Abstract
Perception of environmental cues and adaptation to changing environmental conditions are crucial for survival of sessile organisms like plants. This is even more important for woody perennial species like trees that can occupy a site for thousands of years. We have previously shown that under low nitrogen (LN), poplar trees display a vigorous and long-lasting root growth associated with global transcriptomic reprogramming and an activation of hierarchical genetic networks. Here we use computational analysis to better understand the network among the genes showing distinct chronological patterns of expression during the response. Our analyses confirm the previous findings, define new potential signaling pathways and the possible downstream targets of these signaling events.
Keywords: Signal transduction, biological processes, transcription factors, root specific growth, low nitrogen
In our recent manuscript,1 we reported that poplar root growth was induced by low nitrogen (LN) and sustained for at least 3 weeks. The temporal order of the significantly enriched biological processes suggests the LN activates a wide range of signaling cascades that bolster the long-lasting root growth. To better understand these putative signaling events, we analyzed 1,813 genes involved in the 36 biological processes (BP) that show strict temporal expression patterns associated with distinct growth and developmental phases of the root response (Fig. 3 as shown in 1). These 36 BPs were originally classified into 3 major functional gene ontologies (GO) groups that include signal transduction (5 GO, 331 genes), growth and development (21 GO, 1,298 genes), and root development (10 GO, 184 genes). These 3 groups of BPs are enriched in a temporally consecutive sequence and defined here, to facilitate the description and analyses, as Stage 1, 2, and 3. Stage 1 is chronologically first, occurs very early (within 24 h) and involves LN sensing and signaling events. Stage 2 encompasses genes and processes that are activated as a result of events occurring in Stage 2, takes place later (within 48–96 h) and primarily encompasses activation of cellular growth and proliferation. Stage 3 represents later (weeks) events that include the lateral root growth, proliferation and differentiation.
Because our data suggested of a temporal regulatory cascade involved in the response to LN, we used computational approach to identify the involved putative regulatory factors and their potential interactions. We first used Fisher Exact test to identify statistically significant associations between the expressions of the genes regulated at the 3 stages. We then employed Benjamini and Hochberg False Discovery Rate to correct the resulting p values from the Fisher Exact test, and the dependent gene pairs were then cut-off at threshold p value of 0.05. This resulted in 29, 87, and 16 genes (Table S1) from Stage 1, 2, and 3 respectively that showed statistically significant expression dependency. These genes were used to build a gene regulatory network (GRN) using a bottom-up graphic Gaussian model (GGM) algorithm that we recently developed.2 The overwhelming majority (87/132) of the genes in this network are regulated in Stage 2. Consistent with our hypothesis that this stage involves growth and developmental processes, we found many genes involved in regulation of cell cycle (e.g., cyclins); cell wall biosynthesis and remodeling (e.g., cellulose synthases and expansins); and key regulators of root morphogenesis (e.g., PtaSCR and PtaSHR) (Table S1).
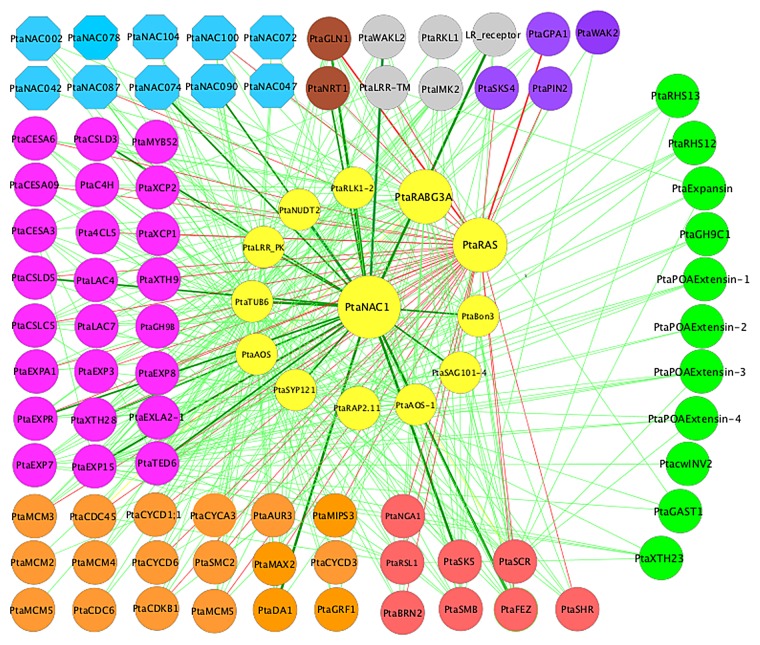
As we showed in our original publication,1 PtaNAC1 (XM_002310652, Potri.007G065400) appears to have a pivotal role in the poplar root response to LN. This new computational analysis confirmed that PtaNAC1 is connected to genes involved in a variety of processes including root development, growth, and nitrogen assimilation (Fig. 1), implying that PtaNAC1 plays important coordinating roles in integration the response at multiple levels. In addition to PtaNAC1, several other genes encoding NAC domain proteins appear to play important roles in the response (Fig. 1). Although involvement of NAC genes in regulation of the response to nitrogen has been almost completely unknown, recent evidence suggests that they play important roles, particularly in relation to modification of root architecture.1,3
Figure 1. GRN involved in the response of poplar roots to LN. Yellow nodes in the center represent genes from Stage 1 and green nodes at the right side contain genes from Stage 3. All other nodes in various colors contain genes from Stage 2. Genes from Stage 2 were classified into different functional types indicated in different colors as follows: cell growth and division (orange), cell wall formation (magenta), transcription factors from the NAC family (deep sky blue), nitrogen metabolism (tan), and root specific growth (red), and dark-violet represents the unclassified genes. PtaNAC1 and PtaRAS connection edges, which are discussed in the text, are presented in bold and green and red color respectively.
The GRN analyses also seem to indicate that the RAS pathway is involved in the signaling events following LN treatment (Fig. 1). For example, a RAS-like protein encoding gene (PtaRAS) differentially regulated at Stage 1, appears to be connected to genes involved in cell division and growth, cell wall formation, and root development (Fig. 1). PtaRAS (XM_002321005, Potri.T106300) is connected to 3 genes encoding NAC transcription factors including PtaFEZ (XM_002331469, Potri.017G082000), PtaBRN2 (BEARSKIN 2) (XM_002329142, Potri.019G066000), and PtaSMB (SOMBRERO) (XM_002316127, Potri.010G176600) (Fig. 1, Table S1). Homologs of these 3 proteins in Arabidopsis are known to govern root-cap development.4,5 PtaRAS also appears to be connected to several genes that are homologs of Arabidopsis genes involved in root radial patterning. These include PtaSHR (SHORT-ROOT) (XM_002332327, Potri.007G063300), PtaSCR (SCARECROW) (XM_002306827, Potri.006G114200), and PtaRSL1 (RHD SIX-LIKE 1) (XM_002305167, Potri.016G076100). Arabidopsis SHR and SCR are plant-specific GRAS family transcription factors that control the radial organization of the root ground tissues.6-8 RSL1 in Arabidopsis thaliana, together with RHD6 (ROOT HAIR DEFECTIVE 6), is required for root-hair cell development and differentiation.9-11
We believe that future unraveling and functional dissection of the GRN networks, suggested by our analysis to be involved in the response to LN, will provide new venues for improvement of nitrogen assimilation and increase of nitrogen use efficiency in agricultural and biofuel crops.
Supplementary Material
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Supplemental Materials
Supplemental materials may be found here: www.landesbioscience.com/journals/psb/article/27211/
References
- 1.Wei H, Yordanov YS, Georgieva T, Li X, Busov V. Nitrogen deprivation promotes Populus root growth through global transcriptome reprogramming and activation of hierarchical genetic networks. New Phytol. 2013;200:483–97. doi: 10.1111/nph.12375. [DOI] [PubMed] [Google Scholar]
- 2.Lu S, Li Q, Wei H, Chang MJ, Tunlaya-Anukit S, Kim H, Liu J, Song J, Sun YH, Yuan L, et al. Ptr-miR397a is a negative regulator of laccase genes affecting lignin content in Populus trichocarpa. Proc Natl Acad Sci U S A. 2013;110:10848–53. doi: 10.1073/pnas.1308936110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vidal EA, Moyano TC, Riveras E, Contreras-López O, Gutiérrez RA. Systems approaches map regulatory networks downstream of the auxin receptor AFB3 in the nitrate response of Arabidopsis thaliana roots. Proc Natl Acad Sci U S A. 2013;110:12840–5. doi: 10.1073/pnas.1310937110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bennett T, van den Toorn A, Sanchez-Perez GF, Campilho A, Willemsen V, Snel B, Scheres B. SOMBRERO, BEARSKIN1, and BEARSKIN2 regulate root cap maturation in Arabidopsis. Plant Cell. 2010;22:640–54. doi: 10.1105/tpc.109.072272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Willemsen V, Bauch M, Bennett T, Campilho A, Wolkenfelt H, Xu J, Haseloff J, Scheres B. The NAC domain transcription factors FEZ and SOMBRERO control the orientation of cell division plane in Arabidopsis root stem cells. Dev Cell. 2008;15:913–22. doi: 10.1016/j.devcel.2008.09.019. [DOI] [PubMed] [Google Scholar]
- 6.Koizumi K, Gallagher KL. Identification of SHRUBBY, a SHORT-ROOT and SCARECROW interacting protein that controls root growth and radial patterning. Development. 2013;140:1292–300. doi: 10.1242/dev.090761. [DOI] [PubMed] [Google Scholar]
- 7.Pauluzzi G, Divol F, Puig J, Guiderdoni E, Dievart A, Périn C. Surfing along the root ground tissue gene network. Dev Biol. 2012;365:14–22. doi: 10.1016/j.ydbio.2012.02.007. [DOI] [PubMed] [Google Scholar]
- 8.Ogasawara H, Kaimi R, Colasanti J, Kozaki A. Activity of transcription factor JACKDAW is essential for SHR/SCR-dependent activation of SCARECROW and MAGPIE and is modulated by reciprocal interactions with MAGPIE, SCARECROW and SHORT ROOT. Plant Mol Biol. 2011;77:489–99. doi: 10.1007/s11103-011-9826-5. [DOI] [PubMed] [Google Scholar]
- 9.Menand B, Yi K, Jouannic S, Hoffmann L, Ryan E, Linstead P, Schaefer DG, Dolan L. An ancient mechanism controls the development of cells with a rooting function in land plants. Science. 2007;316:1477–80. doi: 10.1126/science.1142618. [DOI] [PubMed] [Google Scholar]
- 10.Jang G, Yi K, Pires ND, Menand B, Dolan L. RSL genes are sufficient for rhizoid system development in early diverging land plants. Development. 2011;138:2273–81. doi: 10.1242/dev.060582. [DOI] [PubMed] [Google Scholar]
- 11.Masucci JD, Schiefelbein JW. The rhd6 Mutation of Arabidopsis thaliana Alters Root-Hair Initiation through an Auxin- and Ethylene-Associated Process. Plant Physiol. 1994;106:1335–46. doi: 10.1104/pp.106.4.1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.