Abstract
Plant roots play an important role in uptake of water and nutrients, support of above-ground part and environmental sensing, but the molecular mechanisms underlying the root development are poorly understood in rice. We found that a gene (OsASL1) encoding argininosuccinate lyase is involved in normal root development of rice. OsASL1 cleaves argininosuccinate to arginine and fumarate reversibly, the last step in the arginine biosynthetic pathway. Here, we further characterized OsASL1 in terms of expression pattern, subcellular localization, and arginine effect on the root growth. A detailed expression analysis revealed that 2 transcripts of OsASL1, OsASL1.1 and OsASL1.2, showed different expression patterns; OsASL1.1 was expressed in most organs throughout the whole growth period, whereas OsASL1.2 was mainly expressed in the roots. In contrast to plastid-localized OsASL1.1, OsASL1.2 was localized to the cytosol and nucleus. The short-root phenotype of the mutant was not rescued by exogenous addition of the sodium nitroprusside, a nitric oxide donor, but rescued by an appropriate concentration of Arg. Our results indicate that the subcellular localization was determined by the N terminus of OsASL1 and that appropriate concentration of Arg is required for normal root elongation in rice.
Keywords: rice root, argininosuccinate lyase, expression pattern, arginine, subcellular localization
The basic functions of all plant roots are acquisition of water and nutrients, anchorage of above-ground part, and environmental sensing.1,2 Therefore, a good root system is very important for plant growth. The formation of a root system is a very complex developmental process, which requires many genes.2 Rice has a unique root system, which is composed primarily of seminal root, numerous crown roots that develop from the stem and lateral roots. However, only few genes involved in the root development have been identified. Recently, we isolated a mutant with short root.3 Genetic and molecular analysis showed that the gene (OsASL1) responsible for the short root encodes argininosuccinate lyase cleaving argininosuccinate to arginine and fumarate reversibly at the last enzymatic reaction of arginine (Arg) biosynthesis. These findings indicate that Arg is essential for normal root growth in rice.3
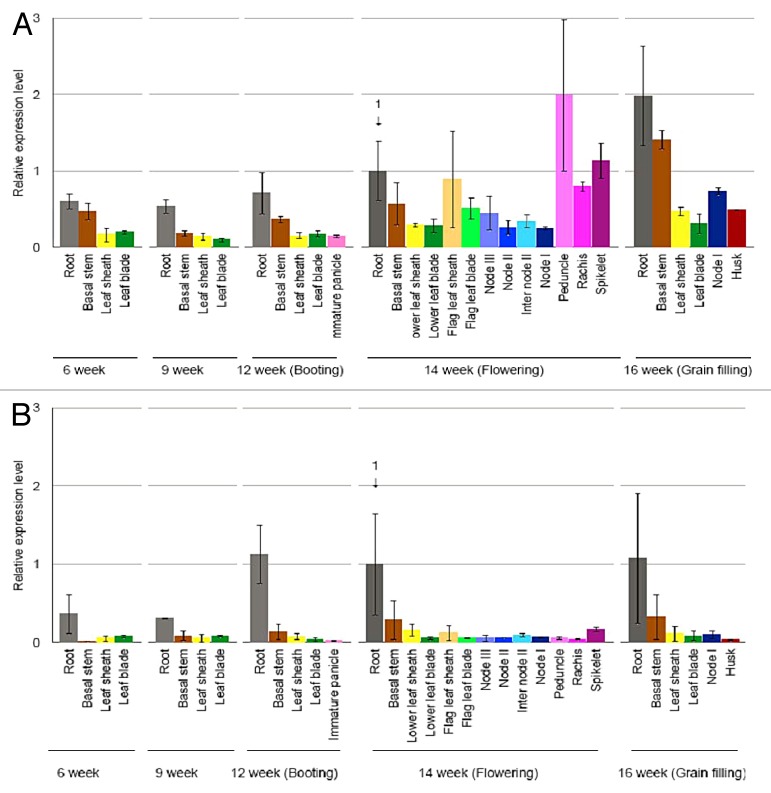
OsASL1 gene has 2 distinct transcripts produced from different transcription start sites of OsASL1, OsASL1.1 and OsASL1.2, respectively. However, yeast assay revealed that only OsASL1.1 has an argininosuccinate lyase activity,3 indicating that the N-terminal region of OsASL1 is important for the enzyme activity or substrate specificity. The OsASL1.1 transcript was detected in both the roots and shoots at the vegetative stage.3 A more detailed analysis of the expression in different organs at different growth stage showed that OsASL1.1 was also expressed in other organs such as flag leaf sheath, peduncle, and spikelet at the reproductive growth stage (Fig. 1A). These results suggest that OsASL1.1 may also play a role in other organs. By contrast, OsASL1.2 was mainly expressed in the roots throughout the whole growth period (Fig. 1B).
Figure 1. Expression pattern of OsASL1.1 and OsASL1.2. Different organs at various stages of rice grown in paddy field were sampled for RNA extraction. The expression was determined by real-time quantitative RT-PCR. Expression of OsASL1.1 (A) and OsASL1.2 (B) relative to the root at flowering stage is shown. HistoneH3 was used as an internal standard. Data are means ± SD of 3 biological replicates.
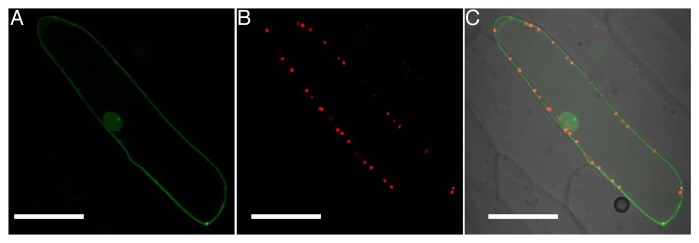
OsASL1.1 is localized to the plastid.3 We also investigated the subcellular localization of OsASL1.2 by co-expressing OsASL1.2-GFP construct with OsFd III-TagRFP, which is a marker protein localized at the plastid,4 in onion epidermal cells. We found that unlike OsASL1.1, OsASL1.2 was not localized to the plastid, but in the nucleus and also likely anchored to the plasma membrane (Fig. 2A-C). The only difference in protein sequence between OsASL1.1 and OsASL1.2 is in the N terminus.3 This region is generally considered to contain most signal peptides of protein localization in the cell, such as signals for mitochondria, chloroplast, and the endoplasmic reticulum.5,6 Our results suggest that the N terminus of OsASL1 determines its subcellular localization (Fig. 2). OsASL1.2 may have different role from OsASL1.1 although further functional analysis of OsASL1.2 is required.
Figure 2. Subcellular localization of OsASL1.2. A fusion of OsASL1.2 and GFP was co-introduced with OsFd III-TagRFP into onion epidermal cells. (A) Fluorescence of GFP; (B) Fluorescence of TagRFP; (C) Merged image of GFP and TagRFP. Scale bars = 20 μm.
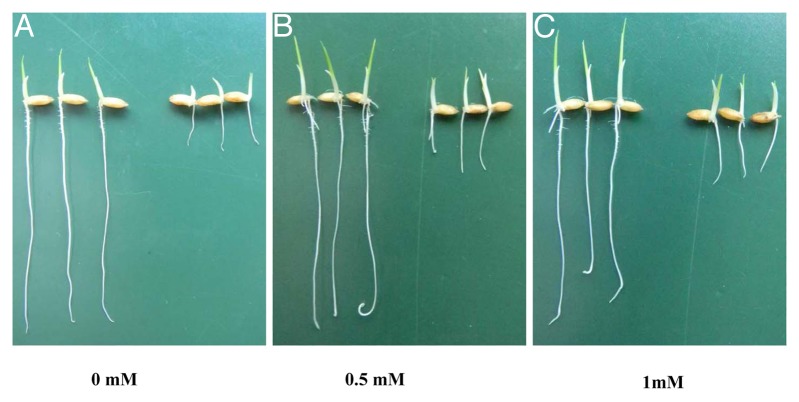
Arg is a precursor for the biosynthesis of nitric oxide (NO), which participates in root development as a signaling molecule in the cell.7,8 To examine whether the short root in the mutant was caused by the lack of NO in the roots, exogenous sodium nitroprusside as a nitric oxide donor was supplied to the growth solution. Addition of sodium nitroprusside at different concentrations did not restore the short-root phenotype of the mutant (Fig. 3A-C). This result suggests that the short-root phenotype in the mutant is not associated with NO signaling pathway.
Figure 3. Effect of exogenous sodium nitroprusside on the primary root growth. Germinated seeds of wild type (left) and mutant (right) were exposed to a 0.5 mM CaCl2 solution containing 0, 0.5, and 1.0 mM sudium nitroprusside as a nitric oxide donor for 5 days.
The short root phenotype could be rescued by supplementation of arginine at 0.1 mM, but not other amino acids,3 indicating that the short-root phenotype results from Arg deficiency. On the other hand, compared with the wild type, free Arg concentration in the roots of the mutant is similar.3 A previous study has reported that a fine-tuned regulatory system controls Arg level at low level in the roots.9 To support this, we tested the effect of different Arg concentrations on the root elongation in both WT and the mutant. At Arg concentration up to 0.1 mM, addition of Arg did not affect the root growth of the WT (Fig. 4A-C), but Arg at 0.05 mM was not able to rescue the short root phenotype. On the other hand, at higher Arg concentration (> 0.2 mM), the root growth of the WT was inhibited although the root growth of the mutant was rescued partially (Fig. 4D-F). These results indicated that although Arg serves not only as an essential amino acid for protein synthesis but also as a precursor for the biosynthesis of many compounds, which are important for the functioning of the cell,7 an appropriate concentration is essential for normal root growth at least in rice.
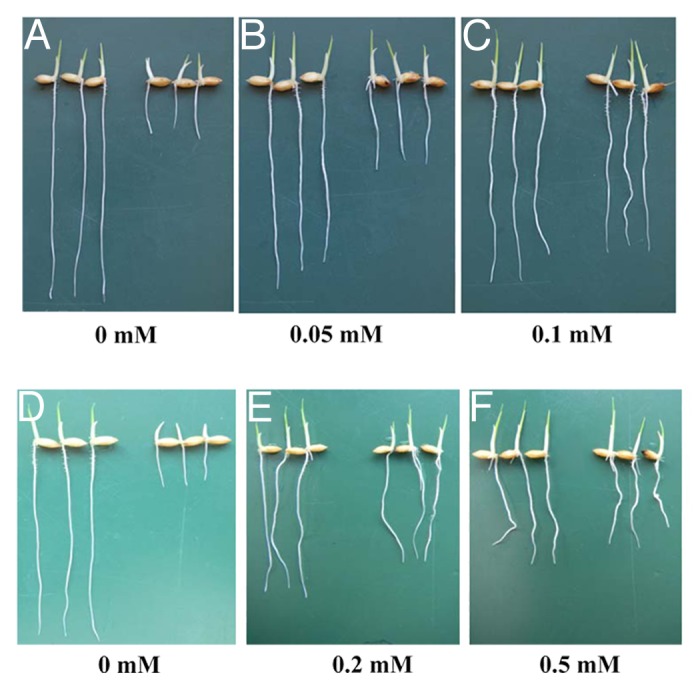
Figure 4. Effect of different Arg concentrations on the primary root growth. (A-F) Germinated seeds of wild type (left) and mutant (right) were exposed for 5 days to a 0.5 mM CaCl2 solution containing 0 (A), 0.05 mM (B), and 0.1mM (C) arginine or 0 (D), 0.2 mM (E), and 0.5 mM (F) arginine.
In summary, our results demonstrated that the subcellular localization of OsASL1.1 and OsASL1.2 was determined by N terminus of OsASL1 and that an appropriate concentration of Arg is essential for normal root elongation in rice.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
The research was supported by a Grant-in-Aid for Scientific Research on Innovative Areas from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (No. 22119002 to J.F.M.) and Ohara foundation for agriculture research.
References
- 1.Coudert Y, Périn C, Courtois B, Khong NG, Gantet P. . Genetic control of root development in rice, the model cereal. Trends Plant Sci 2010; 15:219 - 26; http://dx.doi.org/ 10.1016/j.tplants.2010.01.008; PMID: 20153971 [DOI] [PubMed] [Google Scholar]
- 2.Orman-Ligeza B, Parizot B, Gantet PP, Beeckman T, Bennett MJ, Draye X. . Post-embryonic root organogenesis in cereals: branching out from model plants. Trends Plant Sci 2013; 18:459 - 67; http://dx.doi.org/ 10.1016/j.tplants.2013.04.010; PMID: 23727199 [DOI] [PubMed] [Google Scholar]
- 3.Xia J, Yamaji N, Che J, Shen RF, Ma JF. . Normal root elongation requires arginine produced by argininosuccinate lyase in rice. Plant J 2014; http://dx.doi.org/ 10.1111/tpj.12476; PMID: 24528386 [DOI] [PubMed] [Google Scholar]
- 4.Primavesi LF, Wu H, Mudd EA, Day A, Jones HD. . Visualisation of plastids in endosperm, pollen and roots of transgenic wheat expressing modified GFP fused to transit peptides from wheat SSU RubisCO, rice FtsZ and maize ferredoxin III proteins. Transgenic Res 2008; 17:529 - 43; http://dx.doi.org/ 10.1007/s11248-007-9126-7; PMID: 17710559 [DOI] [PubMed] [Google Scholar]
- 5.Prassinos C, Haralampidis K, Milioni D, Samakovli D, Krambis K, Hatzopoulos P. . Complexity of Hsp90 in organelle targeting. Plant Mol Biol 2008; 67:323 - 34; http://dx.doi.org/ 10.1007/s11103-008-9322-8; PMID: 18368500 [DOI] [PubMed] [Google Scholar]
- 6.Puyaubert J, Denis L, Alban C. . Dual targeting of Arabidopsis holocarboxylase synthetase1: a small upstream open reading frame regulates translation initiation and protein targeting. Plant Physiol 2008; 146:478 - 91; http://dx.doi.org/ 10.1104/pp.107.111534; PMID: 18156294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Morris SM Jr.. . Regulation of enzymes of the urea cycle and arginine metabolism. Annu Rev Nutr 2002; 22:87 - 105; http://dx.doi.org/ 10.1146/annurev.nutr.22.110801.140547; PMID: 12055339 [DOI] [PubMed] [Google Scholar]
- 8.Flores T, Todd CD, Tovar-Mendez A, Dhanoa PK, Correa-Aragunde N, Hoyos ME, Brownfield DM, Mullen RT, Lamattina L, Polacco JC. . Arginase-negative mutants of Arabidopsis exhibit increased nitric oxide signaling in root development. Plant Physiol 2008; 147:1936 - 46; http://dx.doi.org/ 10.1104/pp.108.121459; PMID: 18567826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Frémont N, Riefler M, Stolz A, Schmülling T. . The Arabidopsis TUMOR PRONE5 gene encodes an acetylornithine aminotransferase required for arginine biosynthesis and root meristem maintenance in blue light. Plant Physiol 2013; 161:1127 - 40; http://dx.doi.org/ 10.1104/pp.112.210583; PMID: 23321422 [DOI] [PMC free article] [PubMed] [Google Scholar]