 69, 5878–5882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann K. A., Bass B. L. (2000). Double-stranded RNA adenosine deaminases ADAR1 and ADAR2 have overlapping specificities. Biochemistry 39, 12875–12884 10.1021/bi001383g [DOI] [PubMed] [Google Scholar]
- Li Y., Kappes J. C., Conway J. A., Price R. W., Shaw G. M., Hahn B. H. (1991). Molecular characterization of human immunodeficiency virus type 1 cloned directly from uncultured human brain tissue: identification of replication-competent and -defective viral genomes. J Virol 65, 3973–3985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., George C. X., Patterson J. B., Samuel C. E. (1997). Functionally distinct double-stranded RNA-binding domains associated with alternative splice site variants of the interferon-inducible double-stranded RNA-specific adenosine deaminase. J Biol Chem 272, 4419–4428 10.1074/jbc.272.7.4419 [DOI] [PubMed] [Google Scholar]
- Mahieux R., Gessain A. (2011). HTLV-3/STLV-3 and HTLV-4 viruses: discovery, epidemiology, serology and molecular aspects. Viruses 3, 1074–1090 10.3390/v3071074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez I., Dopazo J., Melero J. A. (1997). Antigenic structure of the human respiratory syncytial virus G glycoprotein and relevance of hypermutation events for the generation of antigenic variants. J Gen Virol 78, 2419–2429 [DOI] [PubMed] [Google Scholar]
- Melcher T., Maas S., Herb A., Sprengel R., Higuchi M., Seeburg P. H. (1996). RED2, a brain-specific member of the RNA-specific adenosine deaminase family. J Biol Chem 271, 31795–31798 10.1074/jbc.271.50.31795 [DOI] [PubMed] [Google Scholar]
- Murphy D. G., Dimock K., Kang C. Y. (1991). Numerous transitions in human parainfluenza virus 3 RNA recovered from persistently infected cells. Virology 181, 760–763 10.1016/0042-6822(91)90913-V [DOI] [PubMed] [Google Scholar]
- O’Hara P. J., Nichol S. T., Horodyski F. M., Holland J. J. (1984). Vesicular stomatitis virus defective interfering particles can contain extensive genomic sequence rearrangements and base substitutions. Cell 36, 915–924 10.1016/0092-8674(84)90041-2 [DOI] [PubMed] [Google Scholar]
- Patterson J. B., Cornu T. I., Redwine J., Dales S., Lewicki H., Holz A., Thomas D., Billeter M. A., Oldstone M. B. (2001). Evidence that the hypermutated M protein of a subacute sclerosing panencephalitis measles virus actively contributes to the chronic progressive CNS disease. Virology 291, 215–225 10.1006/viro.2001.1182 [DOI] [PubMed] [Google Scholar]
- Phuphuakrat A., Kraiwong R., Boonarkart C., Lauhakirti D., Lee T. H., Auewarakul P. (2008). Double-stranded RNA adenosine deaminases enhance expression of human immunodeficiency virus type 1 proteins. J Virol 82, 10864–10872 10.1128/JVI.00238-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rueda P., García-Barreno B., Melero J. A. (1994). Loss of conserved cysteine residues in the attachment (G) glycoprotein of two human respiratory syncytial virus escape mutants that contain multiple A-G substitutions (hypermutations). Virology 198, 653–662 10.1006/viro.1994.1077 [DOI] [PubMed] [Google Scholar]
- Samuel C. E. (2001). Antiviral actions of interferons. Clin Microbiol Rev 14, 778–809 10.1128/CMR.14.4.778-809.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slattery J. P., Franchini G., Gessain A. (1999). Genomic evolution, patterns of global dissemination, and interspecies transmission of human and simian T-cell leukemia/lymphotropic viruses. Genome Res 9, 525–540 [PubMed] [Google Scholar]
- Suspène R., Henry M., Guillot S., Wain-Hobson S., Vartanian J. P. (2005). Recovery of APOBEC3-edited human immunodeficiency virus G→A hypermutants by differential DNA denaturation PCR. J Gen Virol 86, 125–129 10.1099/vir.0.80426-0 [DOI] [PubMed] [Google Scholar]
- Suspène R., Renard M., Henry M., Guétard D., Puyraimond-Zemmour D., Billecocq A., Bouloy M., Tangy F., Vartanian J. P., Wain-Hobson S. (2008). Inversing the natural hydrogen bonding rule to selectively amplify GC-rich ADAR-edited RNAs. Nucleic Acids Res 36, e72 10.1093/nar/gkn295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suspène R., Petit V., Puyraimond-Zemmour D., Aynaud M. M., Henry M., Guétard D., Rusniok C., Wain-Hobson S., Vartanian J. P. (2011). Double-stranded RNA adenosine deaminase ADAR-1-induced hypermutated genomes among inactivated seasonal influenza and live attenuated measles virus vaccines. J Virol 85, 2458–2462 10.1128/JVI.02138-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor D. R., Puig M., Darnell M. E., Mihalik K., Feinstone S. M. (2005). New antiviral pathway that mediates hepatitis C virus replicon interferon sensitivity through ADAR1. J Virol 79, 6291–6298 10.1128/JVI.79.10.6291-6298.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenoever B. R., Ng S. L., Chua M. A., McWhirter S. M., García-Sastre A., Maniatis T. (2007). Multiple functions of the IKK-related kinase IKKϵ in interferon-mediated antiviral immunity. Science 315, 1274–1278 10.1126/science.1136567 [DOI] [PubMed] [Google Scholar]
- Wang Y., Samuel C. E. (2009). Adenosine deaminase ADAR1 increases gene expression at the translational level by decreasing protein kinase PKR-dependent eIF-2α phosphorylation. J Mol Biol 393, 777–787 10.1016/j.jmb.2009.08.070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Zeng Y., Murray J. M., Nishikura K. (1995). Genomic organization and chromosomal location of the human dsRNA adenosine deaminase gene: the enzyme for glutamate-activated ion channel RNA editing. J Mol Biol 254, 184–195 10.1006/jmbi.1995.0610 [DOI] [PubMed] [Google Scholar]
- Wattel E., Vartanian J. P., Pannetier C., Wain-Hobson S. (1995). Clonal expansion of human T-cell leukemia virus type I-infected cells in asymptomatic and symptomatic carriers without malignancy. J Virol 69, 2863–2868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong T. C., Ayata M., Hirano A., Yoshikawa Y., Tsuruoka H., Yamanouchi K. (1989). Generalized and localized biased hypermutation affecting the matrix gene of a measles virus strain that causes subacute sclerosing panencephalitis. J Virol 63, 5464–5468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong T. C., Ayata M., Ueda S., Hirano A. (1991). Role of biased hypermutation in evolution of subacute sclerosing panencephalitis virus from progenitor acute measles virus. J Virol 65, 2191–2199 [DOI] [PMC free article] [PubMed] [Google Scholar]