Abstract
Functional transition of glyoxysomes to leaf peroxisomes is observed in greening cotyledons. Glyoxysomal proteins are rapidly degraded and leaf-peroxisomal proteins are transported into peroxisomes after cotyledons are exposed to light, but the molecular mechanisms underlying these processes remain unclear. We recently discovered that two degradation pathways are involved in the functional transition of peroxisomes using Arabidopsis thaliana. Lon protease 2 (LON2) is responsible for the degradation of glyoxysomal proteins inside peroxisomes, and, in parallel, autophagy eliminates damaged or obsolete peroxisomes. A double mutant defective in both the LON2- and autophagy-dependent degradation pathways accumulated glyoxysomal proteins after the cotyledons became green. Our study also demonstrated that the LON2- and autophagy-dependent pathways are interdependent, with the chaperone function of LON2 suppressing autophagic peroxisome degradation. Moreover, the peptidase domain of LON2 interferes with the suppression of autophagy, indicating that autophagy is regulated by intramolecular modulation between the proteolysis and chaperone functions of LON2.
Keywords: Arabidopsis thaliana, Lon protease, autophagy, chaperone, functional transition, peroxisome
Peroxisomes are single-membrane organelles that are found ubiquitously in eukaryotic cells. Several types of peroxisomes are found in plants, including glyoxysomes and leaf peroxisomes, with functions that are responsive to a variety of environmental and developmental cues.1,2 In etiolated cotyledons, glyoxysomes are responsible for lipid metabolism, which produces sucrose as the energy source for post-germinative seedling growth. Leaf peroxisomes are found in photosynthetic tissues and contain glycolate pathway enzymes that are important for photorespiration. Etiolated cotyledons become green after light exposure and glyoxysomes correspondingly transform into leaf peroxisomes.
Two main hypotheses for the functional transition of peroxisomes have been discussed since the 1970s. The “one-population” theory suggests that peroxisome transition proceeds in a linear and continuous fashion, whereas the “two-population” pathway proposes a discontinuous, two-step mechanism.3 In the one-population hypothesis, glyoxysomes directly transform into leaf peroxisomes and newly synthesized leaf-peroxisomal proteins are transported into peroxisomes at the same time as glyoxysomal proteins are degraded. In the two-population hypothesis, glyoxysomes are eliminated and leaf peroxisomes are developed de novo. In the mid-1980s, immunoelectron microscopic analysis demonstrated that both glyoxysomal and leaf-peroxisomal proteins coexist in a peroxisome during the functional transition.4,5 This indicated that glyoxysomes are directly transformed into leaf peroxisomes, as proposed in the one-population hypothesis. However, the mechanisms underlying the degradation of glyoxysomal proteins remain unclear. Recently, we revealed that two degradation pathways are involved in the functional transition of peroxisomes: Lon protease-dependent protein degradation inside peroxisomes, and selective degradation of peroxisomes via autophagy.6
The aberrant peroxisome morphology 10 (apem10) mutant was isolated based on a characteristic GFP-fluorescence pattern, distinct from that in parental Arabidopsis plants, observed in plants expressing a peroxisome marker, GFP-PTS1. APEM10 encodes Lon protease 2 (LON2). In the apem10 mutant, the number of peroxisomes decreased substantially and peroxisomal proteins accumulated in the cytosol, indicating that LON2 plays an important role in maintaining peroxisome number. Degradation of damaged or obsolete cellular compartments, including peroxisomes, occurs via autophagy in a variety of organisms.7,8,9 We wished to determine whether autophagy was implicated in the reduction in peroxisome number in apem10. Consequently, we analyzed a double mutant derived from crossing apem10 with peroxisome unusual positioning 1 (peup1), in which peroxisome degradation is arrested due to a defective autophagy-related 2 (ATG2) protein.9 No decrease in peroxisome number was observed in the apem10 peup1 double mutant. This indicated that excessive degradation of peroxisomes occurs in apem10 mutants, and that autophagy is suppressed by LON2 in the wild type.
Lon protease is involved in the degradation of oxidized and damaged proteins, and belongs to the AAA+ ATPase superfamily. The AAA+ ATPase domain acts as a molecular chaperone, providing the mechanical power needed to unfold substrate proteins, and likely acts in concert with the N-terminal domain.10,11,12 Plants containing LON2 with a mutation in the ATP-binding site in the AAA+ ATPase domain (LON2[K414A]) were unable to rescue the apem10 phenotype. Conversely, a LON2 variant harboring a mutation in a conserved residue at the active center of the peptidase domain (LON2[S783A]) was able to rescue the apem10 phenotype.6 These results indicated that the chaperone activity of LON2 is required for suppression of autophagy but that the peptidase domain of LON2 is not needed. However, higher numbers of peroxisomes were found in apem10 LON2[S783A] plants than in wild-type plants. This indicates that LON2[S783A] oversuppresses autophagy, suggesting that, in the wild-type LON2 protein, the peptidase domain interferes with the chaperone function to modulate autophagy.6
Glyoxysomal enzymes are rapidly degraded and newly synthesized leaf-peroxisomal enzymes are transported into peroxisomes during the functional transition of glyoxysomes to leaf peroxisomes. Our study revealed that glyoxysomal enzyme levels decrease in apem10 and peup1 mutants during the functional transition, as in the wild type, indicating that single defect in neither the LON2 nor autophagy affected degradation of glyoxysomal enzymes.6 However, glyoxysomal enzymes did accumulate in the apem10 peup1 double mutant,6 indicating that two complementary pathways are involved in the elimination of glyoxysomal enzymes: LON2-mediated protein degradation inside peroxisomes, and peroxisome degradation via autophagy. Glyoxysomal enzymes also accumulated in apem10 LON2[S783A], which concurs with the autophagy-oversuppression phenotype observed in this mutant.6
We propose a new model for the functional transition of peroxisomes (Fig. 1). During the transformation of glyoxysomes to leaf peroxisomes, glyoxysomal enzymes inside the peroxisomes are degraded by LON2, and newly synthesized leaf-peroxisomal enzymes are imported (Fig. 1). Recently, we demonstrated that peroxisomes become oxidized while performing their function, and these peroxisomes are selectively degraded via autophagy.9 In our model, preexisting oxidized glyoxysomes, which are exposed to reactive oxygen species generated in various peroxisomal functions, are selectively eliminated by autophagy in parallel with glyoxysomal enzyme degradation (Fig. 1). During this process, LON2 suppresses autophagy to allow some glyoxysomes to escape degradation. Peroxisomes are rapidly degraded in the absence of LON2, leading to the decreased peroxisome phenotype observed in the apem10 mutant.
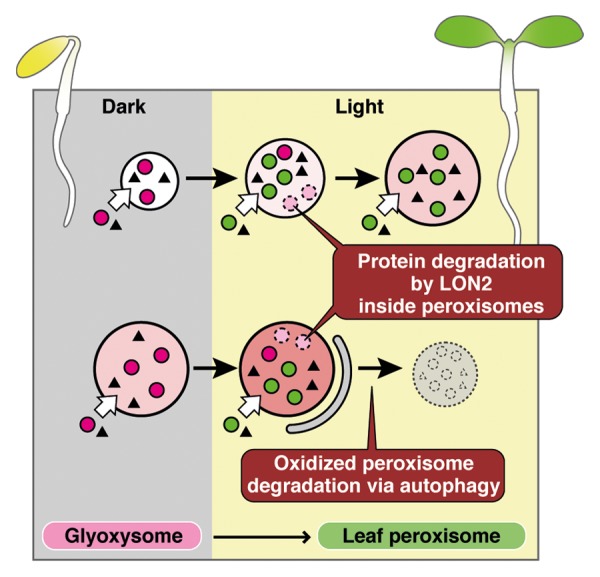
Figure 1. New model proposed for the functional transition of peroxisomes. In etiolated cotyledons, glyoxysomal proteins (magenta circles) are synthesized and transported to peroxisomes. After light is received, cotyledons become green and the glyoxysomes are transformed into leaf peroxisomes. Leaf-peroxisomal proteins (green circles) are transported into leaf peroxisomes, and glyoxysomal proteins are degraded by LON2. In parallel, excess or oxidized peroxisomes are degraded by autophagy. The chaperone function of LON2 suppresses autophagy, allowing some glyoxysomes to successfully transform into leaf peroxisomes. Triangles indicate proteins found in both glyoxysomes and leaf peroxisomes. Increasing red intensity in the peroxisomes represents increasing oxidized levels.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
This study was supported by MEXT KAKENHI to M.N. (grant numbers 22120007, 16085209); to S.M. (no. 18770039); and to S.G.-Y. (no. 25891028); and by JSPS KAKENHI to S.G.-Y. (no. 22–9).
References
- 1.Nishimura M, Takeuchi Y, De Bellis L, Hara-Nishimura I. Leaf peroxisomes are directly transformed to glyoxysomes during senescence of pumpkin cotyledons. Protoplasma. 1993;175:131–7. doi: 10.1007/BF01385011. [DOI] [Google Scholar]
- 2.Kamada T, Nito K, Hayashi H, Mano S, Hayashi M, Nishimura M. Functional differentiation of peroxisomes revealed by expression profiles of peroxisomal genes in Arabidopsis thaliana. Plant Cell Physiol. 2003;44:1275–89. doi: 10.1093/pcp/pcg173. [DOI] [PubMed] [Google Scholar]
- 3.Beevers H. Microbodies in higher plants. Annu Rev Plant Physiol. 1979;30:159–93. doi: 10.1146/annurev.pp.30.060179.001111. [DOI] [Google Scholar]
- 4.Titus DE, Becker WM. Investigation of the glyoxysome-peroxisome transition in germinating cucumber cotyledons using double-label immunoelectron microscopy. J Cell Biol. 1985;101:1288–99. doi: 10.1083/jcb.101.4.1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nishimura M, Yamaguchi J, Mori H, Akazawa T, Yokota S. Immunocytochemical analysis shows that glyoxysomes are directly transformed to leaf peroxisomes during greening of pumpkin cotyledons. Plant Physiol. 1986;81:313–6. doi: 10.1104/pp.81.1.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goto-Yamada S, Mano S, Nakamori C, Kondo M, Yamawaki R, Kato A, Nishimura M. Chaperone and protease functions of LON protease 2 modulate the peroxisomal transition and degradation with autophagy. Plant Cell Physiol. 2014;55:482–96. doi: 10.1093/pcp/pcu017. [DOI] [PubMed] [Google Scholar]
- 7.Ezaki J, Kominami E, Ueno T. Peroxisome degradation in mammals. IUBMB Life. 2011;63:1001–8. doi: 10.1002/iub.537. [DOI] [PubMed] [Google Scholar]
- 8.Lee J, Giordano S, Zhang J. Autophagy, mitochondria and oxidative stress: cross-talk and redox signalling. Biochem J. 2012;441:523–40. doi: 10.1042/BJ20111451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shibata M, Oikawa K, Yoshimoto K, Kondo M, Mano S, Yamada K, Hayashi M, Sakamoto W, Ohsumi Y, Nishimura M. Highly oxidized peroxisomes are selectively degraded via autophagy in Arabidopsis. Plant Cell. 2013;25:4967–83. doi: 10.1105/tpc.113.116947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bissonnette SA, Rivera-Rivera I, Sauer RT, Baker TA. The IbpA and IbpB small heat-shock proteins are substrates of the AAA+ Lon protease. Mol Microbiol. 2010;75:1539–49. doi: 10.1111/j.1365-2958.2010.07070.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gur E, Vishkautzan M, Sauer RT. Protein unfolding and degradation by the AAA+ Lon protease. Protein Sci. 2012;21:268–78. doi: 10.1002/pro.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bartoszewska M, Williams C, Kikhney A, Opaliński Ł, van Roermund CW, de Boer R, Veenhuis M, van der Klei IJ. Peroxisomal proteostasis involves a Lon family protein that functions as protease and chaperone. J Biol Chem. 2012;287:27380–95. doi: 10.1074/jbc.M112.381566. [DOI] [PMC free article] [PubMed] [Google Scholar]


