Abstract
Cuticle secreted on the surface of the epidermis of aerial organs protects plants from the external environment. We recently found that Arabidopsis MIXTA-like R2R3-MYB family members MYB16 and MYB106 regulate cuticle formation in reproductive organs and trichomes. However, the artificial miRNA (amiRNA)-mediated knockdown plants showed no clear phenotypic abnormality in vegetative tissues. In this study, we used RNA interference (RNAi) targeting MYB16 to produce plants with reduced expression of both MYB16 and MYB106. The rosette leaves of RNAi plants showed more severe permeable cuticle phenotypes than the myb106 mutants expressing the MYB16 amiRNA in the previous study. The RNAi plants also showed reduced expression of cuticle biosynthesis genes LACERATA and ECERIFERUM1. By contrast, expression of a gain-of-function MYB16 construct induced over-accumulation of waxy substances on leaves. These results suggest that MYB16 functions as a major regulator of cuticle formation in vegetative organs, in addition to its effect in reproductive organs and trichomes.
Keywords: Transcription factor, cuticle, wax, MIXTA-like protein, Arabidopsis
In plants, the outmost cell layer, or epidermis, differentiates into many types of cells, including leaf pavement cells, stomatal guard cells, trichomes, and petal conical cells. The epidermal cells of the above-ground parts of the plant secrete a cuticle of cutin and wax outside of the cell wall; this cuticle protects plants from biotic and abiotic stresses, confers mechanical strength, regulates water and gas exchange (together with stomata), and prevents organ fusion.1
MIXTA homologs belonging to the R2R3-MYB family subgroup 9 regulate epidermal cell outgrowth of petal conical cells and trichomes. For example, in Antirrhinum majus, the mixta mutant shows alterations in petal color intensity, in which conical cells become flat.2 Other MIXTA homologs in A. majus and petunia promote anticlinal expansion of epidermal cells, as shown by analysis of mutants and overexpression lines.3-5 Overexpression of MYB16, an Arabidopsis MIXTA homolog, induced similar ectopic outgrowths in petals of Arabidopsis and tobacco, suggesting it has similar functions to petunia MYB1 and A. majus MYBML2.4 Another Arabidopsis homolog, MYB106/NOK, regulates trichome maturation, including promotion and limitation of trichome branching.6,7
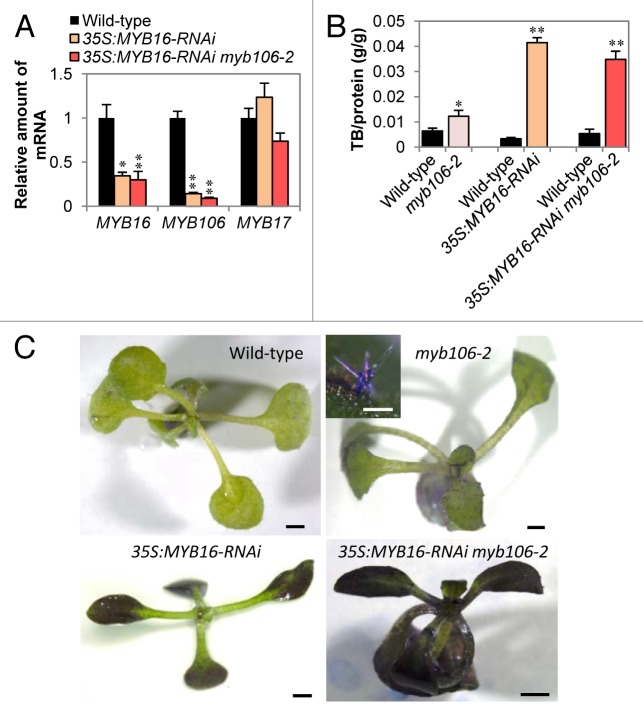
We recently described additional functions of MIXTA-like proteins in cuticle development.8 Expression of MYB106 and MYB16 chimeric repressor fusions (MYB106-SRDX, MYB16-SRDX) under the control of the CaMV 35S promoter induced cuticle deficiencies such as permeable surfaces, reduced epicuticular wax crystals, and loss of nanoridges, in whole plants.8 By contrast, expression of the dominant active form of MYB106 (MYB106-VP16), which has a virus-derived strong transcriptional activation domain VP16,9 induced ectopic formation of nanoridges, which are usually only formed in floral organs, and plate-like wax crystals on rosette leaves.8 These data, together with the results of transcriptome analysis of these transgenic plants and effector reporter assays, revealed that MYB106 positively regulates cuticle formation through activation of the expression of cutin and wax biosynthetic genes and an APETALA2/ETHYLENE RESPONSE FACTOR (AP2/ERF) transcription factor, WAX INDUCER1/SHINE1, which also positively regulates cutin biosynthetic genes.8,10-13 We also analyzed myb106–2 mutants and MYB16 artificial miRNA (MYB16-amiRNA) plants and demonstrated that MYB106 and MYB16 have redundant functions in cuticle nanoridge formation in petal and stamen, and in morphogenesis of petal conical cell and trichome. However, the MYB16-amiRNA myb106–2 double knockdown/out plants did not show a cuticle deficiency in leaves. A leaf phenotype was expected because the MYB16 promoter has activity in leaves and MYB16-SRDX driven by the MYB16 promoter produces an organ fusion phenotype in leaves.8 These data suggest that the amiRNA-mediated suppression of MYB16 was not sufficient, or there is another functionally redundant gene in addition to MYB16 and MYB106. To answer this question, in this study, we produced MYB16 silenced plants by introducing a 35S:MYB16-RNAi construct into wild type and myb106–2 mutants. To construct 35S:MYB16-RNAi, the coding sequence of MYB16 was transferred by Gateway LR reaction, into pHG8-based vector.8,14 qRT-PCR procedure and analysis method were described before.8 3′ untranslated region of MYB106 was amplified by using the specific primers MYB106–3′f 5′-ACCGTCCGATTCCGCGACGA-3′ and MYB106–3′r 5′-ACCGTCCGATTCCGCGACGA-3′.8 5′ untranslated region of MYB16 was amplified by using the specific primers AT5G15310pRB 5′-CGCGGATCCTGTTTTGAGAGCAAAGAAATAAGA-3′8 and AT5G15310pS 5′-AACATTTCCACAACTGTAGCCAAACT-3′. MYB17 was amplified by using the primers 3G61250Rf1 5′-AAAGGTCCTTGGACGCCTGAA-3′ and 3G61250Rr1 5′-TCTTCCCACAGCGAAGTAAACCA-3′. In 35S:MYB16-RNAi and 35S:MYB16-RNAi myb106–2 plants, expression of both MYB16 and MYB106 mRNAs was reduced, but expression of MYB17, which belongs to R2R3-MYB family subgroup 9 and is closest to MYB16 and MYB106, was not affected (Fig. 1A). The high nucleotide sequence similarity between MYB16 and MYB106 likely causes the silencing effect of the 35S:MYB16-RNAi construct on MYB106.
Figure 1. Surface permeability of MYB16 and MYB106 silenced plants. (A) Quantitative RT-PCR analysis of 5′ or 3′ untranslated region of MYB16 and MYB106, respectively, and the coding region of MYB17 in 2-week-old plants (n = 5). Error bars represent standard error. Single and double asterisks indicate P < 0.05 and P < 0.01 in the Welch t-test, respectively. (B) TB uptake per gram protein (n = 8) after staining by 0.05% TB for 2 min. (C) Seedling stained with TB. Bars = 1 mm. The inset panel shows a stained trichome of myb106–2. Bar = 100 μm.
To investigate cuticle permeability in the RNAi plants, we performed toluidine blue (TB) staining.15 Broad regions of leaves in 35S:MYB16-RNAi and 35S:MYB16-RNAi myb106–2 plants were stained by TB, but only the trichome cells in myb106–2 were stained (Fig. 1B and 1C). MYB106 is expressed in trichomes of leaves and the single mutant of MYB106 exhibited abnormalities only in the trichome cells;6,8 therefore, the cuticle deficiency of the leaves in MYB16-RNAi plants is likely to be mainly caused by the suppression of MYB16. We also found that the cuticle related genes LACERATA (LCR) and ECERIFERUM1 (CER1) were also suppressed in the MYB16-RNAi plants. Expression of LCR and CER1 was reduced to approximately half and less than one fifth of wild type, respectively (Fig. 2). LCR encodes a cytochrome P450 monooxygenase CYP86A8 which catalyzes ω-hydration of fatty acids, and is suggested to be involved in the biosynthesis of cutin; indeed, lcr mutants show typical cutin-deficient phenotypes like postgenital organ fusion and hydration of wild type pollen on mutant leaves, which indicates permeable leaf cuticle.16 CER1 protein is a core component of the very-long-chain (VLC) alkane synthesis complex.17 The mutants of CER1 showed reduced wax VLC alkane accumulation and increased cuticle permeability, and CER1 overexpression increased VLC alkane derivatives and reduced permeability, resulting in drought tolerance and increased susceptibility to pathogens.18,19 MYB16-RNAi expression affected the expression of these cutin and wax synthesis genes in leaves, suggesting that MYB16 functions through the regulation of these enzymatic genes in cuticle formation.
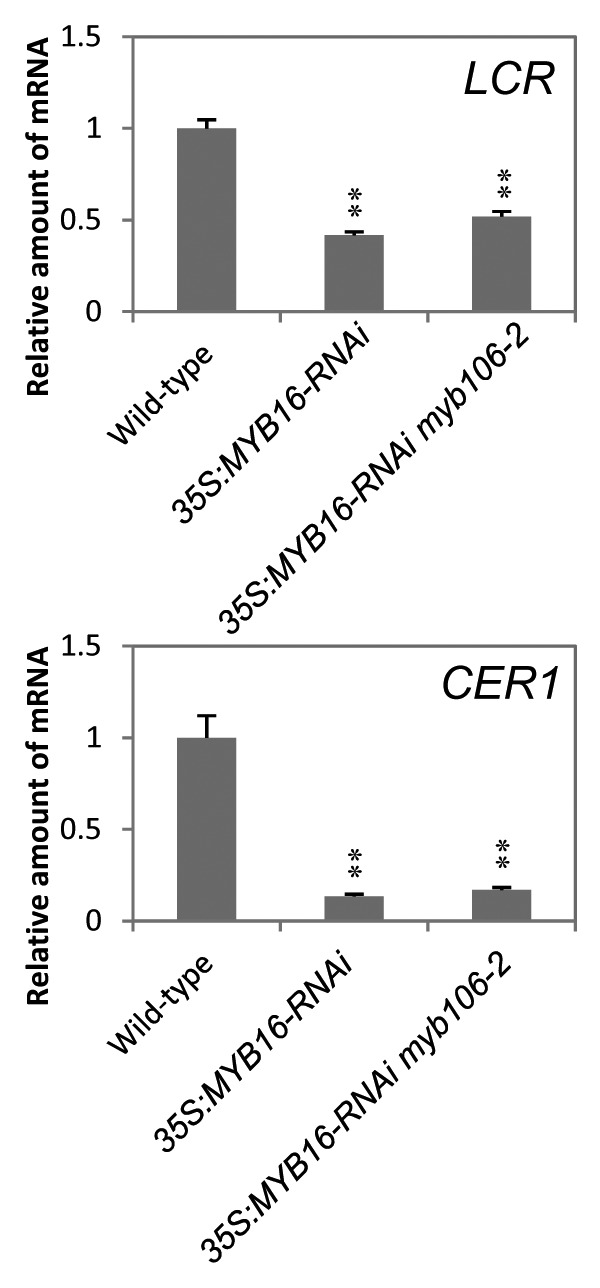
Figure 2. Expression of cuticle-related genes in transgenic plants. qRT-PCR analysis of expression of LCR and CER1 in 2-week-old wild type, 35S:MYB16-RNAi and 35S:MYB16-RNAi myb106–2 plants. Expression level in the wild type is set as 1. Error bars represent SE (n = 5). Double asterisks indicate P < 0.01 in the Welch t-test.
To examine the ability of MYB16 to induce cuticle synthesis, we generated plants expressing the dominant active form of MYB16, in which MYB16 was fused with the VP16 activation domain from herpes simplex virus (35S:MYB16-VP16). The 35S:MYB16-VP16 plants had slightly shiny leaves (Fig. 3A) and scanning electron microscopy revealed over-accumulation of epicuticular wax-like substances (Fig. 3B and 3C), as observed in 35S:MYB106-VP16, 35S:MYB106 and 35S:WIN1 plants.8 MYB16 activated expression from the promoters of WIN1/SHN1 and the cutin biosynthesis gene CYP86A4 in vivo,8 indicating that MYB16 functions as a positive regulator of cuticle formation. Taking these results together, we conclude that MYB16 is a major regulator of cuticle formation in vegetative tissues.
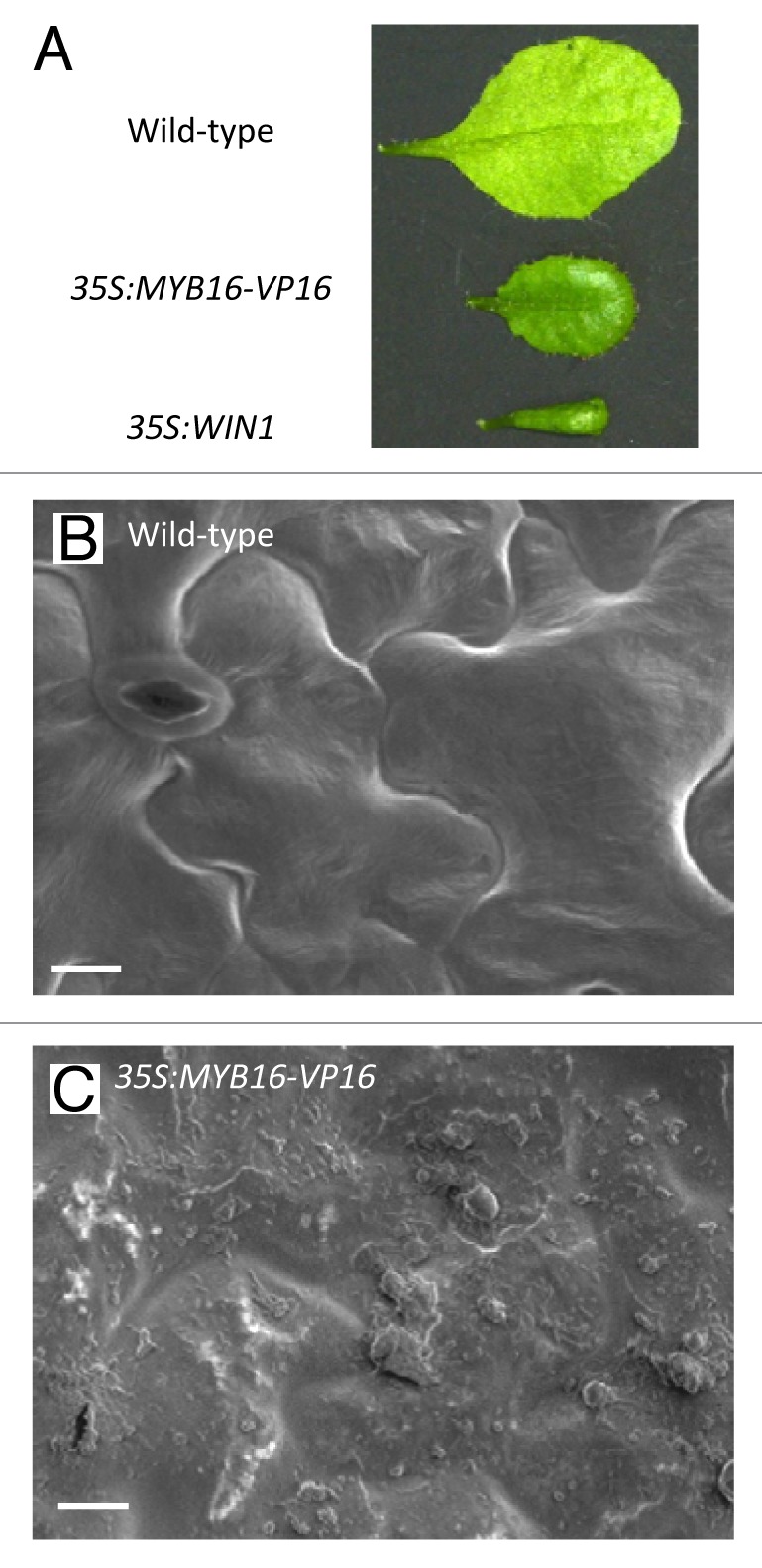
Figure 3. Ectopic accumulation of cuticular substances in 35S:MYB16-VP16 plants. (A) Rosette leaves excised from wild-type, 35S:MYB16-VP16, and 35S:WIN1 plants. (B) and (C) Adaxial surface of rosette leaves of wild-type (B) and 35S:MYB16-VP16 (C) plants observed by scanning electron microscopy. Bars = 10μm.
SHN3, a paralog of WIN1/SHN1, has been suggested to regulate cuticle formation in leaves, based on its expression pattern and ability to induce accumulation of cuticle.10 However, SHN1/2/3 triple amiRNA plants did not show cuticle deficiency in vegetative organs.13 HD-Zip IV family transcription factors specifying epidermal cell identity during various stages in epidermal cell differentiation also regulate some cuticle related genes via the L1-box in their promoters.1,20-22 Mutants of these HD-Zip IV transcription factors showed protodermal and epidermal defects in early developmental stages, or no phenotype due to redundancy.20,21 Cuticle development in leaves after differentiation of epidermal cells requires LCR and CER1, and other cutin and wax synthesizing enzyme and transporter genes (with or without the L1-box in their promoters).23-26 Assessing the involvement of MIXTA-like MYB transcription factors in the regulation of these genes may provide valuable insights into the regulatory mechanism of cuticle development in vegetative organs.
Acknowledgments
We thank the Arabidopsis Biological Resource Center (ABRC) for distributing seeds for the T-DNA-tagged line. We also thank Yuko Takiguchi, Fumie Tobe, Naomi Ujiie, Akiko Kuwazawa, Yukie Kimura, Keiko Kigoshi, and Sumiko Takahashi for their skillful technical assistance. This work was partly supported by the Program for Promotion of Basic and Applied Researches for Innovations in Bio-oriented Industry from the Bio-oriented Technology Research Advancement Institution, Japan (to Mitsuda N).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1.Javelle M, Vernoud V, Rogowsky PM, Ingram GC. Epidermis: the formation and functions of a fundamental plant tissue. New Phytol. 2011;189:17–39. doi: 10.1111/j.1469-8137.2010.03514.x. [DOI] [PubMed] [Google Scholar]
- 2.Noda K, Glover BJ, Linstead P, Martin C. Flower colour intensity depends on specialized cell shape controlled by a Myb-related transcription factor. Nature. 1994;369:661–4. doi: 10.1038/369661a0. [DOI] [PubMed] [Google Scholar]
- 3.Perez-Rodriguez M, Jaffe FW, Butelli E, Glover BJ, Martin C. Development of three different cell types is associated with the activity of a specific MYB transcription factor in the ventral petal of Antirrhinum majus flowers. Development. 2005;132:359–70. doi: 10.1242/dev.01584. [DOI] [PubMed] [Google Scholar]
- 4.Baumann K, Perez-Rodriguez M, Bradley D, Venail J, Bailey P, Jin H, Koes R, Roberts K, Martin C. Control of cell and petal morphogenesis by R2R3 MYB transcription factors. Development. 2007;134:1691–701. doi: 10.1242/dev.02836. [DOI] [PubMed] [Google Scholar]
- 5.Jaffé FW, Tattersall A, Glover BJ. A truncated MYB transcription factor from Antirrhinum majus regulates epidermal cell outgrowth. J Exp Bot. 2007;58:1515–24. doi: 10.1093/jxb/erm020. [DOI] [PubMed] [Google Scholar]
- 6.Jakoby MJ, Falkenhan D, Mader MT, Brininstool G, Wischnitzki E, Platz N, Hudson A, Hülskamp M, Larkin J, Schnittger A. Transcriptional profiling of mature Arabidopsis trichomes reveals that NOECK encodes the MIXTA-like transcriptional regulator MYB106. Plant Physiol. 2008;148:1583–602. doi: 10.1104/pp.108.126979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gilding EK, Marks MD. Analysis of purified glabra3-shapeshifter trichomes reveals a role for NOECK in regulating early trichome morphogenic events. Plant J. 2010;64:304–17. doi: 10.1111/j.1365-313X.2010.04329.x. [DOI] [PubMed] [Google Scholar]
- 8.Oshima Y, Shikata M, Koyama T, Ohtsubo N, Mitsuda N, Ohme-Takagi M. MIXTA-like transcription factors and WAX INDUCER1/SHINE1 coordinately regulate cuticle development in Arabidopsis and Torenia fournieri. Plant Cell. 2013;25:1609–24. doi: 10.1105/tpc.113.110783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sadowski I, Ma J, Triezenberg S, Ptashne M. GAL4-VP16 is an unusually potent transcriptional activator. Nature. 1988;335:563–4. doi: 10.1038/335563a0. [DOI] [PubMed] [Google Scholar]
- 10.Aharoni A, Dixit S, Jetter R, Thoenes E, van Arkel G, Pereira A. The SHINE clade of AP2 domain transcription factors activates wax biosynthesis, alters cuticle properties, and confers drought tolerance when overexpressed in Arabidopsis. Plant Cell. 2004;16:2463–80. doi: 10.1105/tpc.104.022897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Broun P, Poindexter P, Osborne E, Jiang CZ, Riechmann JL. WIN1, a transcriptional activator of epidermal wax accumulation in Arabidopsis. Proc Natl Acad Sci U S A. 2004;101:4706–11. doi: 10.1073/pnas.0305574101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kannangara R, Branigan C, Liu Y, Penfield T, Rao V, Mouille GH, Höfte H, Pauly M, Riechmann JL, Broun P. The transcription factor WIN1/SHN1 regulates Cutin biosynthesis in Arabidopsis thaliana. Plant Cell. 2007;19:1278–94. doi: 10.1105/tpc.106.047076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shi JX, Malitsky S, De Oliveira S, Branigan C, Franke RB, Schreiber L, Aharoni A. SHINE transcription factors act redundantly to pattern the archetypal surface of Arabidopsis flower organs. PLoS Genet. 2011;7:e1001388. doi: 10.1371/journal.pgen.1001388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Helliwell C, Waterhouse P. Constructs and methods for high-throughput gene silencing in plants. Methods. 2003;30:289–95. doi: 10.1016/S1046-2023(03)00036-7. [DOI] [PubMed] [Google Scholar]
- 15.Tanaka T, Tanaka H, Machida C, Watanabe M, Machida Y. A new method for rapid visualization of defects in leaf cuticle reveals five intrinsic patterns of surface defects in Arabidopsis. Plant J. 2004;37:139–46. doi: 10.1046/j.1365-313X.2003.01946.x. [DOI] [PubMed] [Google Scholar]
- 16.Wellesen K, Durst F, Pinot F, Benveniste I, Nettesheim K, Wisman E, Steiner-Lange S, Saedler H, Yephremov A. Functional analysis of the LACERATA gene of Arabidopsis provides evidence for different roles of fatty acid omega -hydroxylation in development. Proc Natl Acad Sci U S A. 2001;98:9694–9. doi: 10.1073/pnas.171285998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bernard A, Domergue F, Pascal S, Jetter R, Renne C, Faure JD, Haslam RP, Napier JA, Lessire R, Joubès J. Reconstitution of plant alkane biosynthesis in yeast demonstrates that Arabidopsis ECERIFERUM1 and ECERIFERUM3 are core components of a very-long-chain alkane synthesis complex. Plant Cell. 2012;24:3106–18. doi: 10.1105/tpc.112.099796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aarts MG, Keijzer CJ, Stiekema WJ, Pereira A. Molecular characterization of the CER1 gene of arabidopsis involved in epicuticular wax biosynthesis and pollen fertility. Plant Cell. 1995;7:2115–27. doi: 10.1105/tpc.7.12.2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bourdenx B, Bernard A, Domergue F, Pascal S, Léger A, Roby D, Pervent M, Vile D, Haslam RP, Napier JA, et al. Overexpression of Arabidopsis ECERIFERUM1 promotes wax very-long-chain alkane biosynthesis and influences plant response to biotic and abiotic stresses. Plant Physiol. 2011;156:29–45. doi: 10.1104/pp.111.172320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Abe M, Takahashi T, Komeda Y. Identification of a cis-regulatory element for L1 layer-specific gene expression, which is targeted by an L1-specific homeodomain protein. Plant J. 2001;26:487–94. doi: 10.1046/j.1365-313x.2001.01047.x. [DOI] [PubMed] [Google Scholar]
- 21.Nakamura M, Katsumata H, Abe M, Yabe N, Komeda Y, Yamamoto KT, Takahashi T. Characterization of the class IV homeodomain-Leucine Zipper gene family in Arabidopsis. Plant Physiol. 2006;141:1363–75. doi: 10.1104/pp.106.077388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu R, Li S, He S, Wassmann F, Yu C, Qin G, Schreiber L, Qu LJ, Gu H. CFL1, a WW domain protein, regulates cuticle development by modulating the function of HDG1, a class IV homeodomain transcription factor, in rice and Arabidopsis. Plant Cell. 2011;23:3392–411. doi: 10.1105/tpc.111.088625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schnurr J, Shockey J, Browse J. The acyl-CoA synthetase encoded by LACS2 is essential for normal cuticle development in Arabidopsis. Plant Cell. 2004;16:629–42. doi: 10.1105/tpc.017608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pighin JA, Zheng H, Balakshin LJ, Goodman IP, Western TL, Jetter R, Kunst L, Samuels AL. Plant cuticular lipid export requires an ABC transporter. Science. 2004;306:702–4. doi: 10.1126/science.1102331. [DOI] [PubMed] [Google Scholar]
- 25.Li Y, Beisson F, Koo AJ, Molina I, Pollard M, Ohlrogge J. Identification of acyltransferases required for cutin biosynthesis and production of cutin with suberin-like monomers. Proc Natl Acad Sci U S A. 2007;104:18339–44. doi: 10.1073/pnas.0706984104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lü S, Zhao H, Parsons EP, Xu C, Kosma DK, Xu X, Chao D, Lohrey G, Bangarusamy DK, Wang G, et al. The glossyhead1 allele of ACC1 reveals a principal role for multidomain acetyl-coenzyme A carboxylase in the biosynthesis of cuticular waxes by Arabidopsis. Plant Physiol. 2011;157:1079–92. doi: 10.1104/pp.111.185132. [DOI] [PMC free article] [PubMed] [Google Scholar]