Abstract
Phosphate (Pi) limitation is one of the major factors negatively impacting crop yield worldwide. Next generation sequencing (NGS) was used to profile the transcriptomes of rice (Oryza sativa) roots and shoots after phosphate starvation and recovery, shedding further light on the complex and dynamic mechanisms involved in Pi homeostasis. The use of NGS also enabled the identification of previously not annotated loci and novel isoforms of genes that are specifically induced by Pi starvation. Furthermore, phosphate re-feeding was observed to have a unique response with a variety of transcription factors and kinases induced in a transient manner. Expression profiles of miRNAs were also assessed upon long-term Pi starvation in roots and shoots revealing several novel miRNAs associated with Pi starvation. Altogether, this study provides key findings regarding Pi homeostasis in plants that will provide a valuable resource for research aimed at generating crops with increased Pi acquisition/use efficiency.
Keywords: phosphate starvation, transcriptome, RNAseq, rice, nutrient starvation
Phosphate (Pi) availability is a major factor determining plant growth and development, thus directly affecting crop productivity. Because of its low mobility and high sorption capacity in the rhizosphere, Pi is one of the least available macronutrient in the soil.1 To overcome this constraint, heavy fertilization has long been used, ultimately leading to environmental pollution.2 In addition, the declining availability of phosphate rock, the main source of Pi fertilizer, has considerably increased the price of Pi fertilizers and thus strengthens the need to develop crops with greater phosphorus efficiency, i.e., crops that can maintain their growth and yield in soils with reduced phosphorus availability. Thus, it is critical to improve our knowledge on the complex mechanisms involved in Pi homeostasis. While previous studies had already examined the effects of Pi starvation on the transcriptome in various species,3-11 recent technologies such as next generation sequencing, now allows capturing the dynamic mechanisms involved in these processes by being able to profile numerous conditions and time points.
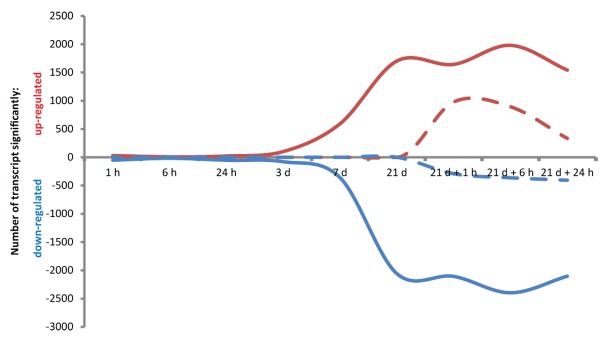
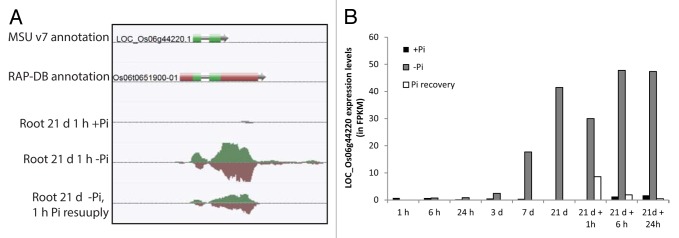
In our recent study, RNA sequencing technology (RNA-seq) was used to profile the transcriptional responses of rice roots and shoots to short- and long-term Pi deprivation as well as to Pi recovery. Covering an extensive period of Pi stress consisting of 9 time points over more than three weeks provided a comprehensive overview of the complex and dynamic mechanisms involved in Pi homeostasis. Interestingly, our data showed that during the first 3 d of Pi deprivation, very little modifications of the root transcriptomes could be observed, with only 101 genes being upregulated after 3 d of Pi starvation (Fig. 1). Indeed, only after a week of Pi deprivation did the number of differentially regulated genes in the roots start to increase, before reaching a plateau after 21 d of Pi deprivation. Among the well characterized phosphate starvation-induced genes (PSI) such as the SPXs genes and high affinity phosphate transporters genes, several highly induced genes previously not associated to Pi homeostasis were also detected. Among these, LOC_Os06g44220, encoding a putative low-temperature and salt-responsive gene not expressed under normal conditions (< 2 FPKM), showed some of the highest induction upon Pi deprivation, reaching more than 40 FPKM in the roots after 21 d of Pi starvation. Yet, very little is known regarding the function of this gene, probably exacerbated by the fact that it is not present on the widely used rice Affymetrix microarrays chip. As a consequence, this study not only provides detailed kinetics of the molecular responses to Pi stress, but also identifies potential novel candidate genes essential for plant responses to Pi starvation. In addition, this work also focused on the less studied effects of short-term Pi re-supply to Pi starved plants, resulting in the identification of numerous genes that are transiently differentially expressed, many of which were transcription factors and kinases. Genes coding for the cupin domain containing proteins (LOC_Os12g05870 and LOC_Os12g05880) were among the most highly induced upon Pi re-supply. Cupins are extremely diverse and include catalytically inactive seed storage proteins, sugar-binding metal-independent epimerases, and metal-dependent enzymes and possess a wide range of activities.12 The Arabidopsis ortholog of these cupins is a germin-like protein (At4g14630), previously shown to be induced in response to NaCl stress.13 Altogether, the identification of these rapid and ephemeral transcriptional modifications provides new elements to further identify regulators of Pi homeostasis.
Figure 1. Number of differentially regulated genes upon Pi deprivation and re-supply at discrete time points in rice roots (FDR < 0.05). Solid line represents differentially expressed genes in response to Pi deprivation while dashed line represent differentially expressed genes in response to Pi resupply. Lines in red represent upregulated genes, while blue lines represent genes downregulated by the various Pi treatments.
While this study generated a detailed overview of the transcriptional responses induced by Pi starvation, it also assessed how conserved these responses were when compared with those observed in Arabidopsis. To do so, a set of conserved Arabidopsis root PSI genes was identified from previous studies.3,4,8 While it is known that the experimental design can greatly affect the transcriptomes of Pi starved plants with limited conservation of PSI genes between studies, we identified a set of 130 core PSI genes in Arabidopsis roots. This set of conserved root PSI genes was then compared with the rice root PSI orthologs, resulting in the identification of 76 genes that were induced by Pi starvation in both rice and Arabidopsis roots. While this result strengthens the fact that the core molecular mechanisms involved in response to Pi starvation are highly conserved in plants, subtle differences exist as exemplified with PHO2. Our work has identified a novel PHO2 isoform, referred to as PHO2.2, that, unlike the original PHO2 isoform, is specifically and highly induced by Pi starvation in roots and shoots. In addition, it was shown that PHO2.2 was more actively translated into protein than the original isoform, thus providing an extra level of complexity in the regulation of one of the key regulators of Pi homeostasis.
Due to increasing evidence of the role of small RNAs in growth, development, stress adaptation and nutrient signaling,14 we generated the first report of miRNAs expression levels upon long-term Pi starvation (21 d) in rice roots and shoots. While this study, confirmed the induction of the well characterized miR399 and miR827, involved in negatively regulating the expression levels of PHO215 and SPX-MFS116, respectively, this work also identified numerous miRNAs previously not associated with Pi starvation such as miR6250, a miRNA previously shown to be induced by arsenite.17 The identification of novel Pi-starvation induced miRNAs, which are potentially also involved in the response to other stresses, e.g., arsenite, may help decipher the complex crosstalks between nutrients, such as the one observed for at-miR827 involved in maintaining nitrate-dependent Pi homeostasis.18
Altogether this work presents a high-resolution genome-wide transcriptome analysis of the responses of rice to Pi starvation and recovery in roots and shoots. In addition, this analysis also identifies numerous potential candidate genes to improve our understanding of Pi homeostasis in plants. (Fig. 2)
Figure 2. Identification of novel potential candidate genes involved in Pi homeostasis. (A) Graphical representation of LOC_Os06 g44220, a gene previously not associated with Pi starvation, but highly induced by Pi deprivation. Screen capture is made from the AnnoJ genome browser (http://www.plantenergy.uwa.edu.au/annoj/Secco_2013.html), representing gene annotations from MSU v7 and the RAP-DB IRGSP-1.00 assemblies as well as RNA expression tracks. (B) Graphical representation of the expression level of LOC_Os06 g44220 during Pi deprivation and Pi re-supply. Expression level is shown in FPKM (for fragment per kilobase of exon per million fragments mapped).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank Oliver Berkowitz for critical reading of the article. This work was supported by the Australian Research Council Super Science Fellowship (FS100100022 to D.S.).
References
- 1.Poirier Y, Bucher M. . Phosphate transport and homeostasis in Arabidopsis. Arabidopsis Book 2002; 1:e0024; http://dx.doi.org/ 10.1199/tab.0024; PMID: 22303200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.MacDonald GK, Bennett EM, Potter PA, Ramankutty N. . Agronomic phosphorus imbalances across the world’s croplands. Proc Natl Acad Sci U S A 2011; 108:3086 - 91; http://dx.doi.org/ 10.1073/pnas.1010808108; PMID: 21282605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Misson J, Raghothama KG, Jain A, Jouhet J, Block MA, Bligny R, Ortet P, Creff A, Somerville S, Rolland N, et al. . A genome-wide transcriptional analysis using Arabidopsis thaliana Affymetrix gene chips determined plant responses to phosphate deprivation. Proc Natl Acad Sci U S A 2005; 102:11934 - 9; http://dx.doi.org/ 10.1073/pnas.0505266102; PMID: 16085708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Morcuende R, Bari R, Gibon Y, Zheng W, Pant BD, Bläsing O, Usadel B, Czechowski T, Udvardi MK, Stitt M, et al. . Genome-wide reprogramming of metabolism and regulatory networks of Arabidopsis in response to phosphorus. Plant Cell Environ 2007; 30:85 - 112; http://dx.doi.org/ 10.1111/j.1365-3040.2006.01608.x; PMID: 17177879 [DOI] [PubMed] [Google Scholar]
- 5.Müller R, Morant M, Jarmer H, Nilsson L, Nielsen TH. . Genome-wide analysis of the Arabidopsis leaf transcriptome reveals interaction of phosphate and sugar metabolism. Plant Physiol 2007; 143:156 - 71; http://dx.doi.org/ 10.1104/pp.106.090167; PMID: 17085508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bustos R, Castrillo G, Linhares F, Puga MI, Rubio V, Pérez-Pérez J, Solano R, Leyva A, Paz-Ares J. . A central regulatory system largely controls transcriptional activation and repression responses to phosphate starvation in Arabidopsis. PLoS Genet 2010; 6:e1001102; http://dx.doi.org/ 10.1371/journal.pgen.1001102; PMID: 20838596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thibaud MC, Arrighi JF, Bayle V, Chiarenza S, Creff A, Bustos R, Paz-Ares J, Poirier Y, Nussaume L. . Dissection of local and systemic transcriptional responses to phosphate starvation in Arabidopsis. Plant J 2010; 64:775 - 89; http://dx.doi.org/ 10.1111/j.1365-313X.2010.04375.x; PMID: 21105925 [DOI] [PubMed] [Google Scholar]
- 8.Woo J, MacPherson CR, Liu J, Wang H, Kiba T, Hannah MA, Wang XJ, Bajic VB, Chua NH. . The response and recovery of the Arabidopsis thaliana transcriptome to phosphate starvation. BMC Plant Biol 2012; 12:62; http://dx.doi.org/ 10.1186/1471-2229-12-62; PMID: 22553952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zheng L, Huang F, Narsai R, Wu J, Giraud E, He F, Cheng L, Wang F, Wu P, Whelan J, et al. . Physiological and transcriptome analysis of iron and phosphorus interaction in rice seedlings. Plant Physiol 2009; 151:262 - 74; http://dx.doi.org/ 10.1104/pp.109.141051; PMID: 19605549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wasaki J, Shinano T, Onishi K, Yonetani R, Yazaki J, Fujii F, Shimbo K, Ishikawa M, Shimatani Z, Nagata Y, et al. . Transcriptomic analysis indicates putative metabolic changes caused by manipulation of phosphorus availability in rice leaves. J Exp Bot 2006; 57:2049 - 59; http://dx.doi.org/ 10.1093/jxb/erj158; PMID: 16720613 [DOI] [PubMed] [Google Scholar]
- 11.Wasaki J, Yonetani R, Kuroda S, Shinano T, Yazaki J, Fujii F, Shimbo K, Yamamoto K, Sakata K, Sasaki T, et al. . Transcriptomic analysis of metabolic changes by phosphorus stress in rice plant roots. Plant Cell Environ 2003; 26:1515 - 23; http://dx.doi.org/ 10.1046/j.1365-3040.2003.01074.x [DOI] [Google Scholar]
- 12.Uberto R, Moomaw EW. . Protein similarity networks reveal relationships among sequence, structure, and function within the Cupin superfamily. PLoS One 2013; 8:e74477; http://dx.doi.org/ 10.1371/journal.pone.0074477; PMID: 24040257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jiang Y, Yang B, Harris NS, Deyholos MK. . Comparative proteomic analysis of NaCl stress-responsive proteins in Arabidopsis roots. J Exp Bot 2007; 58:3591 - 607; http://dx.doi.org/ 10.1093/jxb/erm207; PMID: 17916636 [DOI] [PubMed] [Google Scholar]
- 14.Chiou TJ. . The role of microRNAs in sensing nutrient stress. Plant Cell Environ 2007; 30:323 - 32; http://dx.doi.org/ 10.1111/j.1365-3040.2007.01643.x; PMID: 17263777 [DOI] [PubMed] [Google Scholar]
- 15.Lin WY, Huang TK, Leong SJ, Chiou TJ. . Long-distance call from phosphate: systemic regulation of phosphate starvation responses. J Exp Bot 2013; http://dx.doi.org/ 10.1093/jxb/ert431; PMID: 24368506 [DOI] [PubMed] [Google Scholar]
- 16.Wang C, Huang W, Ying Y, Li S, Secco D, Tyerman S, Whelan J, Shou H. . Functional characterization of the rice SPX-MFS family reveals a key role of OsSPX-MFS1 in controlling phosphate homeostasis in leaves. New Phytol 2012; 196:139 - 48; http://dx.doi.org/ 10.1111/j.1469-8137.2012.04227.x; PMID: 22803610 [DOI] [PubMed] [Google Scholar]
- 17.Liu Q. . Novel miRNAs in the control of arsenite levels in rice. Funct Integr Genomics 2012; 12:649 - 58; http://dx.doi.org/ 10.1007/s10142-012-0282-3; PMID: 22585409 [DOI] [PubMed] [Google Scholar]
- 18.Kant S, Peng M, Rothstein SJ. . Genetic regulation by NLA and microRNA827 for maintaining nitrate-dependent phosphate homeostasis in arabidopsis. PLoS Genet 2011; 7:e1002021; http://dx.doi.org/ 10.1371/journal.pgen.1002021; PMID: 21455488 [DOI] [PMC free article] [PubMed] [Google Scholar]