Abstract
A spontaneous rice mutant, erect leaf1 (elf1–1), produced a dwarf phenotype with erect leaves and short grains. Physiological analyses suggested that elf1–1 is brassinosteroid-insensitive, so we hypothesized that ELF1 encodes a positive regulator of brassinosteroid signaling. ELF1, identified by means of positional cloning, encodes a protein with both a U-box domain and ARMADILLO (ARM) repeats. U-box proteins have been shown to function as E3 ubiquitin ligases; in fact, ELF1 possessed E3 ubiquitin ligase activity in vitro. However, ELF1 itself does not appear to be polyubiquitinated. Mutant phenotypes of 2 more elf1 alleles indicate that the entire ARM repeats is indispensable for ELF1 activity. These results suggest that ELF1 ubiquitinates target proteins through an interaction mediated by ARM repeats. Similarities in the phenotypes of elf1 and d61 mutants (mutants of brassinosteroid receptor gene OsBRI1), and in the regulation of ELF1 and OsBRI1 expression, imply that ELF1 functions as a positive regulator of brassinosteroid signaling in rice.
Keywords: brassinosteroid insensitive, brassinosteroid signaling, dwarf, mutant, rice, U-box ubiquitin E3 ligase
Introduction
Brassinosteroids are endogenous phytohormones that are involved in the regulation of various growth and developmental processes in higher plants.1-3 Brassinosteroids have been shown to enhance various aspects of crop production in rice (Oryza sativa L.), such as disease resistance and abiotic stress tolerance.4,5 Interestingly, overproduction and deficiency of brassinosteroids both increase rice grain yield, although by different mechanisms.6-8 These findings suggest the feasibility of genetic improvement of crop production by the modulation of brassinosteroid function.
Genetic and molecular studies have identified key components of the brassinosteroid signaling pathway, which include membrane receptor kinases (BRI1 and SERKs, including BAK1), intracellular kinases (BIN2 and BSKs) and a phosphatase (BSU1 and PP2A), and nuclear transcription factors (BES1 and BZR1).9,10 Subsequent biochemical studies have revealed many details about signaling events from brassinosteroid perception at the cell surface to gene expression in the nucleus.9,10 In rice, the putative brassinosteroid receptor gene OsBRI1 and its loss-of-function mutants (the d61 mutants) have been identified.11 Although the first 2 d61 mutants identified were weak alleles, 12 alleles with different severities of phenotype have been identified to date.12,13 Two other brassinosteroid-insensitive rice mutants, dlt and lic, have also been identified.14,15 DLT encodes a GRAS-family transcription factor and probably acts downstream of a putative rice BZR1 ortholog. LIC encodes a CCCH-type zinc finger protein and probably acts as an antagonistic transcription factor of a rice BZR1 ortholog. dlt, lic-1, and weak alleles of d61 all showed semi-dwarf, erect-leaf phenotypes, suggesting that such phenotypes are common in brassinosteroid-insensitive rice mutants.
In this study, we characterized a rice mutant, erect leaf1 (elf1–1), having dwarf and erect-leaf phenotypes. This mutant also had a short-grain phenotype similar to that observed for recently identified d61 and brassinosteroid-deficient d2 alleles.13,16 elf1–1 also showed various brassinosteroid-related phenotypes including increased levels of an endogenous bioactive brassinosteroid; therefore, we hypothesized that elf1 mutants have defects in novel brassinosteroid signaling components. The ELF1 gene encodes a U-box-containing E3 ubiquitin ligase, and was found to be identical to the recently identified Taihu Dwarf1 (TUD1) gene.17 Based on these experiments, we concluded that ELF1 functions in brassinosteroid signaling as an ubiquitin E3 ligase.
Results
elf1–1 showed brassinosteroid-insensitive phenotypes
As the result of a large-scale screening of rice mutant collections, we identified several lines that showed the morphological characteristics of brassinosteroid-related mutants, namely, dwarf plant stature and erect leaves. Seven of these mutants were obtained from a Nipponbare library of tissue culture-induced mutations.18 Three of the 7 mutants were alleles of d61,13 which is caused by mutations in the brassinosteroid receptor gene OsBRI1, and another 3 were alleles of d2,16 which is caused by mutations in the brassinosteroid biosynthetic enzyme gene CYP90D2/D2. The remaining mutant, elf1–1, was not an allele of the previously identified brassinosteroid-related mutants, so we further characterized this mutant.
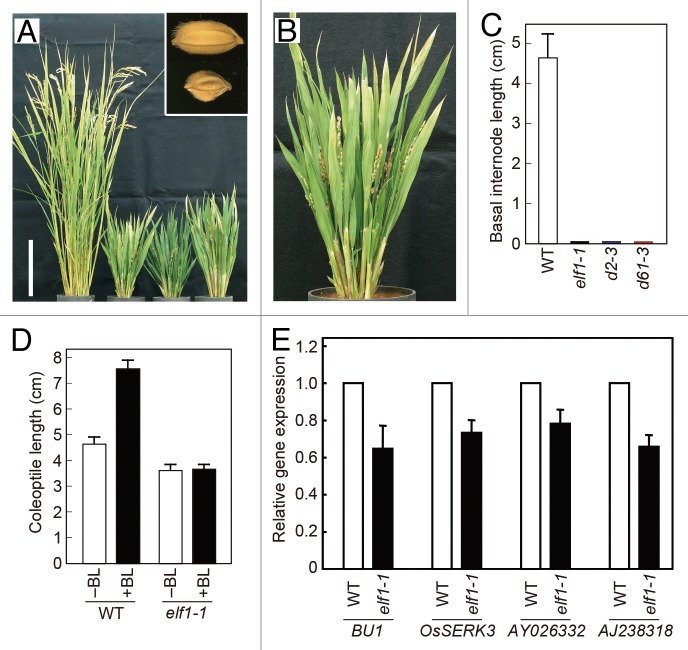
Like the Nipponbare-derived brassinosteroid-related d61 and d2 mutants,13,16 elf1–1 showed a reduction in plant height (to 34% of the wild-type height, n = 10, P < 0.001) as shown in Figure 1A. In wild-type rice, the leaf blade bends away from the vertical axis of the leaf sheath toward the abaxial side, whereas the leaves of elf1–1 were completely erect (Fig. 1A and B). The grains of elf1–1 were visibly shorter and smaller than those of their original strain, Nipponbare (Fig. 1A, inset), similar to the grains of the Nipponbare-derived d61 and d2 mutants.13,16
Figure 1.elf1 mutants showed brassinosteroid-insensitive phenotypes. (A) Comparison of gross morphology between the wild-type (WT) and 3 elf1 mutants. Left to right: Nipponbare (wild-type), elf1–1, elf1–2, and elf1–3. Bar, 20 cm. Inset represents grain morphology of the wild-type (upper) and elf1–1 (lower). (B) Close-up view of elf1–1. (C) Length of the second lower internode of the wild-type, elf1–1, and brassinosteroid-related mutants grown in the dark. Data presented are the means ± SD of 5 plants. (D) Effect of BL treatment on the coleoptile elongation of the wild-type and elf1–1. Data presented are the means ± SD of 5 plants. (E) Quantitative RT–PCR analysis of brassinosteroid-response genes (BU1, OsSERK3, AY026332, and AJ238318). Expression levels were normalized against the values obtained for Histone H3. The value obtained from the wild-type was then normalized to a value of 1.0. Data presented are the means ± SD of 3 biological repeats.
The rice brassinosteroid-insensitive and -deficient mutants show a photomorphogenic phenotype when grown in the dark: the mesocotyl and internodes do not elongate.11,19,20 Thus, this unique phenotype is a good criterion with which to determine whether a novel dwarf mutant is related to brassinosteroids. We grew elf1–1, wild-type, brassinosteroid-deficient (d2–3), and brassinosteroid-insensitive (d61–3) plants in the dark for 2 wk. The basal internodes of the wild-type plants elongated under dark conditions, but elongation did not occur in elf1–1, d2–3, and d61–3 (Fig. 1C). The failure of internode elongation in the dark strongly supported the notion that elf1–1 is a brassinosteroid-related mutant. Next, we measured the effects of brassinolide (BL, a bioactive brassinosteroid) on coleoptile length (Fig. 1D). The coleoptile length of the mutants was not affected by 100 nM BL, whereas the coleoptile length of wild-type plants increased after treatment with 100 nM BL. These results suggested that elf1–1 plants are less sensitive to exogenous BL than are wild-type plants.
To confirm whether the response to brassinosteroids is suppressed in elf1–1 plants at the level of gene regulation, we monitored the expression of brassinosteroid-response genes: BU1, OsSERK3, AY026332, and AJ238318.21-23 The level of BU1 mRNA in elf1–1 seedlings was 65% of that in wild-type seedlings (Fig. 1E). Similarly, the expression levels of OsSERK3, AY026332, and AJ238318 in the elf1–1 seedlings were 66–78% of those in the wild-type seedlings. We previously reported that the steady-state level of BU1 mRNA in Nipponbare-derived mutant of brassinosteroid receptor gene OsBRI1 (d61–1N) was about 68% of that in Nipponbare.13 Among 12 alleles of d61 mutants, d61–1N showed severely dwarfed stature with erect leaves and short grains as elf1–1 (plant height of d61–1N was reduced to 31% of the wild-type height). Similarities in the gross morphology and the decreased level of BU1 expression between elf1–1 and d61–1N suggest that elf1–1 is also a brassinosteroid-insensitive mutant.
Accumulation of bioactive brassinosteroid in elf1–1
We also monitored the expression of 6 brassinosteroid-related genes: OsDWF4, D11, D2, OsCPD1, OsDWF, and OsBRI1. OsDWF4 and D11 encode C-22 hydroxylases (CYP90B2 and CYP724B1, respectively),6 D2 encodes C-23 hydroxylase (CYP90D2),24 OsCPD1 encodes a rice ortholog of C-3 oxidase (CYP90A3),25,26 OsDWF encodes C-6 oxidase (CYP85A1),19 and OsBRI1 encodes the brassinosteroid receptor.11 The expression of these genes is regulated by a homeostatic system that controls bioactive brassinosteroid levels; i.e., expression is increased in brassinosteroid-related mutants and decreased by BL treatment in wild-type.6,11,19,20,25 The level of OsBRI1 mRNA in elf1–1 seedlings was 1.7 times that in wild-type seedlings (Fig. 2A). Similarly, the expression levels of brassinosteroid biosynthetic enzyme genes (except for OsCPD1) in the elf1–1 seedlings were 1.5–1.8 times those in the wild-type seedlings. The smaller increase in OsCPD1 expression in the elf1–1 seedlings (1.2 times that in the wild-type) may be caused by redundancy between the OsCPD1 and OsCPD2 genes.25 The level of CYP85A1 mRNA in elf1–1 seedlings was 1.8 times that in wild-type seedlings (Fig. 2A). In d61–1N, the CYP85A1 expression was increased to 1.3 times that in the wild-type.13 These results indicate that the increased expression of brassinosteroid biosynthetic enzyme genes in elf1–1 was owing to feed-forward upregulation by the homeostatic system. Consistent with these findings, the endogenous content of castasterone, a bioactive brassinosteroid in rice, doubled in the elf1–1 seedlings relative to wild-type (Fig. 2B). The accumulation of bioactive brassinosteroid, accompanied by morphological alterations of the types caused by defects in brassinosteroid function, strongly supports our conclusion that elf1–1 is a brassinosteroid-insensitive mutant and that ELF1 encodes a positive regulator of brassinosteroid signaling in rice.
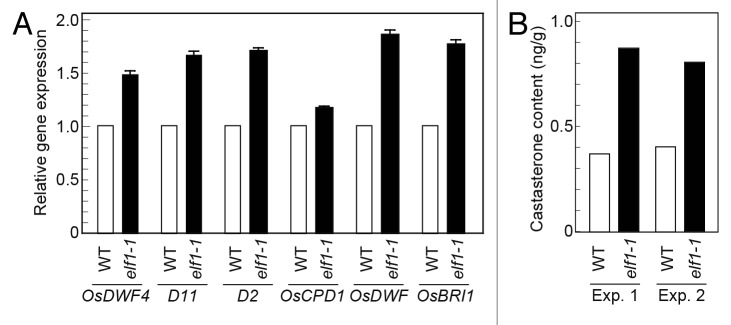
Figure 2. Brassinosteroid biosynthesis was increased in elf1–1 relative to wild-type (WT). (A) Quantitative RT–PCR analysis of brassinosteroid biosynthetic enzyme genes (OsDWF4, D11, D2, OsCPD1, and OsDWF) and receptor gene OsBRI1. Expression levels were normalized against the values obtained for Histone H3. The value obtained from the wild-type was then normalized to a value of 1.0. Data presented are the means ± SD of 3 biological repeats. (B) Endogenous content of castasterone in the wild-type and elf1–1. Two independent experiments were performed.
The ELF1 gene encodes a U-box–containing ubiquitin E3 ligase
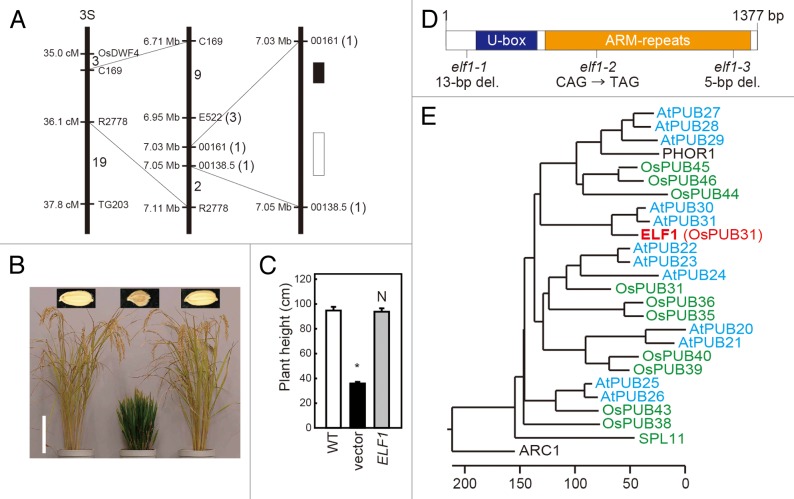
The ELF1 locus was cloned using a map-based chromosome walking procedure and mapped to a ~20-kb region on the short arm of chromosome 3 (Fig. 3A). This region includes 2 predicted open reading frames (locus IDs Os03 g0232600 and Os03 g0232800). By comparing the nucleotide sequences of elf1–1 and the wild-type at each of these loci, we concluded that ELF1 corresponds to Os03 g0232600, which encodes a protein of 459 amino acid residues in a single exon. There was a 13-bp deletion in the Os03 g0232600 locus of elf1–1 relative to wild-type, whereas no difference between elf1–1 and wild-type was detected at the Os03 g0232800 locus. The dwarf and short grain phenotypes of elf1–1 were complemented by the introduction of a 5.3-kb genomic DNA fragment (from positions –3169 to +2167, taking the translation initiation site as +1) containing the entire candidate gene at the Os03 g0232600 locus (Fig. 3B and C) confirmed that the elf1–1 phenotype is caused by a loss-of-function mutation in this predicted ELF1 gene.
Figure 3. Molecular characterization of ELF1. (A) High-resolution linkage and physical map of the ELF1 locus. The horizontal bars represent molecular markers, and the numbers of recombinant plants are indicated on the linkage map. Black and white boxes indicate Os03 g0232600 and Os03 g0232800, respectively. (B) Comparison of gross and seed morphologies between wild-type (left) and elf1–1 mutant transgenic plants containing the empty vector (middle) and a genomic DNA fragment encompassing the entire ELF1 gene (right). Bar, 20 cm. (C) Plant height of wild-type (WT) and elf1–1 mutant transgenic plants containing the empty vector (vector) and a genomic DNA fragment encompassing the entire ELF1 gene (ELF1). Each bar represents the mean ± s.d. of 9 different plants (wild-type) and 3 different plants for each of the 3 independent elf1–1 mutant transgenic lines. Bars labeled with asterisks differ significantly (P < 0.001, Dunnett post-hoc test) from wild-type plants. N, not significant. (D) Molecular structure of the ELF1 and positions of the mutations in the 3 elf1 alleles discovered in the present study. (E) Phylogenetic relationships of PUB proteins. OsPUBs, AtPUBs, and PHOR1 are class III PUB proteins from rice, Arabidopsis, and potato, respectively. SPL11 and ARC1 are class II PUB proteins from rice and rape, respectively.
Sequence comparison suggested that ELF1 contains a typical U-box domain followed by six ARMADILLO (ARM) repeats (Fig. 3D). This domain organization is frequently found in the plant U-box (PUB) protein family, whose members function as E3 ubiquitin ligases.27 The 13-bp deletion in elf1–1 produced a frameshift at Asp-58, resulting in a truncated protein of 77 amino acid residues (Fig. 3D). Therefore, elf1–1 was considered to be a null allele. Based on sequence analysis of ELF1, 2 allelic mutants (elf1–2 and elf1–3) were further identified from our mutant collection (Figs. 1A and 3D). elf1–2 had a single nucleotide substitution (CAG to TAG) that resulted in a substitution of Gln-166 with a stop codon. elf1–3 had a 5-bp deletion that produced a frameshift at Thr-440, resulting in a truncated protein of 458 amino acid residues. The 3 mutant alleles had similar phenotypes, including severely dwarfed stature with erect leaves and short grains, indicates that mutations of ELF1 in elf1–2 and elf1–3 also caused loss of ELF1 protein function in these alleles.
A BLAST search using the whole amino acid sequence of ELF1 revealed the highest sequence similarity with Arabidopsis proteins AtPUB30 and AtPUB31 (63% and 59% identities, respectively). Both AtPUB30 and AtPUB31 were grouped with the class III PUB proteins.27 Phylogenetic analysis of class III PUB proteins from rice, Arabidopsis, and potato revealed that class III PUB proteins fall into several clades containing rice and Arabidopsis proteins, and ELF1 was distantly related to the other rice class III PUB proteins (Fig. 3E).
ELF1 encodes a functional E3 ubiquitin ligase
Because one of the important features of U-box–containing proteins is to function as E3 ubiquitin ligases, and the U-box is essential for the E3 ubiquitin ligase activity of such proteins,28 we tested whether ELF1 also possessed the characteristics of E3 ubiquitin ligase. First, we examined the interactions between ELF1 and the other components of the ubiquitin protein degradation pathway. To test if an intact U-box domain in ELF1 is required for these interactions, we produced intact ELF1 and U-box domain-deleted ELF1 (deleted from Val-65 to Phe-132; ΔU-box) proteins for yeast 2-hybrid assays (Fig. 4A). The intact ELF1 protein interacted with the ubiquitin-conjugating enzyme OsUBC5b (E2 in Figure 4B) and polyubiquitin (Poly-Ub in Figure 4B); however, deletion of the U-box domain in ΔU-box protein greatly reduced these interactions (Fig. 4B).
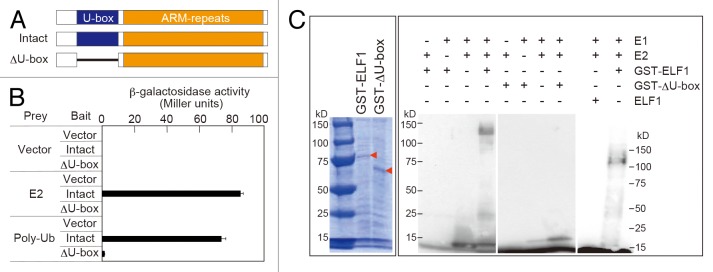
Figure 4. ELF1 functions as an E3 ubiquitin ligase. (A) Structure of intact ELF1 and ΔU-box proteins. (B) U-box domain is necessary for ELF1 to bind ubiquitin-conjugating enzyme (E2) and polyubiquitin (Poly-Ub). (C) Coomassie brilliant blue (CBB)–stained recombinant GST-ELF1 and GST-ΔU-box proteins used for E3 ubiquitin ligase assay (left), and E3 ubiquitin ligase activity of GST-ELF1, GST-ΔU-box, and GST-excised ELF1 proteins (right). The results indicate that GST-ELF1 has U-box-dependent E3 ubiquitin ligase activity in vitro, as seen by the appearance of a high-molecular-weight band corresponding to the ubiquitinated protein.
Next, we wanted to determine whether ELF1 also possesses E3 ubiquitin ligase activity. We expressed in Escherichia coli and affinity-purified the intact ELF1 protein as a glutathione S-transferase (GST) fusion. In the presence of yeast E1 and human E2 Hubc5b, ubiquitination activity was observed for the purified GST-ELF1 fusion protein (Fig. 4C). No clear protein ubiquitination was detected in the absence of E1, E2, or ELF1 (Fig. 4C). In vitro ubiquitination analysis also indicated that the E3 ubiquitin ligase activity was completely abolished in the purified GST-ΔU-box fusion protein (Fig. 4C), suggesting that an intact U-box domain is required for ELF1’s E3 ubiquitin ligase activity. It was noteworthy that GST-tag-excised ELF1 (ELF1 in Figure 4C) did not show auto-ubiquitination, indicating that ELF1 in the GST-ELF1 fusion protein ubiquitinated an amino acid somewhere in the GST-tag region. These results indicate that ELF1 possesses E3 ubiquitin ligase activity, but ELF1 itself does not appear to be polyubiquitinated.
Expression of ELF1 in wild-type and mutant rice
Quantitative reverse-transcription PCR (qRT-PCR) analysis revealed that ELF1 was preferentially expressed in panicles at flowering time, at moderate levels in vegetative shoot apices, leaf sheaths, leaf blades, and elongating internodes, and at the lowest level in roots (Fig. 5A). Previous observations indicated that the expression of brassinosteroid receptor gene OsBRI1 was downregulated by brassinosteroid application and upregulated in brassinosteroid-insensitive and -deficient mutants.14,29 Thus, we examined whether such feedback regulation also occurs with ELF1. qRT-PCR analysis revealed that the expression of ELF1 was slightly decreased by BL treatment (Fig. 5B), whereas it more than doubled in both brassinosteroid-deficient brd1–2 (a loss-of-function mutant of brassinosteroid biosynthetic enzyme gene CYP85A1/OsDWARF)19 and brassinosteroid-insensitive d61–3 (a loss-of-function mutant of OsBRI1),12 as shown in Figure 5C. Interestingly, ELF1 expression also increased in dark-grown wild-type rice (Fig. 5C), as previously reported for OsBRI1.29
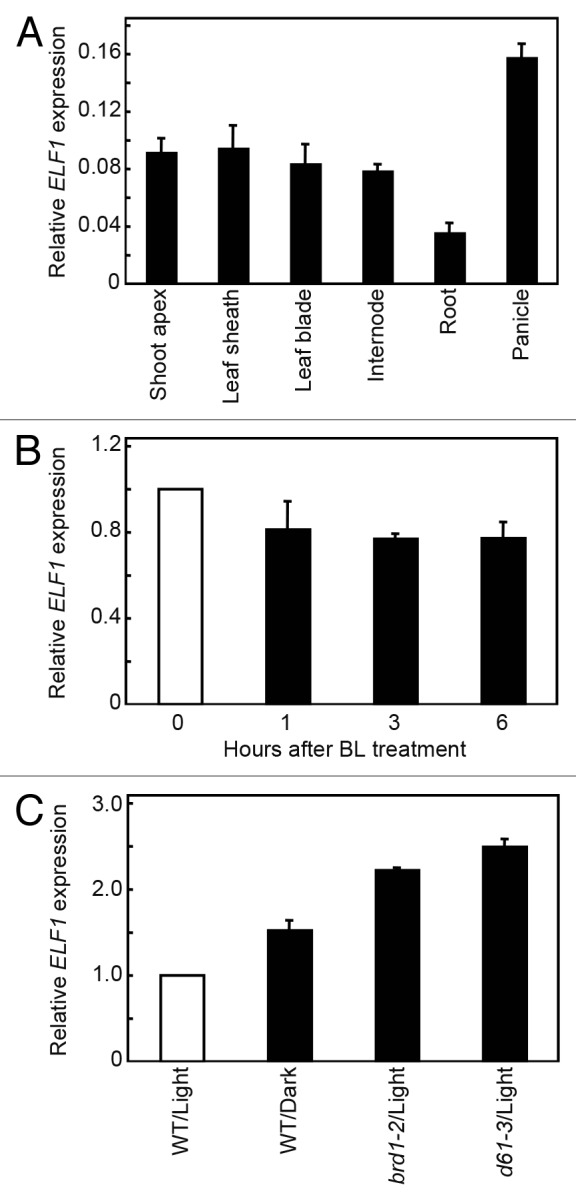
Figure 5. Expression of ELF1 in wild-type and mutant rice. (A) Relative mRNA levels of ELF1 in various organs of wild-type rice. (B) Changes in the expression level of ELF1 in wild-type rice after BL treatment. (C) Wild-type seedlings (WT) were grown under continuous light (Light) or complete darkness (Dark). brd1–2 and d61–3 were grown under continuous light. Expression levels were normalized against the values obtained for Histone H3. In (B) and (C), the value obtained from the wild-type just before the treatment (B) or grown under continuous light (C) was then normalized to a value of 1.0. Data presented are the means ± SD of 3 biological repeats.
Discussion
The gross morphology of the elf1 mutants was very similar to that of the recently identified brassinosteroid-related d61 and d2 mutants,13,16 including reduced plant height (about one-third of the wild-type), erect leaves, and short grains. Consistent with these phenotypes, elf1–1 showed defects in brassinosteroid-related responses such as skotomorphogenesis (etiolation) and coleoptile elongation. Furthermore, upregulation of expression of major brassinosteroid biosynthetic enzyme genes and accumulation of bioactive castasterone were observed in elf1–1 in spite of its brassinosteroid-deficient phenotypes. Thus, we can conclude that elf1 mutants are brassinosteroid-insensitive and that ELF1 encodes a positive regulator of brassinosteroid signaling.
ELF1 encodes a member of the PUB family that contains a typical U-box domain followed by 6 ARM repeats. The U-box is a conserved motif of ~70 amino acids, with characteristics suggesting that it is a structural variant of the RING fold but lacking the signature zinc-binding amino acids.30,31 U-box-containing proteins function as E3 ubiquitin ligases, which indirectly mediate ubiquitin ligation by simultaneously docking both an ubiquitin-loaded E2 enzyme and a targeted substrate.28,32 The ARM repeats are leucine-rich motifs of 42 amino acids that were originally identified in the Drosophila armadillo protein β-catenin, which is involved in a variety of protein–protein interactions.33,34 Because most parts of the ELF1 protein including the U-box domain and ARM repeats are not produced in elf1–1 plants, elf1–1 is considered to be a null allele. The other 2 alleles identified from our mutant collection also showed phenotypes similar to those of elf1–1, suggesting that the entire 6-ARM repeat region is indispensable for ELF1 activity.
There are 64 and 77 PUB proteins in Arabidopsis and rice, respectively, forming several subclasses that presumably play diverse roles.35,36 Two orthologs of ELF1 (AtPUB30 and AtPUB31) are encoded in the Arabidopsis genome, whereas ELF1 is encoded by a single gene in rice. Although the function of 2 Arabidopsis orthologs has not yet been reported, ELF1 is identical to recently reported TUD1.17 The severe alleles of tud1 showed phenotypes similar to those of elf1–1, and the authors demonstrated that recombinant TUD1 protein can interact with the heterotrimeric G protein α subunit known as D1. D1 was originally identified as a signaling component that affects a part of the gibberellin signaling pathway, namely, the induction of α-amylase in the aleurone layer and internode elongation,37-39 although several lines of evidence indicate that D1 also participates in brassinosteroid responses.40,41 Hu et al. (2013)17 suggested that TUD1 directly interacts with and acts downstream of D1 to mediate a brassinosteroid signaling pathway.
In the present study, ELF1 expression was downregulated by the application of brassinosteroid and upregulated in brassinosteroid-insensitive and -deficient mutants. This expression pattern can be explained by the homeostatic system that controls bioactive brassinosteroid levels, and a similar expression pattern was also observed for the brassinosteroid receptor gene OsBRI1.14,29 Similarity in the mutant phenotypes of elf1 and d61 (mutants of OsBRI1), and in the regulation of ELF1 and OsBRI1 expression imply that both ELF1 and OsBRI1 function as positive regulators of brassinosteroid signaling in rice. In addition, ELF1 expression was increased in dark-grown wild-type rice, as previously reported for OsBRI1.29 Genetic evidence has uncovered a role for brassinosteroids in the control of skotomorphogenesis, the process by which the hypocotyls of etiolated seedlings rapidly grow in the dark, enabling them to reach the soil surface. In previous research, brassinosteroid-deficient or -insensitive mutants did not show an elongated phenotype and developed similarly to plants grown in the light.42,43 The defective internode elongation of dark-grown elf1–1 and increased expression of ELF1 in the dark suggest that ELF1, like OsBRI1, functions in skotomorphogenesis. We have demonstrated that ELF1 possesses E3 ubiquitin ligase activity, which requires the U-box domain, but ELF1 itself does not appear to be polyubiquitinated. The importance of the entire 6-ARM repeat region of ELF1 is evident in this study because of the locations of the elf1–2 and -3 mutations. Thus, we consider that ELF1 ubiquitinates target proteins through the interaction mediated by its ARM repeats. We plan to identify target proteins of ELF1 in order to elucidate the functions and mechanisms of ELF1-mediated regulation of brassinosteroid signaling in rice.
Materials and Methods
Plant materials, phytohormone treatments, and gene expression analysis
Seeds of wild-type rice (Oryza sativa L. cv Nipponbare) and mutants were sown on Murashige and Skoog agar medium and grown in a growth chamber at 28 °C under continuous light or complete darkness for 2 wk. Treatment with 100 nM BL was performed as previously described.44 Total RNA extraction, single-strand cDNAs synthesis, and qRT-PCR were performed as previously described.44 Expression levels were normalized against the values obtained for Histone H3, which was used as the internal reference gene. Primer sequences were listed in a previous report.44
Analysis of endogenous brassinosteroid levels
Shoots from wild-type and elf1–1 mutant plants were harvested 4 wk after germination. Purification and quantification of brassinosteroids were performed according to the method described previously.16
Molecular cloning, sequence alignment, phylogenetic tree construction, and complementation test
To map ELF1, linkage analysis was performed using an F2 population of ~4000 plants derived from a cross between elf1–1 (japonica variety) and Kasalath (indica variety). The nucleotide sequence of the ELF1 gene (from positions –3648 to +2353, taking the translation initiation site as +1) from Nipponbare and 3 mutant lines was determined by using an ABI377 sequencer (Applied Biosystems) according to the manufacturer’s instructions, and analyzed by using the Lasergene software program (DNAStar). A BLAST search was performed as previously reported.45 Phylogenetic analysis was performed by using the Lasergene software according to the manufacturer’s instructions. For complementation of the elf1–1 mutation, we amplified the wild-type genomic sequence of ELF1 from –3169 to +2167 (taking the translation initiation site as +1) by PCR and confirmed the sequence of the amplified product. Binary vector construction and rice transformation were performed as previously described.44
Yeast 2-hybrid assay
Full-length ELF1 cDNA and U-box-deleted ELF1 cDNA were inserted into the pGBKT7 vector (Clontech); OsUBC5b (Os04 g0667800) and polyubiquitin (Os06 g0681400) were inserted into the pGADT7 vector (Clontech). β-galactosidase liquid assays were performed according to the manufacturer’s protocol.
Ubiquitin E3 ligase assay
Full-length ELF1 cDNA and U-box-deleted ELF1 cDNA were inserted into the pGEX-6P expression vector (GE Healthcare). Recombinant proteins were purified by GST Purification Modules (GE Healthcare). The in vitro ubiquitinylation assay was performed by using a Ubiquitinylation Kit (Biomol).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
Sakamoto T was supported by Grants-in-Aid for Young Scientists (nos. 19688001 and 24780005) from the Ministry of Education, Culture, Sports, Science, and Technology (MEXT) of Japan. Fujioka S was supported by Grants-in-Aid for Scientific Research (B) (nos. 19380069 and 23380066) from MEXT.
Glossary
Abbreviations:
- ARM
ARMADILLO
- BL
brassinolide
- ELF1
ERECT LEAF1
- PUB
plant U-box
References
- 1.Clouse SD, Sasse JM. Brassinosteroids: essential regulators of plant growth and development. Annu Rev Plant Physiol Plant Mol Biol. 1998;49:427–51. doi: 10.1146/annurev.arplant.49.1.427. [DOI] [PubMed] [Google Scholar]
- 2.Krishna P. Brassinosteroid-mediated stress responses. J Plant Growth Regul. 2003;22:289–97. doi: 10.1007/s00344-003-0058-z. [DOI] [PubMed] [Google Scholar]
- 3.Sasse JM. Physiological action of brassinosteroids: an update. J Plant Growth Regul. 2003;22:276–88. doi: 10.1007/s00344-003-0062-3. [DOI] [PubMed] [Google Scholar]
- 4.Nakashita H, Yasuda M, Nitta T, Asami T, Fujioka S, Arai Y, Sekimata K, Takatsuto S, Yamaguchi I, Yoshida S. Brassinosteroid functions in a broad range of disease resistance in tobacco and rice. Plant J. 2003;33:887–98. doi: 10.1046/j.1365-313X.2003.01675.x. [DOI] [PubMed] [Google Scholar]
- 5.Koh S, Lee SC, Kim MK, Koh JH, Lee S, An G, Choe S, Kim SR. T-DNA tagged knockout mutation of rice OsGSK1, an orthologue of Arabidopsis BIN2, with enhanced tolerance to various abiotic stresses. Plant Mol Biol. 2007;65:453–66. doi: 10.1007/s11103-007-9213-4. [DOI] [PubMed] [Google Scholar]
- 6.Sakamoto T, Morinaka Y, Ohnishi T, Sunohara H, Fujioka S, Ueguchi-Tanaka M, Mizutani M, Sakata K, Takatsuto S, Yoshida S, et al. Erect leaves caused by brassinosteroid deficiency increase biomass production and grain yield in rice. Nat Biotechnol. 2006;24:105–9. doi: 10.1038/nbt1173. [DOI] [PubMed] [Google Scholar]
- 7.Morinaka Y, Sakamoto T, Inukai Y, Agetsuma M, Kitano H, Ashikari M, Matsuoka M. Morphological alteration caused by brassinosteroid insensitivity increases the biomass and grain production of rice. Plant Physiol. 2006;141:924–31. doi: 10.1104/pp.106.077081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu CY, Trieu A, Radhakrishnan P, Kwok SF, Harris S, Zhang K, Wang J, Wan J, Zhai H, Takatsuto S, et al. Brassinosteroids regulate grain filling in rice. Plant Cell. 2008;20:2130–45. doi: 10.1105/tpc.107.055087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim TW, Wang ZY. Brassinosteroid signal transduction from receptor kinases to transcription factors. Annu Rev Plant Biol. 2010;61:681–704. doi: 10.1146/annurev.arplant.043008.092057. [DOI] [PubMed] [Google Scholar]
- 10.Clouse SD. Brassinosteroid signal transduction: from receptor kinase activation to transcriptional networks regulating plant development. Plant Cell. 2011;23:1219–30. doi: 10.1105/tpc.111.084475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yamamuro C, Ihara Y, Wu X, Noguchi T, Fujioka S, Takatsuto S, Ashikari M, Kitano H, Matsuoka M. Loss of function of a rice brassinosteroid insensitive1 homolog prevents internode elongation and bending of the lamina joint. Plant Cell. 2000;12:1591–606. doi: 10.1105/tpc.12.9.1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nakamura A, Fujioka S, Sunohara H, Kamiya N, Hong Z, Inukai Y, Miura K, Takatsuto S, Yoshida S, Ueguchi-Tanaka M, et al. The role of OsBRI1 and its homologous genes, OsBRL1 and OsBRL3, in rice. Plant Physiol. 2006;140:580–90. doi: 10.1104/pp.105.072330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sakamoto T, Kitano H, Fujioka S. Genetic background influences brassinosteroid-related mutant phenotypes in rice. Amer J Plant Sci. 2013;4:212–21. doi: 10.4236/ajps.2013.42028. [DOI] [Google Scholar]
- 14.Tong H, Jin Y, Liu W, Li F, Fang J, Yin Y, Qian Q, Zhu L, Chu C. DWARF AND LOW-TILLERING, a new member of the GRAS family, plays positive roles in brassinosteroid signaling in rice. Plant J. 2009;58:803–16. doi: 10.1111/j.1365-313X.2009.03825.x. [DOI] [PubMed] [Google Scholar]
- 15.Zhang C, Xu Y, Guo S, Zhu J, Huan Q, Liu H, Wang L, Luo G, Wang X, Chong K. Dynamics of brassinosteroid response modulated by negative regulator LIC in rice. PLoS Genet. 2012;8:e1002686. doi: 10.1371/journal.pgen.1002686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sakamoto T, Morinaka Y, Kitano H, Fujioka S. New alleles of rice ebisu dwarf (d2) mutant show both brassinosteroid-deficient and -insensitive phenotypes. Amer J Plant Sci. 2012;3:1699–707. doi: 10.4236/ajps.2012.312208. [DOI] [Google Scholar]
- 17.Hu X, Qian Q, Xu T, Zhang Y, Dong G, Gao T, Xie Q, Xue Y. The U-box E3 ubiquitin ligase TUD1 functions with a heterotrimeric G α subunit to regulate Brassinosteroid-mediated growth in rice. PLoS Genet. 2013;9:e1003391. doi: 10.1371/journal.pgen.1003391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hirochika H, Sugimoto K, Otsuki Y, Tsugawa H, Kanda M. Retrotransposons of rice involved in mutations induced by tissue culture. Proc Natl Acad Sci U S A. 1996;93:7783–8. doi: 10.1073/pnas.93.15.7783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hong Z, Ueguchi-Tanaka M, Shimizu-Sato S, Inukai Y, Fujioka S, Shimada Y, Takatsuto S, Agetsuma M, Yoshida S, Watanabe Y, et al. Loss-of-function of a rice brassinosteroid biosynthetic enzyme, C-6 oxidase, prevents the organized arrangement and polar elongation of cells in the leaves and stem. Plant J. 2002;32:495–508. doi: 10.1046/j.1365-313X.2002.01438.x. [DOI] [PubMed] [Google Scholar]
- 20.Hong Z, Ueguchi-Tanaka M, Umemura K, Uozu S, Fujioka S, Takatsuto S, Yoshida S, Ashikari M, Kitano H, Matsuoka M. A rice brassinosteroid-deficient mutant, ebisu dwarf (d2), is caused by a loss of function of a new member of cytochrome P450. Plant Cell. 2003;15:2900–10. doi: 10.1105/tpc.014712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tanaka A, Nakagawa H, Tomita C, Shimatani Z, Ohtake M, Nomura T, Jiang CJ, Dubouzet JG, Kikuchi S, Sekimoto H, et al. BRASSINOSTEROID UPREGULATED1, encoding a helix-loop-helix protein, is a novel gene involved in brassinosteroid signaling and controls bending of the lamina joint in rice. Plant Physiol. 2009;151:669–80. doi: 10.1104/pp.109.140806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Singla B, Khurana JP, Khurana P. Structural characterization and expression analysis of the SERK/SERL gene family in rice (Oryza sativa) Int J Plant Genomics. 2009;2009:539402. doi: 10.1155/2009/539402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang L, Xu YY, Li J, Powell RA, Xu ZH, Chong K. Transgenic rice plants ectopically expressing AtBAK1 are semi-dwarfed and hypersensitive to 24-epibrassinolide. J Plant Physiol. 2007;164:655–64. doi: 10.1016/j.jplph.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 24.Sakamoto T, Ohnishi T, Fujioka S, Watanabe B, Mizutani M. Rice CYP90D2 and CYP90D3 catalyze C-23 hydroxylation of brassinosteroids in vitro. Plant Physiol Biochem. 2012;58:220–6. doi: 10.1016/j.plaphy.2012.07.011. [DOI] [PubMed] [Google Scholar]
- 25.Sakamoto T, Matsuoka M. Characterization of CONSTITUTIVE PHOTOMORPHOGENESIS AND DWARFISM homologs in rice (Oryza sativa L.) J Plant Growth Regul. 2006;25:245–51. doi: 10.1007/s00344-006-0041-6. [DOI] [Google Scholar]
- 26.Ohnishi T, Godza B, Watanabe B, Fujioka S, Hategan L, Ide K, Shibata K, Yokota T, Szekeres M, Mizutani M. CYP90A1/CPD, a brassinosteroid biosynthetic cytochrome P450 of Arabidopsis, catalyzes C-3 oxidation. J Biol Chem. 2012;287:31551–60. doi: 10.1074/jbc.M112.392720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Azevedo C, Santos-Rosa MJ, Shirasu K. The U-box protein family in plants. Trends Plant Sci. 2001;6:354–8. doi: 10.1016/S1360-1385(01)01960-4. [DOI] [PubMed] [Google Scholar]
- 28.Hatakeyama S, Yada M, Matsumoto M, Ishida N, Nakayama KI. U box proteins as a new family of ubiquitin-protein ligases. J Biol Chem. 2001;276:33111–20. doi: 10.1074/jbc.M102755200. [DOI] [PubMed] [Google Scholar]
- 29.Sakamoto T, Kawabe A, Tokida-Segawa A, Shimizu B, Takatsuto S, Shimada Y, Fujioka S, Mizutani M. Rice CYP734As function as multisubstrate and multifunctional enzymes in brassinosteroid catabolism. Plant J. 2011;67:1–12. doi: 10.1111/j.1365-313X.2011.04567.x. [DOI] [PubMed] [Google Scholar]
- 30.Aravind L, Koonin EV. The U box is a modified RING finger - a common domain in ubiquitination. Curr Biol. 2000;10:R132–4. doi: 10.1016/S0960-9822(00)00398-5. [DOI] [PubMed] [Google Scholar]
- 31.Ohi MD, Vander Kooi CW, Rosenberg JA, Chazin WJ, Gould KL. Structural insights into the U-box, a domain associated with multi-ubiquitination. Nat Struct Biol. 2003;10:250–5. doi: 10.1038/nsb906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jackson PK, Eldridge AG, Freed E, Furstenthal L, Hsu JY, Kaiser BK, Reimann JD. The lore of the RINGs: substrate recognition and catalysis by ubiquitin ligases. Trends Cell Biol. 2000;10:429–39. doi: 10.1016/S0962-8924(00)01834-1. [DOI] [PubMed] [Google Scholar]
- 33.Samuel MA, Salt JN, Shiu SH, Goring DR. Multifunctional arm repeat domains in plants. Int Rev Cytol. 2006;253:1–26. doi: 10.1016/S0074-7696(06)53001-3. [DOI] [PubMed] [Google Scholar]
- 34.Huber AH, Nelson WJ, Weis WI. Three-dimensional structure of the armadillo repeat region of β-catenin. Cell. 1997;90:871–82. doi: 10.1016/S0092-8674(00)80352-9. [DOI] [PubMed] [Google Scholar]
- 35.Wiborg J, O’Shea C, Skriver K. Biochemical function of typical and variant Arabidopsis thaliana U-box E3 ubiquitin-protein ligases. Biochem J. 2008;413:447–57. doi: 10.1042/BJ20071568. [DOI] [PubMed] [Google Scholar]
- 36.Zeng LR, Park CH, Venu RC, Gough J, Wang GL. Classification, expression pattern, and E3 ligase activity assay of rice U-box-containing proteins. Mol Plant. 2008;1:800–15. doi: 10.1093/mp/ssn044. [DOI] [PubMed] [Google Scholar]
- 37.Fujisawa Y, Kato T, Ohki S, Ishikawa A, Kitano H, Sasaki T, Asahi T, Iwasaki Y. Suppression of the heterotrimeric G protein causes abnormal morphology, including dwarfism, in rice. Proc Natl Acad Sci U S A. 1999;96:7575–80. doi: 10.1073/pnas.96.13.7575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ashikari M, Wu J, Yano M, Sasaki T, Yoshimura A. Rice gibberellin-insensitive dwarf mutant gene Dwarf 1 encodes the α-subunit of GTP-binding protein. Proc Natl Acad Sci U S A. 1999;96:10284–9. doi: 10.1073/pnas.96.18.10284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ueguchi-Tanaka M, Fujisawa Y, Kobayashi M, Ashikari M, Iwasaki Y, Kitano H, Matsuoka M. Rice dwarf mutant d1, which is defective in the α subunit of the heterotrimeric G protein, affects gibberellin signal transduction. Proc Natl Acad Sci U S A. 2000;97:11638–43. doi: 10.1073/pnas.97.21.11638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang L, Xu YY, Ma QB, Li D, Xu ZH, Chong K. Heterotrimeric G protein α subunit is involved in rice brassinosteroid response. Cell Res. 2006;16:916–22. doi: 10.1038/sj.cr.7310111. [DOI] [PubMed] [Google Scholar]
- 41.Oki K, Inaba N, Kitagawa K, Fujioka S, Kitano H, Fujisawa Y, Kato H, Iwasaki Y. Function of the α subunit of rice heterotrimeric G protein in brassinosteroid signaling. Plant Cell Physiol. 2009;50:161–72. doi: 10.1093/pcp/pcn182. [DOI] [PubMed] [Google Scholar]
- 42.Clouse SD, Langford M, McMorris TC. A brassinosteroid-insensitive mutant in Arabidopsis thaliana exhibits multiple defects in growth and development. Plant Physiol. 1996;111:671–8. doi: 10.1104/pp.111.3.671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Szekeres M, Németh K, Koncz-Kálmán Z, Mathur J, Kauschmann A, Altmann T, Rédei GP, Nagy F, Schell J, Koncz C. Brassinosteroids rescue the deficiency of CYP90, a cytochrome P450, controlling cell elongation and de-etiolation in Arabidopsis. Cell. 1996;85:171–82. doi: 10.1016/S0092-8674(00)81094-6. [DOI] [PubMed] [Google Scholar]
- 44.Sakamoto T, Morinaka Y, Inukai Y, Kitano H, Fujioka S. Auxin signal transcription factor regulates expression of the brassinosteroid receptor gene in rice. Plant J. 2013;73:676–88. doi: 10.1111/tpj.12071. [DOI] [PubMed] [Google Scholar]
- 45.Sakamoto T, Miura K, Itoh H, Tatsumi T, Ueguchi-Tanaka M, Ishiyama K, Kobayashi M, Agrawal GK, Takeda S, Abe K, et al. An overview of gibberellin metabolism enzyme genes and their related mutants in rice. Plant Physiol. 2004;134:1642–53. doi: 10.1104/pp.103.033696. [DOI] [PMC free article] [PubMed] [Google Scholar]