Abstract
In our previous integrated study combining informatics and molecular biology analyses, we revealed that Arabidopsis small open reading frames (sORFs) predicted by computational analysis have biological functions in morphogenesis. Here, we report that sequences homologous to Arabidopsis sORFs are abundant in intergenic regions of the rice genome. These sequences represent a subset of non-protein-coding DNA, and some are transcribed into mRNA. These results indicate that many sORFs associated with morphogenesis are hidden in the genomes of crop species.
Keywords: small open reading frame, comparative genomics, small peptide, Arabidopsis, rice
In plants, secreted peptides encoded by small coding genes play roles in development, self-incompatibility, pollen tube guidance, and defense responses against insect herbivores, pathogens, and environmental stresses.1-6 Similar to plant hormones, these peptides function as signaling molecules that bind to receptor kinases. Peptide signaling is now recognized to be as important as signaling by classical plant hormones. Plant genomes encode many leucine-rich-repeat receptor kinases, but only a few of the ligand peptides for these receptors have been identified.7,8 It is likely that many sequences encoding hormone-like peptides are hidden in plant genomes. Most hormone-like peptides have been identified in studies on Arabidopsis Genome Initiative (AGI) code genes.9 However, small coding genes (those encoding < 100 amino acids) tend to be missed when AGI code genes are annotated, because small coding sequences are often computed as false-positive predictions.10 To overcome the false-positive prediction problem, we developed computational approaches to predict small open reading frames (sORFs; 30–100 amino acids) with high coding potential, and identified 7,901 coding sORFs in intergenic regions of the Arabidopsis genome.11 Of these, 2,099 sORFs showed significant expression in at least one condition or organ out of 16 organs and 17 environmental conditions.12 Furthermore, approximately 10% (49/473) of the manually selected coding sORFs resulted in various morphological changes when overexpressed in transgenic Arabidopsis plants. These results indicated that sORFs predicted by computational analysis play significant roles in growth and morphogenesis in Arabidopsis.
CLE peptides, a representative class of peptide hormones, were first found in Arabidopsis, and function as key signaling molecules in development.13 Sequences homologous to those encoding Arabidopsis CLEs are conserved in Lotus, Oryza, and Zinnia. Indeed, treatment with CLE peptides induced cell differentiation and meristem maintenance in Lotus, Oryza, and Zinnia.14-16 As well as sequences encoding CLE peptides, those encoding other peptide hormones (ROT4, RTFL/DVL, EPF/EPFL) are also conserved in higher plant species.17-26 The conservation of coding sequences among multiple plant species provides good evidence for the functionality of these peptides.
The aim of this study was to explore the functionality of newly identified homologs of Arabidopsis sORFs in rice. We searched the rice genome for sequences homologous to 49 Arabidopsis sORFs associated with morphogenesis, and examined their RNA expression.
Rice Homologs of Arabidopsis sORFs
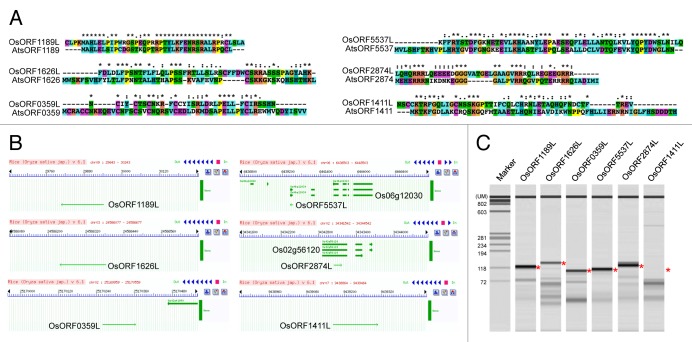
To identify sequences homologous to the Arabidopsis sORFs in rice, we conducted BLAST searches (E-value < 0.01) against the whole Oryza sativa genome. The amino acid sequences encoded by sORFs were aligned by CLUSTALW. Out of 49 AtsORFs, 35 AtsORFs had homologous sequences in the rice genome (IRGSP-1.0). Six of these sORFs are shown in Figure 1 as examples. Previous, we reported that transgenic Arabidopsis plants overexpressing AtsORF1189, 1411, 1626, 0359, 5537, and 2874 showed enlarged rosette leaves, altered leaf color, a seedling-lethal phenotype, small plant size, and altered leaf number, respectively. These results provided evidence for the biological functions of these sORFs.12 The sequences of OsORF1189L and 5537L were very similar to those of AtsORF1189 and 5537, respectively (Fig. 1A), whereas the other rice sORFs homologs showed moderate similarity to their homologous Arabidopsis sORFs. However, when we considered the amino acid sequence similarities between rice and Arabidopsis sORFs, the sequences of OsORF1626L, 2874L, 0359L, and 1411L were very similar to their corresponding sequences in Arabidopsis (Fig. 1A). AtsORF0359 had eight cysteines in the coding sequence; this even number of cysteine residues is often found in functional cysteine-rich peptides such as LURE and EPF/EPFL.2,24 However, an odd number of cysteine residues was conserved between OsORF0359L and AtsORF0359. The start codon of OsORF0359L should be identified to predict this functional sORF accurately.
Figure 1. Molecular features of rice homologs of functional Arabidopsis sORFs. (A) Comparison of amino acid sequences encoded by sORFs between rice and Arabidopsis. Homologous sequences to Arabidopsis sORFs in rice were identified by BLAST searches. Sequences were aligned using CLUSTALW. Asterisk (*) indicates positions with a single fully conserved residue. Colon (:) indicates conservation between groups with strongly similar properties (scoring > 0.5 in Gonnet PAM 250 matrix). Period (.) indicates conservation between groups with weakly similar properties (scoring = < 0.5 in Gonnet PAM 250 matrix). (B) Physical map of rice sORFs on rice genome. DNA sequences for rice sORFs were input into RiceGE database (http://signal.salk.edu/cgi-bin/RiceGE), and physical locations of rice sORFs were identified by BLAST searches. (C) Detection of transcribed rice sORFs. A mixture of cDNAs was synthesized from mRNAs from 40 different samples (22 different organs, and shoots and roots of plants in nine different environmental conditions). Asterisk (*) represents expected PCR product size.
Physical Map of Rice sORFs on Rice Genome
Next, we determined whether the rice sORFs homologs were annotated as coding genes in the rice genome database. Out of 35 sORFs, 22 sORFs were in intergenic regions of the rice genome that have no annotated genes. For example, OsORF1189L, 1626L, 0359L, and 1411L were mapped in intergenic regions (Fig. 1B). This result implies that many sORFs are hidden in the rice genome. The other 13 sORFs overlapped with annotated genes in the rice genome. For example, OsORF2874L and OsORF5537L overlapped with Os02 g56120 and Os06 g12030, respectively (Fig. 1B). Os02 g56120 is annotated as an auxin-responsive AUX/IAA family protein that is involved in IAA signal transduction. However, an orthologous gene has not been identified in the Arabidopsis genome in the PlantPAN database (http://plantpan.mbc.nctu.edu.tw). Of the 13 sORFs, 7 showed a similar trend, indicating that some Arabidopsis sORFs are simply annotated genes in Oryza.
Os06 g12030 encodes a putative glutaredoxin-related protein that functions as an oxidoreductase. In TAIR version 8, AtsORF5537 neighbors AT4G08550, which encodes a putative glutaredoxin-related protein. Therefore, sORF5537 is likely to be a part of a known gene in both Arabidopsis and Oryza. Indeed, the expression pattern of AT4G08550 was quite similar to that of AtsORF5537 in HanaDB-AT (http://evolver.psc.riken.jp/seiken). Of the 13 sORFs, 6 sORFs showed a similar trend to that of sORF5537, indicating that these 6 sORFs are likely to be part of known genes. These results show that comparative genomics analyses are useful for validating sORFs as independent transcriptional units.
Expression Analysis of Rice sORFs
Next, we examined whether the sORFs in rice were transcribed into mRNA. We conducted RT-PCR analyses using a mixture of cDNAs, which were synthesized and prepared from 40 different samples. Out of 35 rice sORFs, 29 were detected as PCR products of the predicted size. Six of these sORFs are shown in Figure 1C. Because 83% (29/35) of the analyzed sORFs were transcribed into RNA in Oryza,12 it is likely that many other sORFs are present in the rice genome and are expressed. In future research, deep transcriptomics and proteomics analyses will be used to identify the transcriptional units of sORFs and to provide evidence of their encoded peptides, respectively.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
This work was supported by the Program for Promotion of Basic and Applied Researches for Innovations in Bio-oriented Industry (BRAIN; M.H-T. and K.H.), and partly funded by Grants-in-Aid for Scientific Research (to M.O., M.H.-T. and K.H.) and by Core Research for Evolutional Science and Technology (CREST) Program “Creation of essential technologies to utilize carbon dioxide as a resource through the enhancement of plant productivity and the exploitation of plant products” of the Japan Science and Technology Agency (JST) (to K.H.).
References
- 1.Fukuda H, Higashiyama T. Diverse functions of plant peptides: entering a new phase. Plant Cell Physiol. 2011;52:1–4. doi: 10.1093/pcp/pcq193. [DOI] [PubMed] [Google Scholar]
- 2.Higashiyama T. Peptide signaling in pollen-pistil interactions. Plant Cell Physiol. 2010;51:177–89. doi: 10.1093/pcp/pcq008. [DOI] [PubMed] [Google Scholar]
- 3.Hirakawa Y, Kondo Y, Fukuda H. Establishment and maintenance of vascular cell communities through local signaling. Curr Opin Plant Biol. 2011;14:17–23. doi: 10.1016/j.pbi.2010.09.011. [DOI] [PubMed] [Google Scholar]
- 4.Matsubayashi Y. Small post-translationally modified Peptide signals in Arabidopsis. Arabidopsis Book. 2011;9:e0150. doi: 10.1199/tab.0150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Matsubayashi Y, Sakagami Y. Peptide hormones in plants. Annu Rev Plant Biol. 2006;57:649–74. doi: 10.1146/annurev.arplant.56.032604.144204. [DOI] [PubMed] [Google Scholar]
- 6.Yamaguchi Y, Huffaker A. Endogenous peptide elicitors in higher plants. Curr Opin Plant Biol. 2011;14:351–7. doi: 10.1016/j.pbi.2011.05.001. [DOI] [PubMed] [Google Scholar]
- 7.Shiu SH, Bleecker AB. Receptor-like kinases from Arabidopsis form a monophyletic gene family related to animal receptor kinases. Proc Natl Acad Sci U S A. 2001;98:10763–8. doi: 10.1073/pnas.181141598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shiu SH, Karlowski WM, Pan R, Tzeng YH, Mayer KF, Li WH. Comparative analysis of the receptor-like kinase family in Arabidopsis and rice. Plant Cell. 2004;16:1220–34. doi: 10.1105/tpc.020834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Arabidopsis Genome I, Arabidopsis Genome Initiative Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;408:796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- 10.Hanada K, Zhang X, Borevitz JO, Li WH, Shiu SH. A large number of novel coding small open reading frames in the intergenic regions of the Arabidopsis thaliana genome are transcribed and/or under purifying selection. Genome Res. 2007;17:632–40. doi: 10.1101/gr.5836207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hanada K, Akiyama K, Sakurai T, Toyoda T, Shinozaki K, Shiu SH. sORF finder: a program package to identify small open reading frames with high coding potential. Bioinformatics. 2010;26:399–400. doi: 10.1093/bioinformatics/btp688. [DOI] [PubMed] [Google Scholar]
- 12.Hanada K, Higuchi-Takeuchi M, Okamoto M, Yoshizumi T, Shimizu M, Nakaminami K, Nishi R, Ohashi C, Iida K, Tanaka M, et al. Small open reading frames associated with morphogenesis are hidden in plant genomes. Proc Natl Acad Sci U S A. 2013;110:2395–400. doi: 10.1073/pnas.1213958110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Betsuyaku S, Sawa S, Yamada M. The Function of the CLE Peptides in Plant Development and Plant-Microbe Interactions. Arabidopsis Book. 2011;9:e0149. doi: 10.1199/tab.0149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ito Y, Nakanomyo I, Motose H, Iwamoto K, Sawa S, Dohmae N, Fukuda H. Dodeca-CLE peptides as suppressors of plant stem cell differentiation. Science. 2006;313:842–5. doi: 10.1126/science.1128436. [DOI] [PubMed] [Google Scholar]
- 15.Okamoto S, Shinohara H, Mori T, Matsubayashi Y, Kawaguchi M. Root-derived CLE glycopeptides control nodulation by direct binding to HAR1 receptor kinase. Nat Commun. 2013;4:2191. doi: 10.1038/ncomms3191. [DOI] [PubMed] [Google Scholar]
- 16.Suzaki T, Yoshida A, Hirano HY. Functional diversification of CLAVATA3-related CLE proteins in meristem maintenance in rice. Plant Cell. 2008;20:2049–58. doi: 10.1105/tpc.107.057257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Narita NN, Moore S, Horiguchi G, Kubo M, Demura T, Fukuda H, Goodrich J, Tsukaya H. Overexpression of a novel small peptide ROTUNDIFOLIA4 decreases cell proliferation and alters leaf shape in Arabidopsis thaliana. Plant J. 2004;38:699–713. doi: 10.1111/j.1365-313X.2004.02078.x. [DOI] [PubMed] [Google Scholar]
- 18.Wen J, Lease KA, Walker JC. DVL, a novel class of small polypeptides: overexpression alters Arabidopsis development. Plant J. 2004;37:668–77. doi: 10.1111/j.1365-313X.2003.01994.x. [DOI] [PubMed] [Google Scholar]
- 19.Hara K, Kajita R, Torii KU, Bergmann DC, Kakimoto T. The secretory peptide gene EPF1 enforces the stomatal one-cell-spacing rule. Genes Dev. 2007;21:1720–5. doi: 10.1101/gad.1550707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hara K, Yokoo T, Kajita R, Onishi T, Yahata S, Peterson KM, Torii KU, Kakimoto T. Epidermal cell density is autoregulated via a secretory peptide, EPIDERMAL PATTERNING FACTOR 2 in Arabidopsis leaves. Plant Cell Physiol. 2009;50:1019–31. doi: 10.1093/pcp/pcp068. [DOI] [PubMed] [Google Scholar]
- 21.Uchida N, Lee JS, Horst RJ, Lai HH, Kajita R, Kakimoto T, Tasaka M, Torii KU. Regulation of inflorescence architecture by intertissue layer ligand-receptor communication between endodermis and phloem. Proc Natl Acad Sci U S A. 2012;109:6337–42. doi: 10.1073/pnas.1117537109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sugano SS, Shimada T, Imai Y, Okawa K, Tamai A, Mori M, Hara-Nishimura I. Stomagen positively regulates stomatal density in Arabidopsis. Nature. 2010;463:241–4. doi: 10.1038/nature08682. [DOI] [PubMed] [Google Scholar]
- 23.Kondo T, Kajita R, Miyazaki A, Hokoyama M, Nakamura-Miura T, Mizuno S, Masuda Y, Irie K, Tanaka Y, Takada S, et al. Stomatal density is controlled by a mesophyll-derived signaling molecule. Plant Cell Physiol. 2010;51:1–8. doi: 10.1093/pcp/pcp180. [DOI] [PubMed] [Google Scholar]
- 24.Takata N, Yokota K, Ohki S, Mori M, Taniguchi T, Kurita M. Evolutionary relationship and structural characterization of the EPF/EPFL gene family. PLoS One. 2013;8:e65183. doi: 10.1371/journal.pone.0065183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Combier JP, Küster H, Journet EP, Hohnjec N, Gamas P, Niebel A. Evidence for the involvement in nodulation of the two small putative regulatory peptide-encoding genes MtRALFL1 and MtDVL1. Mol Plant Microbe Interact. 2008;21:1118–27. doi: 10.1094/MPMI-21-8-1118. [DOI] [PubMed] [Google Scholar]
- 26.Floyd SK, Bowman JL. The ancestral developmental tool kit of land plants. Int J Plant Sci. 2007;168:1–35. doi: 10.1086/509079. [DOI] [Google Scholar]