Abstract
The jasmonate (JA) plant hormones regulate responses to biotic and abiotic stress and aspects of plant development, including male fertility in Arabidopsis thaliana. The bHLH-type transcription factor JA-ASSOCIATED MYC2-LIKE1 (JAM1) negatively regulates JA signaling and gain-of-function JAM1 transgenic plants have impaired JA-mediated male fertility. Here we report that JAM2 and JAM3, 2 bHLHs closely related to JAM1, also act as transcriptional repressors. Moreover, overexpression of JAM2 and JAM3 also results in reduced male fertility. These results suggest that JAM1, JAM2, and JAM3 act redundantly as negative regulators of JA-mediated male fertility.
Keywords: bHLH, transcriptional repressor, jasmonate, fertility, protein-protein interaction
Jasmonates (JAs) are lipid-derived hormones that regulate plant responses to stresses such as wounding and herbivore attack and also act in various developmental processes, including fertility.1-3 Arabidopsis plants impaired in JA biosynthesis or signaling exhibit defective pollen maturation, delayed anther dehiscence and shortened filament elongation; these defects result in male sterility.4-8 MYB transcription factors, such as MYB21 and MYB24, affect JA-mediated male fertility.9-13 JA induces the expression of these MYB genes in flowers by the COI1-mediated JA signaling pathway.12We previously reported that JA-ASSOCIATED MYC2-LIKE1 (JAM1) acts as a negative regulator of JA signaling and that JAM1 gain-of-function transgenic plants exhibit JA-insensitive male sterility.14Sasaki-Sekimoto et al.15 reported that JAM1 and the closely related JAM2 and JAM3 have redundant functions in JA responses, including JA-mediated root elongation, anthocyanin accumulation, and the expression of JA-responsive genes. However, the functional role of JAM2 and JAM3 in JA signaling and male fertility remained to be determined.
To address their functional roles in male fertility, we first tested whether JAM2 and JAM3 have transcriptional repression activity similar to JAM114 by performing reporter-effector transient expression assays using the luciferase (LUC) reporter driven by the constitutive cauliflower mosaic virus (CaMV) 35S promoter with 5 repeats of the GAL4 binding sequence (Fig. 1). The effector constructs with the protein coding region of JAM2 or JAM3 fused to the GAL4 DNA binding domain (GAL4-JAM2 and GAL4-JAM3) clearly repressed LUC activity, indicating that these bHLHs act as transcriptional repressors. JAM2 and JAM3 have comparable transcriptional repression activities to that of JAM1. Also similar to JAM1, the acidic region corresponding to the activation domain of MYC2 is not conserved in either JAM2 or JAM3.14,16 These results are consistent with the previous report that JAM2 and JAM3 (bHLH013 and bHLH003) did not exhibit transcriptional activation activity.17
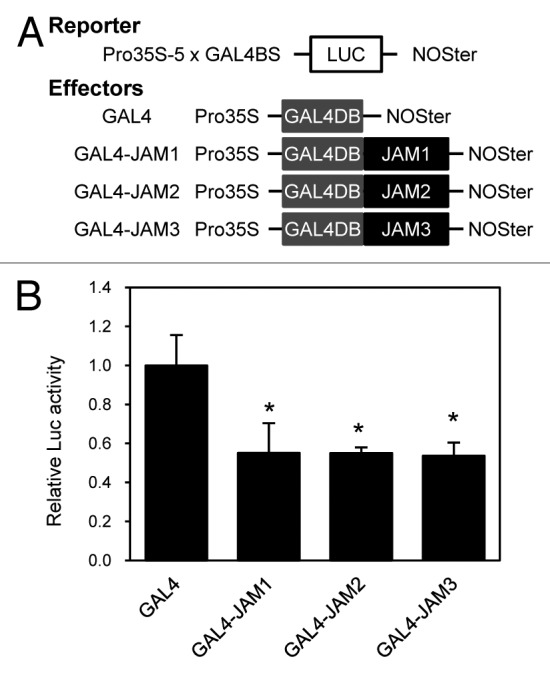
Figure 1. Repression activity of JAM2 and JAM3. (A) Schematic representation of the constructs. (B) Transient expression assays for the GAL4DB fused JAM2 and JAM3 constructs shown in (A). The value for GAL4DB alone was set to 1, and relative values are shown. Error bars indicate SD of results from 3 replicates. Asterisks indicate significant difference from GAL4 (Student t-test, p < 0. 05).
Although JAM1, JAM2, and JAM3 were reported to have redundant functions in JA signaling,15 the functional role of JAM2 and JAM3 in JA-mediated male fertility remained unclear. To test whether JAM2 and JAM3 are involved in male fertility, we made transgenic plants that express JAM2 or JAM3 fused to GFP under the control of the CaMV 35S promoter (35S:JAM2/3-GFP). 35S:JAM2/3-GFP plants exhibited reduced fertility (Fig. 2A and B), as also observed in 35S:JAM1-GFP plants.14 The control 35S:GFP plants that highly express GFP alone exhibited normal fertility, indicating that reduced fertility of 35S:JAM2/3-GFP plants results from overexpression of JAM2 and JAM3, respectively. 35S:JAM2-GFP plants exhibited more severe reduction in fertility than 35S:JAM3-GFP plants, even though JAM3-GFP expression was higher than JAM2-GFP (Fig. 2C and D), suggesting that JAM2 appears to be more active than JAM3 in flowers. Consistent with these observations, the expression levels of MYB21 were clearly downregulated in 35S:JAM2-GFP plants compared with 35S:JAM3-GFP plants (Fig. 2E).
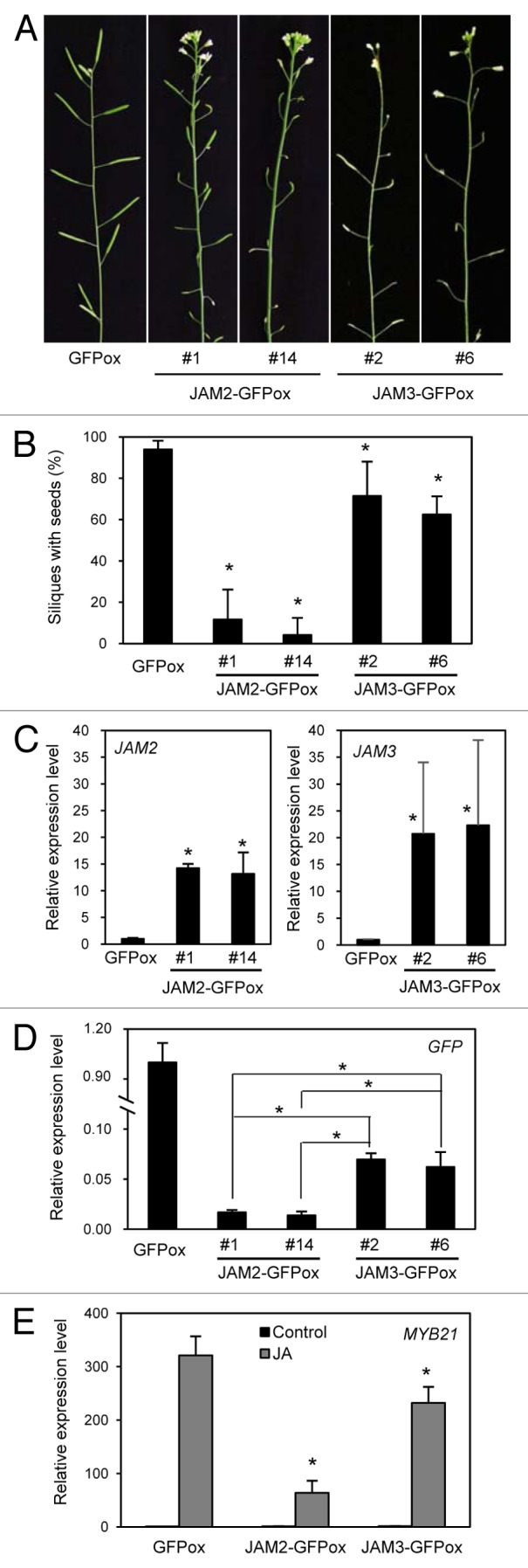
Figure 2. Reduced fertility of JAM2- and JAM3-overexpressing plants. (A) Inflorescences of 6-week-old transgenic plants expressing GFP, JAM2-GFP #1 and #14, and JAM3-GFP #2 and #6 (left to right). (B) Ratio of siliques with seeds to total silique number. Error bars indicate SD of 4 independent lines. Asterisks indicate significant difference from GFP-expressing plants (Student t-test, p < 0. 05). (C) Relative expression levels of JAM2 and JAM3 in inflorescences. The value for plants transgenically expressing GFP alone (and thus expressing JAM2 and JAM3 only from the endogenous loci) was set to 1, and the relative values are shown. Error bars represent SD of results from 3 biological replicates. Asterisks indicate significant difference from GFP-expressing plants (Student t-test, p < 0. 05). (D) Relative expression levels of GFP in inflorescence. Relative values and SD were calculated as in C. Asterisks indicate significant difference (Student t-test, p < 0. 05). (E) Relative expression levels of MYB21 with (gray bar) and without (black bar) MeJA treatment. Seedlings grown for 8 d on MS medium were treated with 50 μM MeJA for 1 h. The value for plants expressing GFP without JA treatment was set to 1, and the relative values are shown. Error bars represent SD of results from 3 biological replicates. Asterisks indicate significant difference from GFP-expressing plants (Student t-test, p < 0. 05).
The bHLH-type transcription factors bind DNA by forming homo- or heterodimers; for example, MYC2, MYC3 and MYC4 bind each other and form heterodimers.17 We analyzed dimerization between JAM1, JAM2, JAM3 and MYC2 by yeast two-hybrid assays and found that JAM1 interacts with JAM3 but not with JAM1 or JAM2, and JAM2 interacts with JAM2 and JAM3 (Fig. 3). None of them interacted with MYC2 when MYC2 was used as prey protein. Activation activity of MYC2 was too strong to use as a bait protein in our yeast two-hybrid system. These results indicate that JAM transcription factors form heterodimers with each other, but not with the MYC2 activator.
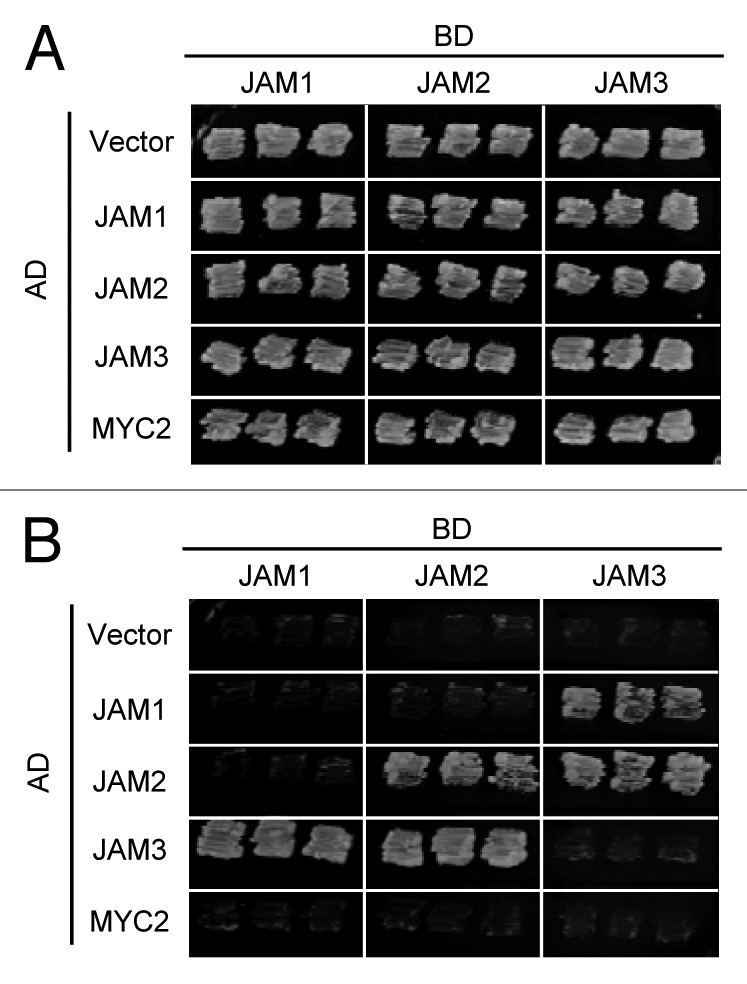
Figure 3. Yeast two-hybrid analysis of JAMs and MYC2. GAL4-JAMs and LexA-JAMs or MYC2 were used as bait and prey, respectively. Yeast cells co-transformed with bait and prey were selected and subsequently grown on media lacking Leu and Trp (A), or on selective media lacking His, Leu, and Trp (B). The concentration of 3-amino-1,2,4-triazole was 2 mM. Empty vector for the prey plasmids was used as a negative control.
We demonstrated here that JAM2 and JAM3 act as transcriptional repressors and regulate JA mediated male fertility. Because JAM1, JAM2, and JAM3 function as negative regulators of JA signaling,15 the effect on fertility in JAM2/3-overexpressing plants likely results from impairment of male reproductive organ development through negative regulation of JA signaling, as observed in JAM1-overexpressing plants.14 In addition, the high conservation of the amino acid sequences of the bHLH domains in JAM1, JAM2, and JAM314 suggests that they have similar DNA binding preferences and downregulate the expression of MYB genes, such as MYB21, that are involved in JA-mediated male organ development (Fig. 2E).9-13 It remains to be clarified whether those MYBs are direct targets of JAMs. Our results show that JAM1, JAM2, and JAM3 redundantly and negatively regulate JA signaling in flowers.
Although the 3 JAMs act redundantly, our results revealed differences among them. For example, the yeast two-hybrid assays showed selective interactions between JAMs. Also, Sasaki-Sekimoto et al.15 reported that JAM1 and JAM2 but not JAM3 are JA-inducible genes. JAM3 may form heterodimers with JAM1 or JAM2 when JA induces the expression of JAM1 or JAM2. In addition, quite recently Song et al.18 demonstrated that bHLH17/JAM1 and bHLH3/JAM3 are nuclear proteins whereas bHLH13/JAM2 localizes both in cytosol and nucleus. Variation in the severity of reduced fertility in JAM-overexpressing plants may arise from intracellular localizations and the combinations of JAMs in repressor complexes. The Arabidopsis genome contains 12 JAZs, 3 MYCs, and 3 JAMs. Various combinations of these factors may regulate each JA-mediated response. Thus, it will be necessary to analyze the combinations to fully understand the regulatory mechanisms of JA signaling.
Materials and Methods
Preparation of constructs
The coding sequences of JAM2 and JAM3 without the stop codons were amplified from an Arabidopsis cDNA library with specific primers containing attB1 or attB2 sequences (attB1-JAM2-F; 5′-GGGGACAAGT TTGTACAAAA AAGCAGGCTC CATGAATATT GGTCGCCTAG TGTG-3′, attB2-JAM2-R; 5′-GGGGACCACT TTGTACAAGA AAGCTGGGTC TCTACCTGAT GATGTTCTTG ACT-3′, attB1-JAM3-F; 5′-GGGGACAAGT TTGTACAAAA AAGCAGGCTC CATGGGTCAA AAGTTTTGGG AGA-3′, attB2-JAM3-R; 5′-GGGGACCACT TTGTACAAGA AAGCTGGGTC CTGTGATAGA GAGGCAAGGA GCT-3′), and the resultant DNA fragments were cloned into pDONR207 (Invitrogen) by BP clonase reaction (Invitrogen, 11789-020). Entry clones for JAM1 and MYC2 without the stop codon were prepared previously.14 Genes cloned into pDONR207 were transferred to modified effector plasmids for expression of the GAL4-fused protein in plant cells, modified pBTM116 for bait and modified pVP16S114 for prey, by LR clonase reaction (Invitrogen, 11791-020). The reporter plasmid for the transient expression assay was described previously.19To prepare plasmids expressing JAM2 and JAM3 fused with GFP at the C terminus, DNA fragments encoding JAM2 and JAM3 were amplified from an Arabidopsis cDNA library with appropriate primers (JAM2-GFP-F; 5′-GATGAATATT GGTCGCCTAG TGT-3′, JAM2-GFP-R; 5′- TCTACCTGAT GATGTTCTTG AC-3′, JAM3-GFP-F; 5′-GATGGGTCAA AAGTTTTGGG AGA-3′, JAM3-GFP-R; 5′-CTGTGATAGA GAGGCAAGG-3′) and inserted into the SmaI site of p35SGFP. The region corresponding to the transgene was transferred to the pBCKH plant expression vector20 by LR clonase reaction (Invitrogen, 11791-020).
Transient expression assay and yeast two-hybrid analysis
Transient expression assay and yeast two-hybrid analysis were performed as described previously.14
RNA Extraction and Quantitative RT-PCR
RNA extraction and quantitative RT-PCR were performed as described previously.14 Gene-specific primers used for RT-PCR were as follows: JAM2-F; 5′-CTTGTTGGGA GACGCGGTTT-3′, JAM2-R; 5′-CTCTCTCTGC TTCCATGACC TTTA-3′, JAM3-F; 5′-GAAATCAGTG TTTGGTGGGT CTGA-3′, JAM3-R; 5′-CAAGACTCAG CTGTCGACCA A-3′, GFP-F; 5′-CGACCACATG AAGCAGCACG-3′, GFP-R; 5′-TGAAGTCGAT GCCCTTCAGC-3′, MYB21-F; 5′- AAGTAGTGGA GGTTCGGGAT CA-3′, MYB21-R; 5′- CCGTGGTTGG CGATATAGTT GA-3′, PP2AA3-F; 5′-GACCAAGTGA ACCAGGTTAT TGG-3′, PP2AA3-R; 5′-TACTCTCCAG TGCCTGTCTT CA-3′. Relative amounts of transcripts were calculated by an absolute quantification method, with the PP2AA3 gene as an internal control. At least 3 biological replicates were included in each experiment.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Glossary
Abbreviations:
- JA
jasmonate
- bHLH
basis helix-loop-helix
- JAM
JA-ASSOCIATED MYC2-LIKE
- COI1
CORONATINE INSENSITIVE1
- LUC
luciferase
- CaMV
cauliflower mosaic virus
- GFP
green fluorescent protein
- RT-PCR
reverse transcriptional polymerase chain reaction
References
- 1.Devoto A, Turner JG. Regulation of jasmonate-mediated plant responses in arabidopsis. Ann Bot. 2003;92:329–37. doi: 10.1093/aob/mcg151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Balbi V, Devoto A. Jasmonate signalling network in Arabidopsis thaliana: crucial regulatory nodes and new physiological scenarios. New Phytol. 2008;177:301–18. doi: 10.1111/j.1469-8137.2007.02292.x. [DOI] [PubMed] [Google Scholar]
- 3.Wasternack C, Hause B. Jasmonates: biosynthesis, perception, signal transduction and action in plant stress response, growth and development. An update to the 2007 review in Annals of Botany. Ann Bot (Lond) 2013;111:1021–58. doi: 10.1093/aob/mct067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Feys B, Benedetti CE, Penfold CN, Turner JG. Arabidopsis mutants selected for resistance to the phytotoxin coronatine are male sterile, insensitive to methyl jasmonate, and resistant to a bacterial pathogen. Plant Cell. 1994;6:751–9. doi: 10.1105/tpc.6.5.751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stintzi A, Browse J. The Arabidopsis male-sterile mutant, opr3, lacks the 12-oxophytodienoic acid reductase required for jasmonate synthesis. Proc Natl Acad Sci U S A. 2000;97:10625–30. doi: 10.1073/pnas.190264497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ishiguro S, Kawai-Oda A, Ueda J, Nishida I, Okada K. The DEFECTIVE IN ANTHER DEHISCIENCE gene encodes a novel phospholipase A1 catalyzing the initial step of jasmonic acid biosynthesis, which synchronizes pollen maturation, anther dehiscence, and flower opening in Arabidopsis. Plant Cell. 2001;13:2191–209. doi: 10.1105/tpc.010192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Park JH, Halitschke R, Kim HB, Baldwin IT, Feldmann KA, Feyereisen R. A knock-out mutation in allene oxide synthase results in male sterility and defective wound signal transduction in Arabidopsis due to a block in jasmonic acid biosynthesis. Plant J. 2002;31:1–12. doi: 10.1046/j.1365-313X.2002.01328.x. [DOI] [PubMed] [Google Scholar]
- 8.Chung HS, Howe GA. A critical role for the TIFY motif in repression of jasmonate signaling by a stabilized splice variant of the JASMONATE ZIM-domain protein JAZ10 in Arabidopsis. Plant Cell. 2009;21:131–45. doi: 10.1105/tpc.108.064097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mandaokar A, Thines B, Shin B, Lange BM, Choi G, Koo YJ, Yoo YJ, Choi YD, Choi G, Browse J. Transcriptional regulators of stamen development in Arabidopsis identified by transcriptional profiling. Plant J. 2006;46:984–1008. doi: 10.1111/j.1365-313X.2006.02756.x. [DOI] [PubMed] [Google Scholar]
- 10.Song S, Qi T, Huang H, Ren Q, Wu D, Chang C, Peng W, Liu Y, Peng J, Xie D. The Jasmonate-ZIM domain proteins interact with the R2R3-MYB transcription factors MYB21 and MYB24 to affect Jasmonate-regulated stamen development in Arabidopsis. Plant Cell. 2011;23:1000–13. doi: 10.1105/tpc.111.083089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mandaokar A, Browse J. MYB108 acts together with MYB24 to regulate jasmonate-mediated stamen maturation in Arabidopsis. Plant Physiol. 2009;149:851–62. doi: 10.1104/pp.108.132597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reeves PH, Ellis CM, Ploense SE, Wu MF, Yadav V, Tholl D, Chételat A, Haupt I, Kennerley BJ, Hodgens C, et al. A regulatory network for coordinated flower maturation. PLoS Genet. 2012;8:e1002506. doi: 10.1371/journal.pgen.1002506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cheng H, Song S, Xiao L, Soo HM, Cheng Z, Xie D, Peng J. Gibberellin acts through jasmonate to control the expression of MYB21, MYB24, and MYB57 to promote stamen filament growth in Arabidopsis. PLoS Genet. 2009;5:e1000440. doi: 10.1371/journal.pgen.1000440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nakata M, Mitsuda N, Herde M, Koo AJ, Moreno JE, Suzuki K, Howe GA, Ohme-Takagi M. A bHLH-type transcription factor, ABA-INDUCIBLE BHLH-TYPE TRANSCRIPTION FACTOR/JA-ASSOCIATED MYC2-LIKE1, acts as a repressor to negatively regulate jasmonate signaling in arabidopsis. Plant Cell. 2013;25:1641–56. doi: 10.1105/tpc.113.111112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sasaki-Sekimoto Y, Jikumaru Y, Obayashi T, Saito H, Masuda S, Kamiya Y, Ohta H, Shirasu K. Basic Helix-Loop-Helix Transcription Factors JASMONATE-ASSOCIATED MYC2-LIKE1 (JAM1), JAM2, and JAM3 Are Negative Regulators of Jasmonate Responses in Arabidopsis. Plant Physiol. 2013;163:291–304. doi: 10.1104/pp.113.220129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fernández-Calvo P, Chini A, Fernández-Barbero G, Chico JM, Gimenez-Ibanez S, Geerinck J, Eeckhout D, Schweizer F, Godoy M, Franco-Zorrilla JM, et al. The Arabidopsis bHLH transcription factors MYC3 and MYC4 are targets of JAZ repressors and act additively with MYC2 in the activation of jasmonate responses. Plant Cell. 2011;23:701–15. doi: 10.1105/tpc.110.080788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Niu Y, Figueroa P, Browse J. Characterization of JAZ-interacting bHLH transcription factors that regulate jasmonate responses in Arabidopsis. J Exp Bot. 2011;62:2143–54. doi: 10.1093/jxb/erq408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Song S, Qi T, Fan M, Zhang X, Gao H, Huang H, Wu D, Guo H, Xie D. The bHLH subgroup IIId factors negatively regulate jasmonate-mediated plant defense and development. PLoS Genet. 2013;9:e1003653. doi: 10.1371/journal.pgen.1003653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hiratsu K, Ohta M, Matsui K, Ohme-Takagi M. The SUPERMAN protein is an active repressor whose carboxy-terminal repression domain is required for the development of normal flowers. FEBS Lett. 2002;514:351–4. doi: 10.1016/S0014-5793(02)02435-3. [DOI] [PubMed] [Google Scholar]
- 20.Mitsuda N, Hiratsu K, Todaka D, Nakashima K, Yamaguchi-Shinozaki K, Ohme-Takagi M. Efficient production of male and female sterile plants by expression of a chimeric repressor in Arabidopsis and rice. Plant Biotechnol J. 2006;4:325–32. doi: 10.1111/j.1467-7652.2006.00184.x. [DOI] [PubMed] [Google Scholar]


