Abstract
Post-translational modifications (PTMs) such as phosphorylation, ubiquitination, and sumoylation play significant roles in regulating abscisic acid (ABA) signaling. The targets for PTM are usually transcriptional regulators such as Abscisic acid Insensitive 5 (ABI5). PTM regulate ABI5 stability as well as activity. The abundance of ABI5 is tightly controlled by the ubiquitination-26S proteasome system. E3 ubiquitin ligases such as KEG negatively regulate ABA signaling by promoting ABI5 ubiquitination and subsequent degradation by the 26S proteasome. In our recent study we demonstrated that, in the absence of ABA, KEG-mediated turnover of ABI5 occurs within the cytoplasm. Whereas ubiquitination promotes ABI5 degradation, sumoylation prohibits degradation of the transcription factor. While phosphorylation has been shown to regulate ABI5 activity, our studies and others suggest that the phosphorylation status of ABI5 does not play a significant role in modulating ABI5 turnover.
Keywords: abscisic acid, abscisic acid insensitive 5, ubiquitination, sumoylation, phosphorylation
The phytohormone abscisic acid (ABA) regulates many aspects of plant growth and development as well as response to abiotic and biotic stresses.1,2 During seed development, ABA regulates maturation, dormancy, and germination. ABA mediates responses to abiotic stresses, such as drought, cold, salinity, and UV radiation, as well as pathogen attack.2 Adverse environmental conditions result in increased ABA production and perception, which initiate signaling events that culminate in changes in gene expression required for stress tolerance. Cellular responses to ABA are mediated by a suite of transcription factors including members of the B3, APETALA2- (AP2), and basic leucine zippers (bZIPs) domain families.2-5 One of the most studied ABA-responsive transcription factors is Abscisic Acid Insensitive (ABI) 5. ABI5 is 1 of 13 functionally and structurally related bZIP proteins, referred to as ABA response element binding (AREB) or ABA-responsive promoter elements (ABRE) binding factors (ABFs), which mediate cellular responses to ABA.4-6 Each AREB/ABF protein contains 4 conserved domains (named C1-C4) and a DNA binding bZIP domain within the carboxyl terminal (Fig. 1A).5 AREB/ABF are ABA- and/or stress-inducible genes and the encoded proteins function as homo- or hetero-dimers that bind to ABRE elements found in the regulatory regions of many stress-related genes.7,8
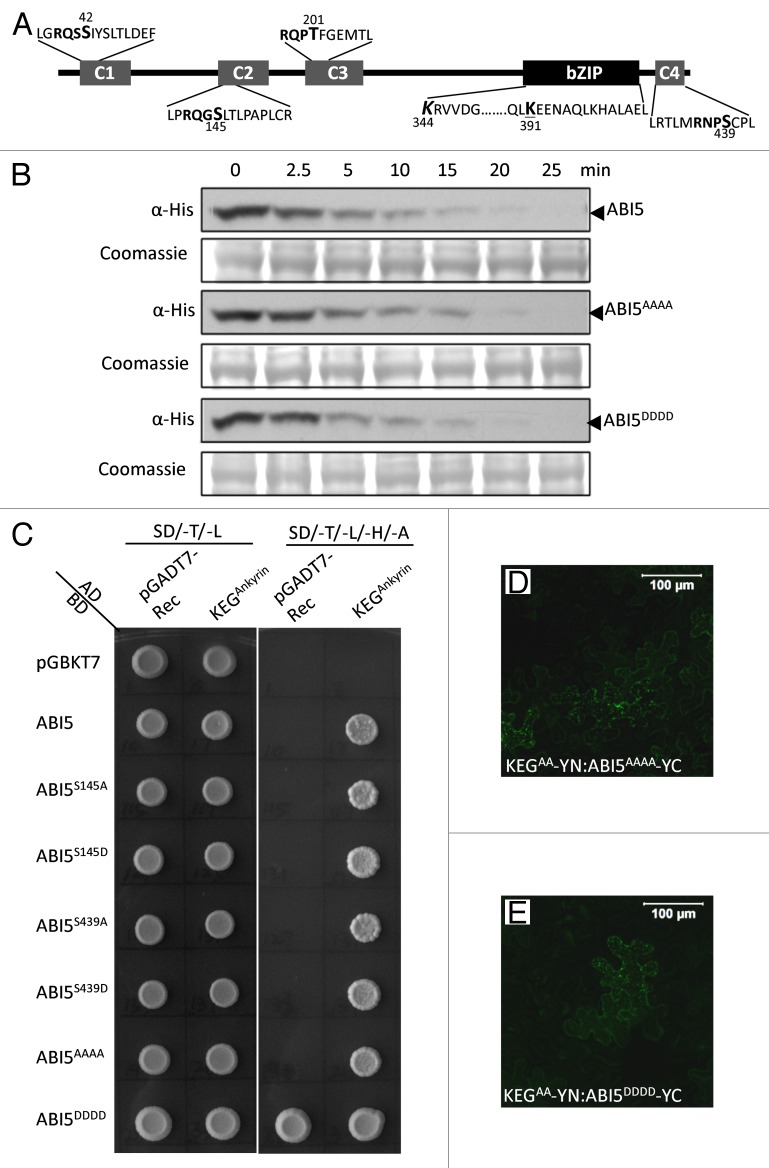
Figure 1. Phosphorylation does not impact ABI5 turnover and interactions with KEG E3 ligase. (A) Schematic representation of ABI5 showing the conserved regions, C1 - C4, and basic leucine zipper (bZIP) domain. Lysine (K) residues linked to ubiquitination (K344, bold and italics) and sumoylation (K391, bold and underlined) are highlighted. Phosphorylation sites within each conserved region (C1-C4) are shown in bold. Phosphoamino acids (serine [S] and threonine [T]) that were changed to alanine (A) or asparagine (D) are indicated (larger font and numbered). (B) Cell-free protein degradation assays. Phosphorylation sites in all 4 conserved domains (ABI5AAAA and ABI5DDDD) of ABI5 were mutated to A or D. Recombinant Flag-His tagged ABI5, ABI5AAAA and ABI5DDDD were incubated with protein extracts prepared from wild type Arabidopsis seedlings. Samples were taken at the indicated times and the level of ABI5 proteins determined by western blotting using His antibodies. Coomassie staining was used to confirm equal loading. (C) Yeast-2-hybrid experiments showing interactions between KEG and a series of ABI5 phosphomutants. Phosphorylation sites in the C2 (ABI5S145A and ABI5S145D), C4 (ABI5S439A and ABI5S439D), or all 4 conserved domains (ABI5AAAA and ABI5DDDD) of ABI5 were mutated to A or D. ABI5 cDNA constructs were fused to the GAL4 binding domain (BD) and KEG cDNA encoding for the ankyrin repeats (KEGAnkyrin) was fused to the GAL4 activating domain (AD). Interaction was analyzed by growth on selection medium without Trp, Leu, His, and Ade (SD/-T/-L/-H/-A). ABI5DDDD produced a high level of autoactivation. The pGADT7 and pGBKT7-Rec empty vectors were used as negative controls. (D-E) BiFC analysis in tobacco epidermal cells. Fluorescence indicates interactions between KEGAA-YN (RING mutant lacking E3 ligase activity) and ABI5AAAA-YC (D) or ABI5DDDD-YC (E). Bars = 100 μm.
ABI5 was identified via a forward genetic screen for mutations that rendered seeds/seedlings insensitivity to the inhibitory effects of ABA.9,10 Subsequently, overexpression of ABI5 was found to confer hypersensitivity to ABA.11 ABI5 is expressed in various tissue types at varying levels throughout the plant’s life cycle. In the developing embryo, ABI5 levels gradually increase, with highest levels occurring in mature seeds.8 Post-germination, ABI5 levels are low to non-detectable. Low levels of expression occur in vegetative tissues such as flowers, siliques, root tips, and leaf veins.8 ABA-mediated increase in ABI5 levels occur mainly within a narrow developmental time frame from 1 to 2 days post-germination.12 During this time frame, ABA promotes the accumulation of ABI5 via 2 mechanisms, increased transcription and reduced proteolysis. Transcription factors ABI3 and ABI4 as well as ABI5 are required for ABA induced ABI5 expression.8,13 Post-germination, in unstressed conditions, low levels of ABI5 are maintained via degradation of the transcription factor by the 26S proteasome, a large multi-catalytic protease complex.12,14,15 Upon exposure to stress conditions such as drought or salinity or elevated levels of ABA, proteasome-dependent turnover decreases and ABI5 becomes more stable.12,15 The accumulated ABI5 then induces expression of ABA-responsive genes required for growth inhibition and stress tolerance.11,16
Proteasomal degradation of ABI5 is dependent upon the ubiquitination pathway. Ubiquitination is the covalent attachment of ubiquitin, a small, compact and highly conserved protein, to select substrates. Attachment occurs via an isopeptide bond between a C-terminal glycine of the ubiquitin and a lysine (K) residue in the substrate protein. Ubiquitin conjugation requires the sequential action of 3 enzymes: the ubiquitin activating enzyme (E1) which activates ubiquitin, the ubiquitin conjugating enzyme (E2), which accepts the activated ubiquitin forming an E2-ubiquitin intermediate, and the substrate recruiting ubiquitin ligase (E3), which facilitates transfer of ubiquitin from the E2 to the selected protein. For targeting to the 26S proteasome, the conjugation process is repeated to generate a polyubiquitin chain using K48 of ubiquitin to create ubiquitin-ubiquitin linkages.17 The importance of ubiquitination in regulating ABA signaling is exemplified by the number of ubiquitin ligases that are found to be involved in plant responses to ABA.18,19 Ubiquitin ligases which directly target ABI5 include Keep on Going (KEG), a single subunit RING-type E3, which interacts with the E2 and ABI5 via its RING domain and ankyrin repeats, respectively.20 Post-germination, in the absence of ABA or stress, KEG is required to maintain low levels of ABI5 so as to ensure early seedling establishment.20,21 Loss of KEG function leads to accumulation of ABI5, hypersensitivity to ABA, and post-germinative growth arrest.20 We recently demonstrated that KEG, which is localized to the cytoplasm and trans-Golgi network/early endosome, targets nuclear-localized ABI5 for proteasomal degradation within the cytoplasm.22,23 KEG-mediated degradation requires interaction with the C3 domain of ABI5 as well as a carboxyl terminal K344 (Fig. 1A).23 Loss of the KEG-interacting C3 domain or K344 in conjunction with a nuclear localization signal (NLS) results in accumulation of ABI5 in the cytoplasm.23 KEG-mediated degradation of ABI5 is postulated to prohibit nuclear localization of the transcription factor so as to further inhibit ABA signaling. We have previously shown that ABA treatment promotes KEG ubiquitination and subsequent degradation, which would then allow for accumulation and transport of ABI5 into the nucleus and activation of the transcription factor via phosphorylation.21
Protein ubiquitination is reversible. Deubiquitinating enzymes (DUBs) are proteases that release ubiquitin moieties from modified proteins prior to degradation, thus preventing substrate turnover. The Arabidopsis genome is predicted to encode for numerous DUBs; however, only a few have been characterized.24 These include ubiquitin-specific protease (UBP) 3 and UBP4, which are essential for pollen maturation; UBP26, which is involved in seed development; and UBP15, which is required for apical dominance and flowering.25,26 Although DUBs have been linked to tolerance of abiotic stresses such as salt and heavy metal, enzymes that regulate ABA signaling or target ABI5 for deubiqutination have not yet been identified.27,28
Protein ubiquitination and subsequent degradation is modulated by other reversible PTMs such as phosphorylation and sumoylation. Phosphorylation is the addition of a phosphate group to tyrosine (Y), serine (S), or threonine (T) residues of a substrate protein. Families of kinases and phosphatases catalyze the addition or removal, respectively, of phosphate groups from different proteins. As one of the most prevalent forms of PTM, it is not surprising that there is interplay between phosphorylation and ubiquitination. Cross-talk between the 2 modifications can occur in many different ways. For example, phosphorylation of a substrate may facilitate recognition by the E3 ligase, or phosphorylation may directly activate an E3 ligase. In many cases, phosphorylation is a perquisite for ubiquitin-dependent proteasomal degradation. One well-studied example is IκBalpha, where phosphorylation by IκB kinase (IKK) results in recognition of IκBalpha by the F-box protein E3RSIκB, a substrate receptor for a Cullin-based E3 complex.29 IKK-mediated phosphorylation leads to IκBalpha polyubiquitination and targeting to the proteasome for degradation.30 ABI5 has 4 conserved phosphorylation sites, each found within 1 of 4 conserved regions, C1-C4 (Fig. 1A). Phosphorylation of ABI5 is accomplished by members of the Sucrose Non-fermenting1-related protein kinases (SnRK) 2 and CBL-interacting protein kinase (CIPK) families.31,32 Serine/Threonine Protein Phosphatase 6 (PP6) was recently shown to dephosphorylate ABI5 and negatively regulate ABA signaling.33 Phosphorylation of ABI5 is required for activation of the transcription factor.34 However, phosphorylation does not seem to modulate ABI5 stability, at least in the absence of ABA (Fig. 1). If phosphorylation is required prior to protein degradation, then replacing ABI5 phosphoamino acids with alanine, which is expected to prohibit phosphorylation, should stabilize the transcription factor. However, in cell free degradation assays, the turnover of a mutant version of the transcription factor with all 4 phosphoamino acids (S42, S145, T201, and S439) replaced with alanine (ABI5AAAA) was similar to that of wild type ABI5 (Fig. 1B). Also, changing all 4 phosphoamino acids to asparagine (D), which mimics constitutive phosphorylation, did not affect degradation of the phosphomutant (ABI5DDDD) compared with the wild type transcription factor (Fig. 1B). Similarly, Wang et al. (2013) reported that changing 3 of the 4 phosphoamino acids (S42, S145, and T201) to alanine did not alter ABI5 abundance.35 In addition, altering the phosphoamino acids did not interfere with the ability of ABI5 to interact with KEG in yeast 2-hybrid assays or during bimolecular fluorescence complementation (BiFC) analysis (Fig. 1C to E). Compared with wild-type ABI5, single mutations within the C2 (ABI5S145A and ABI5S145D) or C4 (ABI5S439A and ABI5S439D) region or loss of all 4 phosphorylation sites (ABI5AAAA) did not alter ABI5 interaction with KEG ankyrin repeats (KEG497–829) in yeast (Fig. 1C). Unfortunately, ABI5 with all 4 phosphorylation sites changed to D (ABI5DDDD) produced a high level of autoactivation, and prohibited study of this ABI5 form in yeast (Fig. 1C). BiFC assays were performed using Agrobacterium-mediated transient expression in tobacco leaf epidermal cells. Full-length KEG containing a non-functional RING E3 ligase domain (KEGAA) was used so as to prohibit substrate ubiquitination and degradation upon protein-protein interactions. BiFC fluorescence signals were observed following co-expression of KEGAA fused to the amino (N)-terminal YFP fragment (KEGAA-YN) with ABI5AAAA or ABI5DDDD fused to the carboxyl (C)-terminal YFP fragment (ABI5AAAA –YC or ABI5DDDD-YC) indicating that the YFP fluorophore was reconstituted due to the interaction between KEGAA and the ABI5 phosphomutants (Fig. 1D and 1E). Overall, phosphorylation does not seem to modulate ABI5 interaction with KEG or influence ABI5 turnover. However, the possibility that one or more of the phosphoamino acids may be required for modulating ABI5 abundance in the presence of ABA, so as to attenuate signaling, still remains to be determined.
Sumoylation is another reversible PTM that modulates ABI5 stability. The process involves the covalent attachment of small ubiquitin-related modifier (SUMO) to a lysine residue on a selected substrate.35 Similar to ubiquitin conjugation, sumoylation requires the sequential action of 3 enzymes, E1 activating enzymes, E2 conjugating enzymes, and E3 ligases. Deconjugation is accomplished by specific proteases that cleave SUMO from substrates.35 Sumoylation has been shown to antagonize ubiquitin-dependent protein degradation. Continuing with the example of IκBalpha, ubiquitination promotes proteasome-mediated degradation, whereas sumo modification stabilizes the protein.36 In Arabidopsis, SUMO conjugation utilizes an E1 heterodimer consisting of SUMO-activating enzyme 1(SAE1) and SAE2 and a single E2 SUMO-conjugating enzyme (SCE1).37 Few Arabidopsis SUMO E3s have been identified including SIZ1 [for SAP (scaffold attachment factor, acinus, protein inhibitor of activated signal transducer, and activator of transcription) and Miz1 (Ms32-interacting zinc finger) domain] and High Ploidy2 (HPY2)/Methyl Methane Sulphonate Sensitivity21 (MMS21).37 A role for SUMO in regulating of ABI5 abundance is evidenced by the low levels of ABI5 observed in siz1 seedlings compared with wild type before and after ABA treatment.38 Sumoylation of ABI5 requires K391. Overexpression of a mutant ABI5 harboring a K391 to arginine (R) mutation in abi5–4 background results in hypersensitivity to ABA, which suggests that, in addition to stabilizing AB15, SUMO modification negatively regulates ABA signaling.38 Thus, sumoylation is thought to maintain a degradation-resistant inactive pool of ABI5 in the absence of the hormone. Interestingly, ABI5 sumoylation site (K391) is within the same domain as the lysine (K344) residue required for KEG-dependent turnover of the transcription factor (Fig. 1A). In the cases of IκBalpha, SUMO modification has been shown to compete with ubiquitination for a common lysine residue.36 This does not seem to be the case for ABI5, however, whether or not the proximity of the 2 sites allows for cross-talk between sumoylation and ubiquitination remains to be determined. However, K391 as a site for ubiquitination still remains a possibility that needs to be pursued.
Reversible post-translational modifications such as phosphorylation, ubiquitination, and sumoylation play significant roles in regulating ABA signaling. The use of PTMs allows cells to rapidly and efficiently turn on and off signaling events in accordance with their external environment or in response to perception of specific stimuli. By targeting transcriptional regulators such as ABI5, PTMs can precisely regulate transcriptional activity and effectively tailor cellular responses required for stress tolerance.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
The authors would like to thank Wendy Lyzenga for comments on the manuscript. Stone SL is supported by grants from the Natural Sciences and Engineering Research Council of Canada (NSERC).
References
- 1.Cutler SR, Rodriguez PL, Finkelstein RR, Abrams SR. Abscisic acid: emergence of a core signaling network. Annu Rev Plant Biol. 2010;61:651–79. doi: 10.1146/annurev-arplant-042809-112122. [DOI] [PubMed] [Google Scholar]
- 2.Finkelstein R. Abscisic Acid synthesis and response. Arabidopsis Book. 2013;11:e0166. doi: 10.1199/tab.0166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bensmihen S, Giraudat J, Parcy F. Characterization of three homologous basic leucine zipper transcription factors (bZIP) of the ABI5 family during Arabidopsis thaliana embryo maturation. J Exp Bot. 2005;56:597–603. doi: 10.1093/jxb/eri050. [DOI] [PubMed] [Google Scholar]
- 4.Finkelstein R, Gampala SS, Lynch TJ, Thomas TL, Rock CD. Redundant and distinct functions of the ABA response loci ABA-INSENSITIVE(ABI)5 and ABRE-BINDING FACTOR (ABF)3. Plant Mol Biol. 2005;59:253–67. doi: 10.1007/s11103-005-8767-2. [DOI] [PubMed] [Google Scholar]
- 5.Bensmihen S, Rippa S, Lambert G, Jublot D, Pautot V, Granier F, Giraudat J, Parcy F. The homologous ABI5 and EEL transcription factors function antagonistically to fine-tune gene expression during late embryogenesis. Plant Cell. 2002;14:1391–403. doi: 10.1105/tpc.000869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Finkelstein RR, Lynch TJ. The Arabidopsis abscisic acid response gene ABI5 encodes a basic leucine zipper transcription factor. Plant Cell. 2000;12:599–609. doi: 10.1105/tpc.12.4.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Arroyo A, Bossi F, Finkelstein RR, León P. Three genes that affect sugar sensing (abscisic acid insensitive 4, abscisic acid insensitive 5, and constitutive triple response 1) are differentially regulated by glucose in Arabidopsis. Plant Physiol. 2003;133:231–42. doi: 10.1104/pp.103.021089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brocard IM, Lynch TJ, Finkelstein RR. Regulation and role of the Arabidopsis abscisic acid-insensitive 5 gene in abscisic acid, sugar, and stress response. Plant Physiol. 2002;129:1533–43. doi: 10.1104/pp.005793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Finkelstein RR. Mutations at two new Arabidopsis ABA response loci are similar to the abi3 mutations. Plant J. 1994;5:765–71. doi: 10.1046/j.1365-313X.1994.5060765.x. [DOI] [Google Scholar]
- 10.Lopez-Molina L, Chua N-H. A null mutation in a bZIP factor confers ABA-insensitivity in Arabidopsis thaliana. Plant Cell Physiol. 2000;41:541–7. doi: 10.1093/pcp/41.5.541. [DOI] [PubMed] [Google Scholar]
- 11.Brocard IM, Lynch TJ, Finkelstein RR. Regulation and role of the Arabidopsis abscisic acid-insensitive 5 gene in abscisic acid, sugar, and stress response. Plant Physiol. 2002;129:1533–43. doi: 10.1104/pp.005793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lopez-Molina L, Mongrand S, Chua NH. A postgermination developmental arrest checkpoint is mediated by abscisic acid and requires the ABI5 transcription factor in Arabidopsis. Proc Natl Acad Sci U S A. 2001;98:4782–7. doi: 10.1073/pnas.081594298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lopez-Molina L, Mongrand S, McLachlin DT, Chait BT, Chua NH. ABI5 acts downstream of ABI3 to execute an ABA-dependent growth arrest during germination. Plant J. 2002;32:317–28. doi: 10.1046/j.1365-313X.2002.01430.x. [DOI] [PubMed] [Google Scholar]
- 14.Smalle J, Kurepa J, Yang P, Emborg TJ, Babiychuk E, Kushnir S, Vierstra RD. The pleiotropic role of the 26S proteasome subunit RPN10 in Arabidopsis growth and development supports a substrate-specific function in abscisic acid signaling. Plant Cell. 2003;15:965–80. doi: 10.1105/tpc.009217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Piskurewicz U, Jikumaru Y, Kinoshita N, Nambara E, Kamiya Y, Lopez-Molina L. The gibberellic acid signaling repressor RGL2 inhibits Arabidopsis seed germination by stimulating abscisic acid synthesis and ABI5 activity. Plant Cell. 2008;20:2729–45. doi: 10.1105/tpc.108.061515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Uno Y, Furihata T, Abe H, Yoshida R, Shinozaki K, Yamaguchi-Shinozaki K. Arabidopsis basic leucine zipper transcription factors involved in an abscisic acid-dependent signal transduction pathway under drought and high-salinity conditions. Proc Natl Acad Sci U S A. 2000;97:11632–7. doi: 10.1073/pnas.190309197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Komander D, Rape M. The ubiquitin code. Annu Rev Biochem. 2012;81:203–29. doi: 10.1146/annurev-biochem-060310-170328. [DOI] [PubMed] [Google Scholar]
- 18.Lyzenga WJ, Stone SL. Abiotic stress tolerance mediated by protein ubiquitination. J Exp Bot. 2012;63:599–616. doi: 10.1093/jxb/err310. [DOI] [PubMed] [Google Scholar]
- 19.Liu H, Stone SL. E3 ubiquitin ligases and abscisic acid signaling. Plant Signal Behav. 2011;6:344–8. doi: 10.4161/psb.6.3.13914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stone SL, Williams LA, Farmer LM, Vierstra RD, Callis J. KEEP ON GOING, a RING E3 ligase essential for Arabidopsis growth and development, is involved in abscisic acid signaling. Plant Cell. 2006;18:3415–28. doi: 10.1105/tpc.106.046532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu H, Stone SL. Abscisic acid increases Arabidopsis ABI5 transcription factor levels by promoting KEG E3 ligase self-ubiquitination and proteasomal degradation. Plant Cell. 2010;22:2630–41. doi: 10.1105/tpc.110.076075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gu Y, Innes RW. The KEEP ON GOING protein of Arabidopsis recruits the ENHANCED DISEASE RESISTANCE1 protein to trans-Golgi network/early endosome vesicles. Plant Physiol. 2011;155:1827–38. doi: 10.1104/pp.110.171785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu H, Stone SL. Cytoplasmic degradation of the Arabidopsis transcription factor abscisic acid insensitive 5 is mediated by the RING-type E3 ligase KEEP ON GOING. J Biol Chem. 2013;288:20267–79. doi: 10.1074/jbc.M113.465369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vierstra RD. The ubiquitin-26S proteasome system at the nexus of plant biology. Nat Rev Mol Cell Biol. 2009;10:385–97. doi: 10.1038/nrm2688. [DOI] [PubMed] [Google Scholar]
- 25.Liu Y, Wang F, Zhang H, He H, Ma L, Deng XW. Functional characterization of the Arabidopsis ubiquitin-specific protease gene family reveals specific role and redundancy of individual members in development. Plant J. 2008;55:844–56. doi: 10.1111/j.1365-313X.2008.03557.x. [DOI] [PubMed] [Google Scholar]
- 26.Doelling JH, Phillips AR, Soyler-Ogretim G, Wise J, Chandler J, Callis J, Otegui MS, Vierstra RD. The ubiquitin-specific protease subfamily UBP3/UBP4 is essential for pollen development and transmission in Arabidopsis. Plant Physiol. 2007;145:801–13. doi: 10.1104/pp.106.095323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhao J, Zhou H, Li X. UBIQUITIN-SPECIFIC PROTEASE16 interacts with a HEAVY METAL ASSOCIATED ISOPRENYLATED PLANT PROTEIN27 and modulates cadmium tolerance. Plant Signal Behav. 2013;8:e25680. doi: 10.4161/psb.25680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sridhar VV, Kapoor A, Zhang K, Zhu J, Zhou T, Hasegawa PM, Bressan RA, Zhu JK. Control of DNA methylation and heterochromatic silencing by histone H2B deubiquitination. Nature. 2007;447:735–8. doi: 10.1038/nature05864. [DOI] [PubMed] [Google Scholar]
- 29.Yaron A, Hatzubai A, Davis M, Lavon I, Amit S, Manning AM, Andersen JS, Mann M, Mercurio F, Ben-Neriah Y. Identification of the receptor component of the IkappaBalpha-ubiquitin ligase. Nature. 1998;396:590–4. doi: 10.1038/25159. [DOI] [PubMed] [Google Scholar]
- 30.Chen Z, Hagler J, Palombella VJ, Melandri F, Scherer D, Ballard D, Maniatis T. Signal-induced site-specific phosphorylation targets I κ B alpha to the ubiquitin-proteasome pathway. Genes Dev. 1995;9:1586–97. doi: 10.1101/gad.9.13.1586. [DOI] [PubMed] [Google Scholar]
- 31.Weiner JJ, Peterson FC, Volkman BF, Cutler SR. Structural and functional insights into core ABA signaling. Curr Opin Plant Biol. 2010;13:495–502. doi: 10.1016/j.pbi.2010.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lyzenga WJ, Liu H, Schofield A, Muise-Hennessey A, Stone SL. Arabidopsis CIPK26 interacts with KEG, components of the ABA signalling network and is degraded by the ubiquitin-proteasome system. J Exp Bot. 2013;64:2779–91. doi: 10.1093/jxb/ert123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dai M, Xue Q, Mccray T, Margavage K, Chen F, Lee JH, Nezames CD, Guo L, Terzaghi W, Wan J, et al. The PP6 phosphatase regulates ABI5 phosphorylation and abscisic acid signaling in Arabidopsis. Plant Cell. 2013;25:517–34. doi: 10.1105/tpc.112.105767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang Y, Li L, Ye T, Lu Y, Chen X, Wu Y. The inhibitory effect of ABA on floral transition is mediated by ABI5 in Arabidopsis. J Exp Bot. 2013;64:675–84. doi: 10.1093/jxb/ers361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Geiss-Friedlander R, Melchior F. Concepts in sumoylation: a decade on. Nat Rev Mol Cell Biol. 2007;8:947–56. doi: 10.1038/nrm2293. [DOI] [PubMed] [Google Scholar]
- 36.Desterro JM, Rodriguez MS, Hay RT. SUMO-1 modification of IkappaBalpha inhibits NF-kappaB activation. Mol Cell. 1998;2:233–9. doi: 10.1016/S1097-2765(00)80133-1. [DOI] [PubMed] [Google Scholar]
- 37.Miura K, Hasegawa PM. Sumoylation and other ubiquitin-like post-translational modifications in plants. Trends Cell Biol. 2010;20:223–32. doi: 10.1016/j.tcb.2010.01.007. [DOI] [PubMed] [Google Scholar]
- 38.Miura K, Lee J, Jin JB, Yoo CY, Miura T, Hasegawa PM. Sumoylation of ABI5 by the Arabidopsis SUMO E3 ligase SIZ1 negatively regulates abscisic acid signaling. Proc Natl Acad Sci U S A. 2009;106:5418–23. doi: 10.1073/pnas.0811088106. [DOI] [PMC free article] [PubMed] [Google Scholar]