Abstract
Mitogen-Activated Protein Kinase (MAPK) genes encode proteins that mediate various signaling pathways associated with biotic and abiotic stress responses in eukaryotes. The MAPK genes form a 3-tier signal transduction cascade between cellular stimuli and physiological responses. Recent identification of soybean MAPKs and availability of genome sequences from other legume species allowed us to identify their MAPK genes. The main objectives of this study were to identify MAPKs in 3 legume species, Lotus japonicus, Medicago truncatula, and Phaseolus vulgaris, and to assess their phylogenetic relationships. We used approaches in comparative genomics for MAPK gene identification and named the newly identified genes following Arabidopsis MAPK nomenclature model. We identified 19, 18, and 15 MAPKs and 7, 4, and 9 MAPKKs in the genome of Lotus japonicus, Medicago truncatula, and Phaseolus vulgaris, respectively. Within clade placement of MAPKs and MAPKKs in the 3 legume species were consistent with those in soybean and Arabidopsis. Among 5 clades of MAPKs, 4 founder clades were consistent to MAPKs of other plant species and orthologs of MAPK genes in the fifth clade-"Clade E" were consistent with those in soybean. Our results also indicated that some gene duplication events might have occurred prior to eudicot-monocot divergence. Highly diversified MAPKs in soybean relative to those in 3 other legume species are attributable to the polyploidization events in soybean. The identification of the MAPK genes in the legume species is important for the legume crop improvement; and evolutionary relationships and functional divergence of these gene members provide insights into plant genome evolution.
Keywords: Glycine max, Lotus japonicus, Medicago truncatula, Phaseolus vulgaris, Legume genomics, MAPK, MAPK evolution, MAPKK, functional divergence, gene duplication
Background
Gene members of Mitogen-Activated Protein Kinase (MAPK) family encode proteins involved in signaling pathways associated with biotic and abiotic stress responses in all eukaryotes. The MAPK gene members are identified in a limited number of plant species, and they are yet to be identified in a majority of plant species. The MAPK gene family consists of 3 functionally linked subfamilies known as MAPK ( = MPK), MAPKK ( = MEK or MAP2K), and MAPKKK ( = MEKK), initiating a cascade of signaling for various biological responses.1-3 The MAPK gene diversity within each subfamily varies widely across species. For example, the number of MAPKs and MAPKKs are 5 and 1 in Chlamydomonas, 6 and 6 in Sachharomyces, 21 and 11 in Populus,4 16 and 12 in Brachypodium,5 20 and 10 in Arabidopsis,1,2 38 and 11 in Glycine,6 and 15 and 8 in Oryza,4 respectively.
MAPK gene members are associated with various stress related plant signaling mechanisms. MAPK3 and MAPK6 in Arabidopsis are found to mediate signals in response to MAMPs (Microbe-Associate Molecular Pattern) such as flagellin (flg22) and chitin elicitors7,8 and control cell death along with the pathogenic responses by producing (ROS) and indole-derived phytoalexin (camalexin).9,10 Orthologs of these 2 genes in rice are involved in fungal resistance. However, their upstream MAPKK pathways and complete roles are not fully understood.11 Acting as an upstream MAPKK, the ortholog of AtMAPKK4 in rice is associated with OsMAPK3 [Phytozome accession: LOC_Os03 g17700].12 Phosphorylation of OsMAPKK4 is undertaken by 6 upstream MAPKKKs: [Phytozome: LOC_Os01g50370, LOC_Os05g46760, LOC_Os01g50400, LOC_Os01g50410, LOC_Os01g50420, and LOC_Os05g46750] prompting the pathways that regulate myriads of stress responses including pathogen, insect, drought, salinity, flood, and cold.13
MAPK signaling modules are also involved in developmental processes in eukaryotes. Some of these modules are well studied in yeast. For example, in budding yeast, MAPKs regulate spore formation and spore wall assembly14,15; phosphorylation of ste12 (mating-specific transcription factors) by Fus3 MAPK16,17 is activated by pheromone responsive upstream-MAPK cascade18 and activation of Fus3 triggers phosphorylation of FAR1 for the G1 arrest in cell cycle, probably through inhibition of G1 cyclins.19 Under a nitrogen-deficit environment, a budding yeast alters morphological growth pattern (i.e., occurrence of pseudohyphal growth), while under glucose-deficit environment, invasive growth,20 also called filamentous growth, occurs.21 This filamentous growth in budding yeast is enhanced by various MAP kinases including Kss1, and negatively regulated by Fus3.20,22,23 In yeast, MAPKs are also involved in the maintenance of cell wall integrity and adaptability to changing osmotic potential,24 allowing the cell wall to grow continually under hyper-osmolarity conditions through the initiation of High Osmolarity Glycerol (HOG) MAPK pathway.25 MAPKK known as Ste7 that targets Kss1 and Fus3 in yeast,26 is found to be vital for the pathways associated with mating17,23 and filamentous growth.20,23 Identification of MAPK genes in Arabidopsis lead to increasing instances of MAPK studies in other plant species as their genomes became available. One of such recent studies has used Arabidopsis MAPKs as reference sequences to identify their orthologs in rice and poplar,4 and we used a similar approach to identify MAPK gene members in soybean.6 MAPK genes in other legume species are yet to be identified, and their evolutionary history and functional divergence need to be investigated in order to enhance research on stress signaling and legume-microbe interaction.
Legumes can grow in nitrogen-deficit soil because of their ability to fix atmospheric nitrogen through a symbiotic association with Rhizobia in root nodules. Previous studies have shown that the root nodule development in legumes involves various kinase activities.27,28 One another study showed that a gene called TDY1, believed to encode a MAPK homolog, was involved in root nodulation and root-tip development in Medicago.29 One of the well-studied MAPKs in legumes is MAPK4 ortholog in soybean: this gene is involved in negative regulation of defense mechanism while in positive regulation of growth and development.30 MAPKs in legumes are known to involve in various signaling responses that involve root organogenesis?for example, in Lupinus albus, activation of MAPKs such as SIMK and SAMK are required for infection of Bradyrhizobia sp. or other compatible microbes to establish symbiotic interactions.31 During biological nitrogen fixation in legumes, the mechanisms of signal transduction pathway that follows the exchange of signals between legume (e.g., flavonoids) and Rhizobia (e.g., nod factors) is being unraveled (reviewed in ref.32): transcription factors belonging to NIN, GRAS, and ERF families are involved in the immediate responses such as ion fluxes, CCaMK mediated signaling, and subsequent activation of MAPK genes (reviewed in28). It is also worth noting that there are similarities in the early plant response to infection by pathogenic and symbiotic bacteria.33,34 Recent studies27,28 have predicted the potential roles played by various kinase genes in the root nodulation process.
With the recent identification of soybean MAPKs6 and increasing availability of genomic sequences from other legume species, it is now possible to identify gene members of MAPK and MAPKK subfamilies in Lotus, Medicago, and Phaseolus. In our previous study,6 we identified 38 MAPKs, 11 MAPKKs, and 150 MAPKKKs in soybean, the plant species with the highest number of MAPKs published to date. The main objectives of this study were to identify MAPKs and MAPKKs in the genome of Lotus japonicus, Medicago truncatula, and Phaseolus vulgaris and assess their evolutionary relationships. The insights from this study would contribute to enhance our understanding of the evolution of these genes in legumes, and also functional characterization of MAPK genes with potential legume specific regulatory roles would help in legume crop improvement.
Results
We identified 19, 18, and 15 MAPK members in the genomes of Lotus japonicus (Lj), Medicago truncatula (Mt), and Phaseolus vulgaris (Pv), respectively. Similarly, we identified 7, 4, and 9 MAPKK members in Lj, Mt, and Pv, respectively. Maximum Parsimony (MP) and Maximum Likelihood (ML) analyses of each data set yielded phylogenies with similar topologies but with slightly variable bootstrap support (BS). The model test performed showed that the Jones-Taylor-Thornton (JTT) model with discrete Gamma distribution and Invariant sites (G+I) was the best-fit evolutionary model for both MAPK and MAPKK protein sequences.
MAPKs
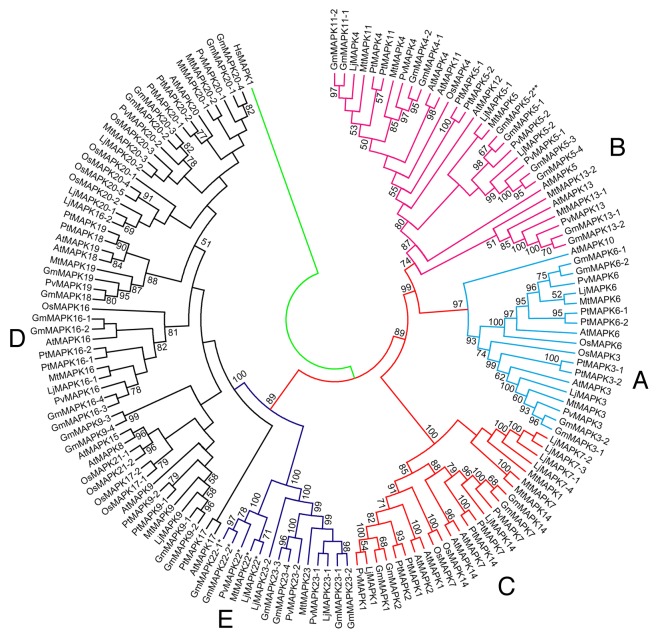
Newly identified MAPKs in Lj, Mt, and Pv along with the published MAPKs in Gm and 3 non-legume members (At, Os, and Pt), and their phylogenetic relationships are shown in Figure 1 and Figures S1 and S2. Maximum Parsimony cladogram (Fig. S1) and Maximum Likelihood phylogram (Fig. S2) were similar in topologies. The amino acid sequence length for the identified MAPK genes ranged from 205 to 590, 340 to 615, and 368 to 613 in Lj, Mt, and Pv, respectively. The MAPK genes from all 3 genomes displayed similar evolutionary patterns forming 5 clades. Among these clades, 4 clades (A, B, C, and D; see Fig. 1) in all 3 species were consistent to those in Gm, At, Pt, and Os. The 5 clades were moderately to strongly supported (BS support for clade A, B, C, D, and E in ML tree were 84, 75, 97, 99, and 100, respectively). The MAPK gene members in clade A, B, C, and E had TEY motif, whereas those in clade D had TDY motif. The clade-wide placement of MAPK orthologs with specific motifs was consistent to Arabidopsis MAPKs.1 The MAPK genes in clade E were exclusively from legume species where 2 of the MAPK gene members from Glycine and one from Lotus, Medicago, and Phaseolus each had a TQY motif. The MAPK gene members in each clade also showed similar gene structure (Fig. S3) across the species with similar distribution of exon and introns (for example, the number of exons in clade E members were highly multiplied, whereas the gene members in clade C had fewer exons). Duplicated MAPK genes occurred in all 4 legume species. For some MAPK orthologs of Arabidopsis, the number of paralogs in soybean was twice as many as those in other legume species. For example, the orthologs of MAPK5, MAPK9, MAPK16, and MAPK20 had 4 copies in soybean, whereas other legume species had either one or 2 copies. However, in some cases, such as in LjMAPK7, we found more copies in Lj than their counterparts in Gm and other legume species. As shown in Figure 1, we did not find the orthologs of AtMAPK2, AtMAPK8, AtMAPK10, AtMAPK11, AtMAPK12, AtMAPK13, AtMAPK15, AtMAPK17, AtMAPK18, AtMAPK19, and OsMAPK21 in Lotus. Similarly, orthologs of AtMAPK1, AtMAPK2, AtMAPK8, AtMAPK10, AtMAPK12, AtMAPK15, AtMAPK17, AtMAPK18, and OsMAPK21 were absent in Medicago. AtMAPK2, AtMAPK8, AtMAPK9, AtMAPK10, AtMAPK11, AtMAPK12, AtMAPK14, AtMAPK15, AtMAPK17, AtMAPK18, and OsMAPK21 were absent in Phaseolus. The expression analyses of MAPK gene members across species also indicated that majority of gene members in Lotus, Medicago and Phaseolus had high gene expression values (Figs. 2, 3 and 4). Gene members such as LjMAPK7?4, MtMAPK20?3 and PvMAPK5-2 were among the least expressed genes.
Figure 1. Maximum Likelihood analysis of MAPKs of 4 legumes (Soybean, Lotus, Medicago, and Phaseolus) and their orthologs in 3 non-legumes (Arabidopsis, rice, and poplar) The values above the branches are bootstrap support of 100 replicates. The JTT+G+I evolutionary model was employed to perform Maximum Likelihood analysis. The members with phosphorylation motif TEY are nested in clade A, B, and C, those with the TDY motif in clade D, and members with legume specific TQY (denoted by *), and one of the gene members of soybean with TVY (denoted by **) motif in clade E and B, respectively. The MAPK gene models were accepted for phylogenetic analysis using protein sequences of Serine/Threonine kinase subfamily having conserved Aspartate and Lysine residues in their catalytic domain with (D[L/I/V]K) motif and TXY phosphorylation motif in their activation loop.
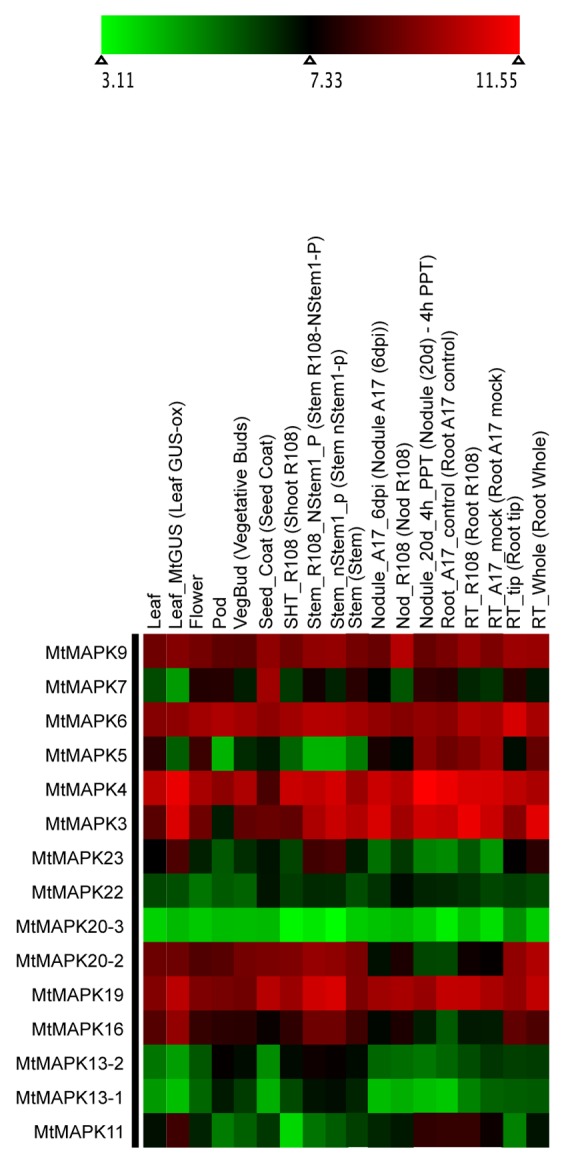
Figure 2. Heatmap visualization of LjMAPKs. Log 2-based value was employed to construct the heatmap for LjMAPKs gene expression in different tissues.

Figure 3. Heatmap visualization of MtMAPKs. Log 2-based value was employed to construct the heatmap for MtMAPKs gene expression in different tissues.
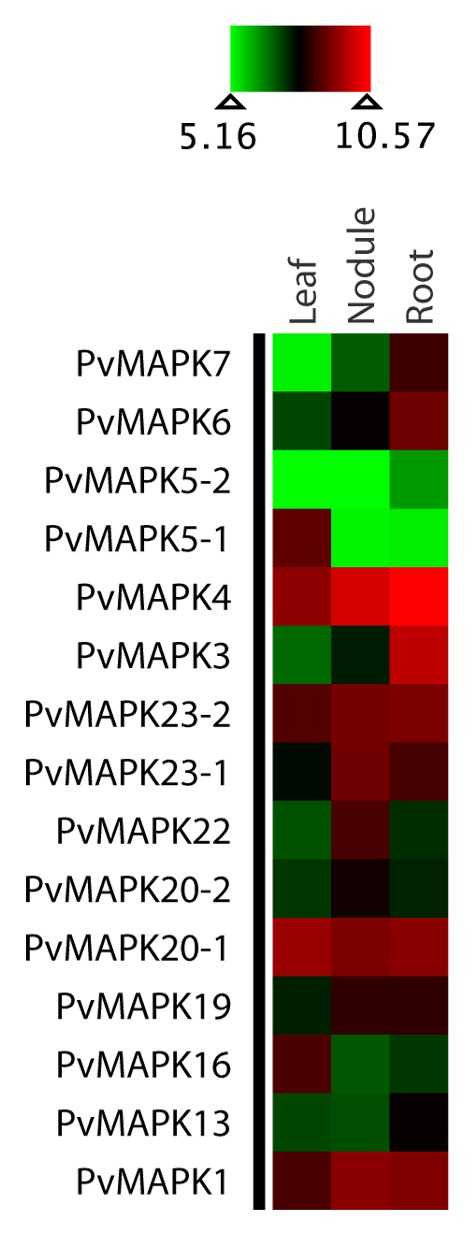
Figure 4. Heatmap visualization of PvMAPKs. Log 2-based value was employed to construct the heatmap for PvMAPKs gene expression in different tissues.
MAPKKs
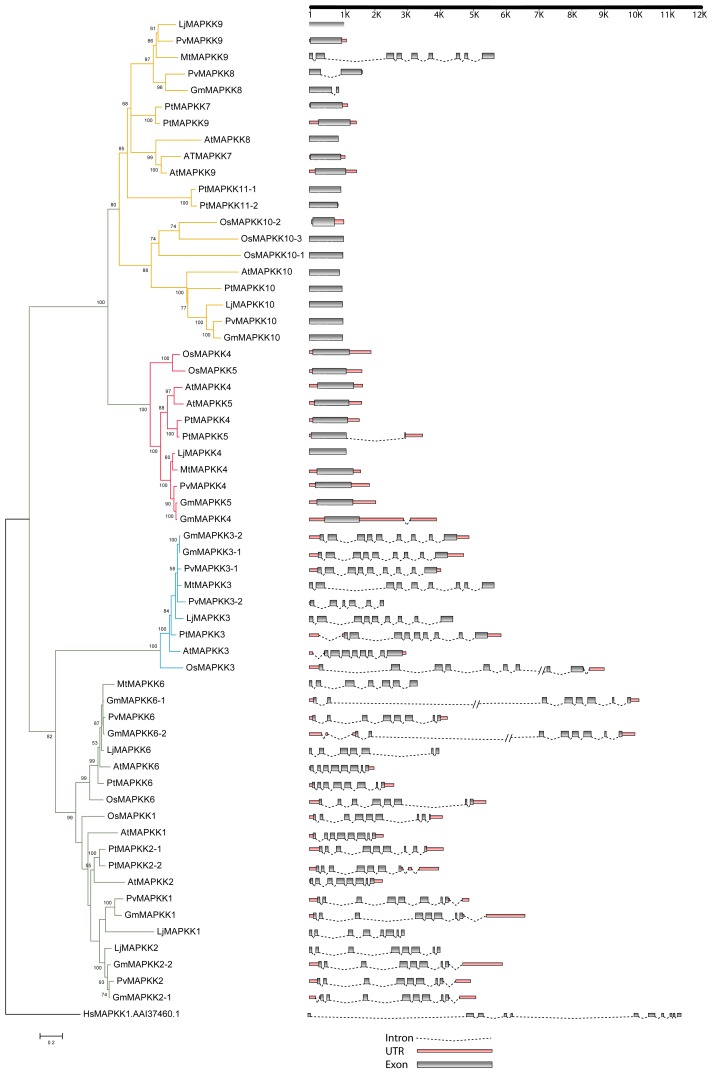
Newly identified MAPKK genes in Lotus japonicas, Medicago truncatula, and Phaseolous vulgaris are shown in Figure 5 and Figures S4 and S5. The MAPKK gene members formed 4 distinct clades (clades A, B, C, and D with BS support of 78, 100, 98, and 57, respectively) in each species consistent to those in Arabidopsis.1 The amino acid sequence length for the identified MAPKK genes ranged from 335 to 526, 366 to 519, and 253 to 519 in Lotus, Medicago, and Phaseolus, respectively. We did not find orthologs of AtMAPKK5, AtMAPKK7, and AtMAPKK8 in Lotus. Similarly, we did not find the orthologs of AtMAPKK1, AtMAPKK2, AtMAPKK5, AtMAPKK7, AtMAPKK8, and AtMAPKK10 in Medicago and those of AtMAPKK5 and AtMAPKK7 in Phaseolus. The expression profile for MAPKK gene members in Lotus, Medicago and Phaseolus indicated the majority of the gene members had higher expression as compared with differentially expressed MAPKs in soybean6 (Figs. 6, 7 and 8). The MAPKK10 orthologs in Glycine,6 Lotus, and Phaseolus had no or minimal expression in any of the compared tissues, while the orthologs of MAPKK4 and MAPKK5 had high expression values in both Lotus and Medicago. However, in the case of Phaseolus, MAPKK2 and MAPKK3 have comparatively higher expression, including little expression of MAPKK8 in root, nodule, and leaf (Fig. 8).
Figure 5. A) Maximum Likelihood analysis of GmMAPKKs and their orthologs in Arabidopsis, poplar, and rice. In the ML phylogram, the values above the branches are bootstrap support of 100 replicates. The JTT+G+I evolutionary model was employed to perform Maximum Likelihood analysis. The MAPKK gene models were accepted for phylogenetic analysis using dual-specificity protein kinases having conserved Aspartate and Lysine residues in their catalytic domain with (D[L/I/V]K) motif and S-X5-T phosphorylation motif along their activation loop. Gene models showing intron/exon lengths (right panel) were mapped onto the phylogram. B) Maximum Likelihood analysis of GmMAPKKs and their orthologs in Arabidopsis, poplar, and rice. In the ML phylogram, the values above the branches are bootstrap support of 100 replicates. The JTT+G+I evolutionary model was employed to perform Maximum Likelihood analysis. The MAPKK gene models were accepted for phylogenetic analysis using dual-specificity protein kinases having conserved Aspartate and Lysine residues in their catalytic domain with (D[L/I/V]K) motif and S-X5-T phosphorylation motif along their activation loop. Gene models showing intron/exon lengths (right panel) were mapped onto the phylogram.
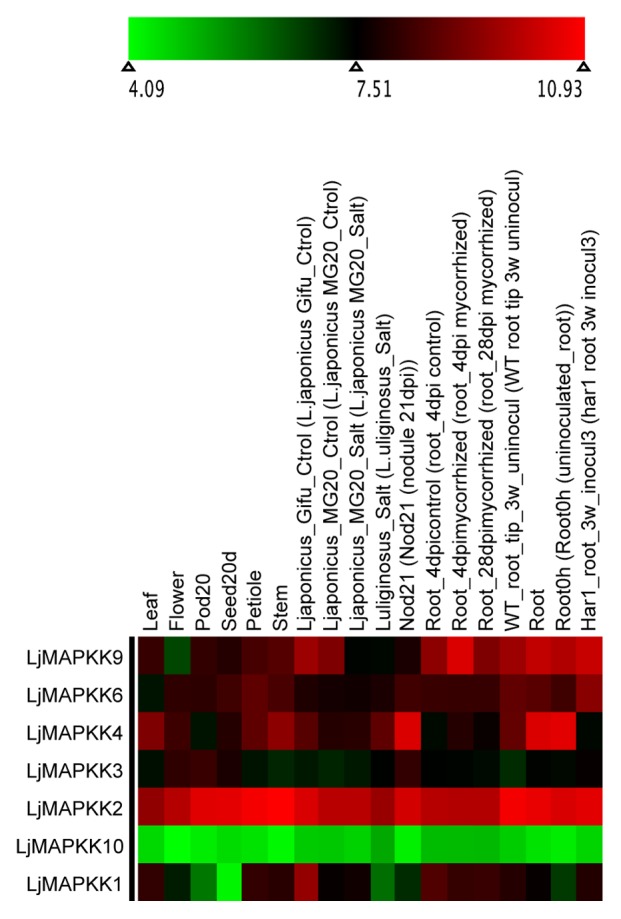
Figure 6. Heatmap visualization of LjMAPKKs. Log 2-based value was employed to construct the heatmap for LjMAPKKs gene expression in different tissues.
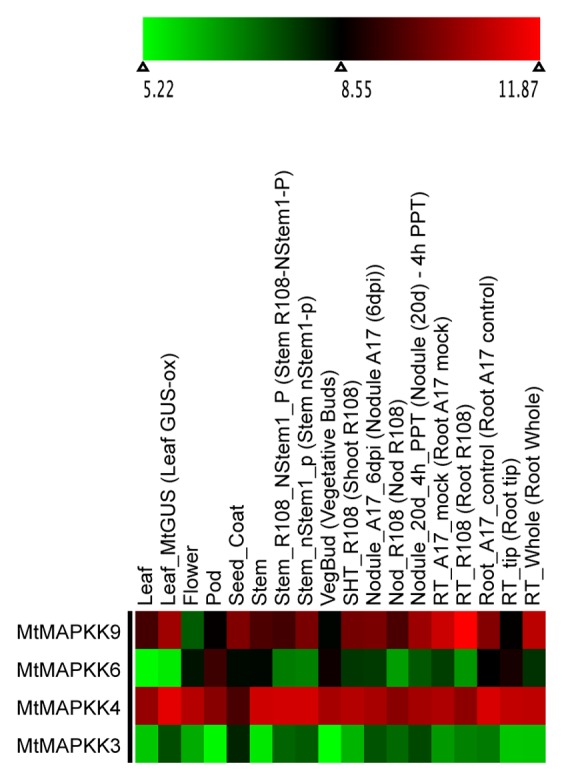
Figure 7. Heatmap visualization of MtMAPKKs. Log 2-based value was employed to construct the heatmap for MtMAPKKs gene expression in different tissues.
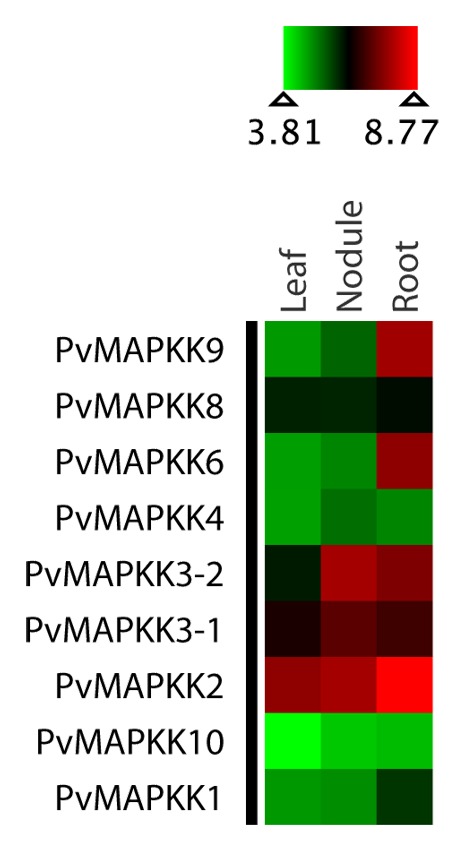
Figure 8. Heatmap visualization of PvMAPKKs.Log 2-based value was employed to construct the heatmap for MtMAPKKs gene expression in different tissues.
Discussion
Identification and evolutionary history of MAPK members
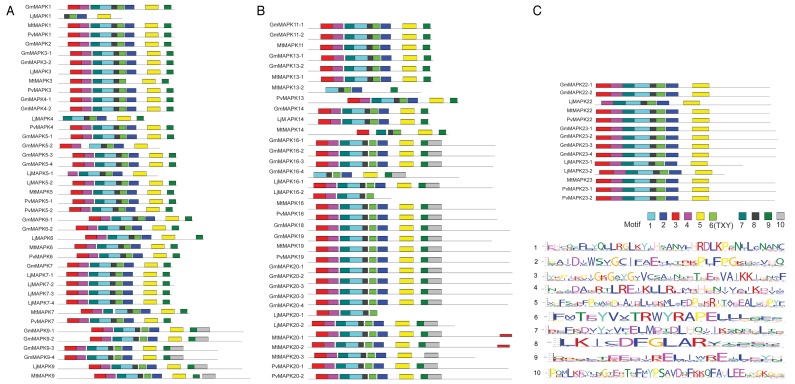
The diversity of MAPKs differed across species with less than half in Lotus, Medicago, and Phaseolus compared with that in Glycine. The number of MAPKKs was relatively similar in Lotus and Phaseolus, while only 4 MAPKKs were identified in Medicago. Number of paralogs of some of the MAPK genes in the 3 legume species was almost similar. However, the orthologs of these genes in soybean have retained approximately twice as many duplicate copies. The larger genome size of soybean (1115 Mb, 2n = 40)35 compared with other legumes (Lj is 472Mb, [2n = 12],36 Mt is ca. 500 Mb, [2n = 32],37 and Pv is 625 Mb, [2n = 22]38) contains proportionately higher number of the MAPK genes. MEME analysis for the approximate sequence pattern including logos of protein motifs related to MAPK specific ?signals? showed the conservation of domains throughout the gene members of each subfamily and subgroups (Fig. 9A-C and 10). Some of these conserved domains as predicted by Pfam and PROSITE for MAPK and MAPKKs included protein kinase catalytic domain, MAPK conserved site (specific to MAPKs), serine/threonine-active site, and ATP binding site.
Figure 9. A. Predicted domain structure of GmMAPKs, LjMAPKs, MtMAPKs, and PvMAPKs. Conserved domain structures as predicted by MEME analysis of GmMAPKs, LjMAPKs, MtMAPKs, and PvMAPKs. Ten different sites were analyzed for the prediction of conserved domain structures. Each stack height in the logos for 10 different predicted motifs represents the sequence conservation, which is measured in bits. The height of each residue within the stack indicates the frequency of corresponding amino acid competing for that position. B. Predicted domain structure of GmMAPKs, LjMAPKs, MtMAPKs, and PvMAPKs. Conserved domain structures as predicted by MEME analysis of GmMAPKs, LjMAPKs, MtMAPK,s and PvMAPKs. Ten different sites were analyzed for the prediction of conserved domain structures. Each stack height in the logos for 10 different predicted motifs represents the sequence conservation, which is measured in bits. The height of each residue within the stack indicates the frequency of corresponding amino acid competing for that position. C. Predicted domain structure of GmMAPKs, LjMAPKs, MtMAPKs, and PvMAPKs. Conserved domain structures as predicted by MEME analysis of GmMAPKs, LjMAPKs, MtMAPKs, and PvMAPKs. Ten different sites were analyzed for the prediction of conserved domain structures. Each stack height in the logos for 10 different predicted motifs represents the sequence conservation, which is measured in bits. The height of each residue within the stack indicates the frequency of corresponding amino acid competing for that position.
Figure 10. Predicted domain structure of GmMAPKKs, LjMAPKKs, MtMAPKKs, and PvMAPKKs. Conserved domain structures as predicted by MEME analysis of GmMAPKKs, LjMAPKKs, MtMAPKKs, and PvMAPKKs. Ten different sites were analyzed for the prediction of conserved domain structures. Each stack height in the logos for 10 different predicted motifs represents the sequence conservation, which is measured in bits. The height of each residue within the stack indicates the frequency of corresponding amino acid competing for that position.
Evolutionary processes resulting into duplication of individual gene, polyploidization of genome, or even segmental or chromosomal duplications could contribute to the functional complexities of the organism.39,40 The soybean genome has undergone at least 2 rounds of whole genome duplications35 resulting into its genome expansion. Duplication of genes could have occurred through any of unequal crossing over, retroposition and chromosomal or genome duplication.40 In addition to the GmMAPKs with 2 copies, the MAPK with 4 paralogs such as MAPK5, MAPK9, MAPK16, and MAPK20 in soybean are at least twice as many as in Lj, Mt, Pv, At, Pt, and Os. Orthologs of MAPK 7 and MAPK14 were consistently present in both legume and non-legume species, and they were more diversified in Lotus and Brachypodium5 than in other species. The orthologs of MAPK7 in Lotus were found to have undergone tandem duplications yielding 4 copies as these paralogs are tightly linked in chromosome 4. In Phaseolus however, we were able to find only one copy of MAPK ortholog of MAPK7/MAPK14 paralogs. Loss of functionally redundant duplicate genes could be one of the measures to reduce the nitrogen and phosphorus expenses in plants during DNA synthesis, especially in plants with higher nitrogen requisition such as legumes. This conservation of nitrogen and phosphorus through genome reduction has been reported in free-living prokaryotes.41 We were also able to recover the orthologs of MAPK3 and MAPK6 from the genomes of all 3 species, and not surprisingly their orthologs in soybean had 2 paralogs each. Occurrence of at least 2 copies of MAPK5, MAPK20 and MAPK23 (exception: MtMAPK20) in Lj, Mt, and Pv and 4 copies in Gm indicates that the expansion of these genes occurred before the divergence of these species. The emergence of paralogous copies of MAPK23 orthologs in Gm, Lj, Mt, and Pv are inferred to have resulted from the genome duplication that occurred before the divergence of legume species. Two additional copies of GmMAPK23 located in the same chromosome (GmMAPK23?3 and GmMAPK23?4 in chromosome 16) are inferred to have resulted from tandem duplication.6 We found new clade of MAPK members in legume, with possible legume specific functions, which offers novel research areas to investigate. MAPK gene members with TQY motif in legume specific clade are also unique to nematodes such as Caenorhabditis elegans [GenBank: NP_494947.2], Caenorhabditis brenneri [GenBank: EGT51072.1], Brugia malayi [GenBank: XP_001896626.1], Loa loa [GenBank: XP_003140630.1], Caenorhabditis remanei [GenBank: XP_003109019.1], and Caenorhabditis briggsae [GenBank: XP_002630655.1]. The gene members with TQY motif including majority of the gene members in clade E also showed high gene expression values. Studies have shown that nematodes such as C. elegans are able to identify the bacteria for their food needs,42,43 and they mediate in establishing legume-rhizobia symbiotic interactions.44 This presence of the MAPK members with TQY motif in distantly related organisms is intriguing evolutionary event that warrants further investigation.
Phylogenetic reinterpretation of evolutionarily related MAPK models
Although the Arabidopsis MAPK model1 provides a systematic nomenclature to be adopted and expanded to newly sequenced genomes, the paralogous/orthologous status of the duplicated genes could not be interpreted using this model. Later, the MAPK nomenclature model was redesigned in the study of rice and poplar MAPKs4 to elucidate the evolutionary relationships among the duplicated genes. Nomenclature covering gene duplication event would be appropriate to the genomes of species that have evolved by genome duplications. One of such genomes is Arabidopsis thaliana itself,45 although it is commonly treated as a diploid species. In this study, we also reinterpreted the evolutionary relationship of the Arabidopsis MAPK genes that have been previously studied. Some of the MAPK gene members that we infer paralogous in Arabidopsis are: AtMAPK1 and AtMAPK2, AtMAPK4 and AtMAPK11, AtMAPK7 and AtMAPK14, AtMAPK8 and AtMAPK15, AtMAPK18 and AtMAPK19 in MAPK subfamily and AtMAPKK1 and AtMAPKK2, AtMAPKK4 and AtMAPKK5 and AtMAPKK7, AtMAPKK8 and AtMAPKK9 in MAPKK subfamily. This interpretation of AtMAPKs and the expansion of soybean MAPKs furnish an intriguing array of inferences about the evolutionary processes/relationships among duplicated genes. In legumes, most of these genes within each clade retain a higher level of sequence identity (more than 70% in most cases) indicating not only divergent evolution and their emergence from common ancestry, but also their high level of structural and functional conservation as inferred for their orthologs in Arabidopsis.1 The gene structure (Fig. S3) for MAPK orthologs in both legumes and non-legumes share a similar pattern, for example, the gene members in clade C have very few numbers of introns as compared with gene members in other clades. The gene members in clade E also share the similar distribution of exons throughout the species. Likewise, as discussed in the study of Arabidopsis4 and soybean,6 MAPKK gene members in clade C and clade D showed highly conserved pattern of intron/exon distribution with the presence of only one exon (see Fig. 5). However, in the case of very few gene members such as MtMAPKK9, PvMAPKK8, or GmMAPKK8 are presumably under the influence of structural divergence with varying number of exons.
Functional divergence
Orthologs of various MAPK gene members across legume species exhibited functional divergence. MAPK genes in soybean with a relatively higher number of duplicates (paralogs) compared with those in Lotus, Medicago, and Phaseolus may be under the influence of selection pressure as inferred from gene expression values which needs further evaluation using selection pressure analysis. Ancient polyploidization of the soybean genome must have led to diversification of genes, functional divergence, as well as redundancy: As expected of polyploidization, many duplicated gene members may have undergone neofunctionalization, subfunctionalization, hypofunctionalization, or non-functionalization as discussed in some literature.39,46 Almost all MAPK genes in Lotus and Medicago (except LjMAPK7?4 and MtMAPK20?3) have high expression (Figs. 2, 3, 4 and Figs. 6, 7, 8), whereas their orthologs and duplicate copies in soybean6 are differentially expressed in different tissues. Many MAPK genes (particularly one or more duplicates of GmMAPK5, GmMAPK9, GmMAPK11, GmMAPK16) in soybean have low to null gene expression values. Similarly, gene expression analysis also indicates that the paralogs of MAPKK10 and MAPKK8 in legumes have no or very low gene expression values consistent with the study on their orthologs in rice, poplar, and Arabidopsis.4 Orthologs of MAPKK8 and MAPKK10 in Medicago, however, could not be recovered. As in previous findings on rice, poplar, and Arabidopsis,4 orthologs of MAPKK10 in all legumes lack a complete activation loop (S/TxxxxxS/T), and whether these genes in some species are about to undergo extinction and they are lost in other requires further investigation.
Conclusions
Identification and phylogenetic analyses of MAPKs and MAPKKs in 4 legume species have revealed strongly supported clades consistent to the founder clades in Arabidopsis. The numbers of MAPKs were 19, 18, and 15, and that of MAPKKs were 7, 4, and 9 in Lotus japonicas (Lj), Medicago truncatula (Mt), and Phaseolus vulgaris (Pv) genomes, respectively. Comparative genomics of MAPKs in 4 legumes (Glycine, Lotus, Medicago, and Phaseolus) along with their orthlogs in 3 non-legume species [Arabidopsis thaliana (At), Poplar trichocarpa (Pt), and Oryza sativa (Os)] allowed us to: 1) recover a potential legume specific clade, 2) infer interesting evolutionary relationships among genes, and 3) assess evolutionary processes/events about expansion or reduction of these genes through genome duplication or chromosomal rearrangement. Our results indicate the expansion of GmMAPK families in soybean relative to Lotus, Medicago, and Phaseolus due to the ancient genome duplications and recent chromosomal rearrangements specific to soybean genomes. Expression profiles based on transcriptomic data indicates differential expression patterns among duplicate genes within soybean as opposed to more constant expression patterns of these genes within each of Lotus japonicus, Medicago truncatula, and Phaseolus vulgaris. Diversification and expansion of the MAPK gene families through gene/genome duplication may result into functional novelties. Identification of legume specific clade and MAPK members with TQY motif in all legume species is central to our findings, which need further investigation.
Materials and Methods
Identification of MAPK gene members in Lotus, Medicago, and Phaseolus
Hidden Markov Model (HMM) based searches were performed to identify MAPK and MAPKKs in Lotus japonicas (Lj), Medicago truncatula (Mt), and Phaseolus vulgaris (Pv) using 20 Arabidopsis MAPK and 10 MAPKK aligned protein sequences. HMM profile was built using HMM version 3.0 (http://hmmer.janelia.org/) in our Linux platform, and homology searches were performed against the protein data set of Mt, Pv downloaded from (phytozome.net), and Lj downloaded from (http://www.kazusa.or.jp/lotus). The putative MAPK and MAPKK protein sequences within the inclusion threshold of e-value 0.01 were selected for the BLOSUM based alignment in ClustalW version 2.048 as explained in.4 The results were accepted only if they possessed consensus for MAPK-related motifs and conserved domains.1,2 MEME (Multiple Expectation-maximization for Motif Elicitation) was used to predict similarities among protein sequences and visualize conserved motifs in specific subdomains of MAPK, MAPKK, and MAPKKK employing 2 conditions: 1) ideal motif width was set to be between 6 and 50, and 2) the motif search was set to identify 10 motif regions.49 Previously identified MAPKs from non-legume species (Arabidopsis, TAIR; http://arabidopsis.org,2,3 rice, and poplar4) were used to perform comparative study along with their orthologs in the legume species.
Phylogenetic analysis, intron-exon structure, and expression data
Sequence identity greater than 40% was used to detect homology among the protein sequences.50-52 Homology confidence among the protein sequences was established using Maximum Likelihood estimates of pairwise genetic distances and phylogenetic placements. Phylogenetic placements along with the high sequence identity (> 40%) among the genes were used to define paralogous status of the duplicated genes and to assign a number to the identified MAPK and MAPKK gene members of Lotus, Medicago, and Phaseolus (Table S1). Both parametric (ML) and non-parametric (MP) methods of phylogenetic analyses were performed to infer the orthologous and paralogous relationships. Maximum Likelihood analyses were performed using the best-fit evolutionary model integrated in the program MEGA5.2.2.53 Branch supports were estimated using bootstrap support54 of 2000 and 100 replications for MP and ML analyses, respectively. Human MAP Kinase sequences HsMAPK1 [GenBank: NP_002736.3] and HsMAPKK1 [GenBank: AAI37460.1] were used as outgroup. The gene structure (with intron/exon distribution) of all MAPK gene members included in the analyses were created using the programs Geneious55 and FancyGene.57 In order to investigate functional divergence in duplicated gene members, we downloaded RNaseq expression data of Glycine from RNaseq atlas (57; www.soybase.org), of Lotus and Medicago from LegumeIP (http://plantgrn.noble.org/LegumeIP/), Affymetrix microarray data of Phaseolus from (http://www.ncbi.nlm.nih.gov/geo),58 and that of Arabidopsis from MPSS database (_ENREF_5859; http://mpss.udel.edu/; accessed May 12, 2012). Mayday workbench60 was used to create a heatmap visualization of expression data. For the heatmap visualization, normalized gene expression at different growth conditions, tissues, and at different stress levels were log 2- based to explore functional divergence across the species.
Supplementary Material
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Author Contributions
Neupane A performed data mining, in silico and phylogenetic analyses, investigated the gene family structure and their functional/evolutionary relationships through comparative genomics and drafted the manuscript. Co-first author Nepal MP conceived, designed, and coordinated the original project, supervised data analyses, and helped in drafting the manuscript. Benson BV, Piya S, and MacArthur KJ contributed useful discussion and helped in data mining, drawing gene models, and expression data analysis.
Acknowledgments
This research was supported by South Dakota Soybean Research and Promotion Council (SDSRPC), Center for Excellence in Drought Tolerance Research (CEDTR), South Dakota Agricultural Experiment Station (SDAES), and Faculty Start up fund to Nepal MP from the Department of Biology and Microbiology of South Dakota State University.
References
- 1.Ichimura K, Shinozaki K, Tena G, Sheen J, Henry Y, Champion A, Kreis M, Zhang SQ, Hirt H, Wilson C, et al. MAPK Group. Mitogen-activated protein kinase cascades in plants: a new nomenclature. Trends Plant Sci. 2002;7:301–8. doi: 10.1016/S1360-1385(02)02302-6. [DOI] [PubMed] [Google Scholar]
- 2.Jonak C, Okrész L, Bögre L, Hirt H. Complexity, cross talk and integration of plant MAP kinase signalling. Curr Opin Plant Biol. 2002;5:415–24. doi: 10.1016/S1369-5266(02)00285-6. [DOI] [PubMed] [Google Scholar]
- 3.Tena G, Asai T, Chiu WL, Sheen J. Plant mitogen-activated protein kinase signaling cascades. Curr Opin Plant Biol. 2001;4:392–400. doi: 10.1016/S1369-5266(00)00191-6. [DOI] [PubMed] [Google Scholar]
- 4.Hamel LP, Nicole MC, Sritubtim S, Morency MJ, Ellis M, Ehlting J, Beaudoin N, Barbazuk B, Klessig D, Lee J, et al. Ancient signals: comparative genomics of plant MAPK and MAPKK gene families. Trends Plant Sci. 2006;11:192–8. doi: 10.1016/j.tplants.2006.02.007. [DOI] [PubMed] [Google Scholar]
- 5.Chen L, Hu W, Tan S, Wang M, Ma Z, Zhou S, Deng X, Zhang Y, Huang C, Yang G, et al. Genome-wide identification and analysis of MAPK and MAPKK gene families in Brachypodium distachyon. PLoS ONE. 2012;7:e46744. doi: 10.1371/journal.pone.0046744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Neupane A, Nepal MP, Piya S, Subramanian S, Rohila JS, Reese RN, Benson BV. Identification, nomenclature, and evolutionary relationships of mitogen-activated protein kinase (MAPK) genes in soybean. Evol Bioinform Online. 2013;9:363–86. doi: 10.4137/EBO.S12526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Asai T, Tena G, Plotnikova J, Willmann MR, Chiu WL, Gomez-Gomez L, Boller T, Ausubel FM, Sheen J. MAP kinase signalling cascade in Arabidopsis innate immunity. Nature. 2002;415:977–83. doi: 10.1038/415977a. [DOI] [PubMed] [Google Scholar]
- 8.Miya A, Albert P, Shinya T, Desaki Y, Ichimura K, Shirasu K, Narusaka Y, Kawakami N, Kaku H, Shibuya N. CERK1, a LysM receptor kinase, is essential for chitin elicitor signaling in Arabidopsis. Proc Natl Acad Sci USA. 2007;104:19613–8. doi: 10.1073/pnas.0705147104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ren D, Liu Y, Yang KY, Han L, Mao G, Glazebrook J, Zhang S. A fungal-responsive MAPK cascade regulates phytoalexin biosynthesis in Arabidopsis. Proc Natl Acad Sci USA. 2008;105:5638–43. doi: 10.1073/pnas.0711301105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ren D, Yang H, Zhang S. Cell death mediated by MAPK is associated with hydrogen peroxide production in Arabidopsis. J Biol Chem. 2002;277:559–65. doi: 10.1074/jbc.M109495200. [DOI] [PubMed] [Google Scholar]
- 11.Kishi-Kaboshi M, Okada K, Kurimoto L, Murakami S, Umezawa T, Shibuya N, Yamane H, Miyao A, Takatsuji H, Takahashi A, et al. A rice fungal MAMP-responsive MAPK cascade regulates metabolic flow to antimicrobial metabolite synthesis. Plant J. 2010;63:599–612. doi: 10.1111/j.1365-313X.2010.04264.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ding X, Richter T, Chen M, Fujii H, Seo YS, Xie M, Zheng X, Kanrar S, Stevenson RA, Dardick C, et al. A rice kinase-protein interaction map. Plant Physiol. 2009;149:1478–92. doi: 10.1104/pp.108.128298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jung KH, Cao P, Seo YS, Dardick C, Ronald PC. The Rice Kinase Phylogenomics Database: a guide for systematic analysis of the rice kinase super-family. Trends Plant Sci. 2010;15:595–9. doi: 10.1016/j.tplants.2010.08.004. [DOI] [PubMed] [Google Scholar]
- 14.Krisak L, Strich R, Winters RS, Hall JP, Mallory MJ, Kreitzer D, Tuan RS, Winter E. SMK1, a developmentally regulated MAP kinase, is required for spore wall assembly in Saccharomyces cerevisiae. Genes Dev. 1994;8:2151–61. doi: 10.1101/gad.8.18.2151. [DOI] [PubMed] [Google Scholar]
- 15.Wagner M, Briza P, Pierce M, Winter E. Distinct steps in yeast spore morphogenesis require distinct SMK1 MAP kinase thresholds. Genetics. 1999;151:1327–40. doi: 10.1093/genetics/151.4.1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Breitkreutz A, Boucher L, Tyers M. MAPK specificity in the yeast pheromone response independent of transcriptional activation. Curr Biol. 2001;11:1266–71. doi: 10.1016/S0960-9822(01)00370-0. [DOI] [PubMed] [Google Scholar]
- 17.Elion EA, Satterberg B, Kranz JE. FUS3 phosphorylates multiple components of the mating signal transduction cascade: evidence for STE12 and FAR1. Mol Biol Cell. 1993;4:495–510. doi: 10.1091/mbc.4.5.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Herskowitz I. MAP kinase pathways in yeast: for mating and more. Cell. 1995;80:187–97. doi: 10.1016/0092-8674(95)90402-6. [DOI] [PubMed] [Google Scholar]
- 19.Chang F, Herskowitz I. Identification of a gene necessary for cell cycle arrest by a negative growth factor of yeast: FAR1 is an inhibitor of a G1 cyclin, CLN2. Cell. 1990;63:999–1011. doi: 10.1016/0092-8674(90)90503-7. [DOI] [PubMed] [Google Scholar]
- 20.Chen RE, Thorner J. Function and regulation in MAPK signaling pathways: lessons learned from the yeast Saccharomyces cerevisiae. Biochim Biophys Acta 2007; 1773:1311-40; [DOI] [PMC free article] [PubMed]
- 21.Madhani HD, Fink GR. The riddle of MAP kinase signaling specificity. Trends Genet. 1998;14:151–5. doi: 10.1016/S0168-9525(98)01425-5. [DOI] [PubMed] [Google Scholar]
- 22.Truckses DM, Garrenton LS, Thorner J. Jekyll and Hyde in the microbial world. Science. 2004;306:1509–11. doi: 10.1126/science.1104677. [DOI] [PubMed] [Google Scholar]
- 23.Sabbagh W, Jr., Flatauer LJ, Bardwell AJ, Bardwell L. Specificity of MAP kinase signaling in yeast differentiation involves transient versus sustained MAPK activation. Mol Cell. 2001;8:683–91. doi: 10.1016/S1097-2765(01)00322-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Levin DE. Cell wall integrity signaling in Saccharomyces cerevisiae. Microbiol Mol Biol Rev. 2005;69:262–91. doi: 10.1128/MMBR.69.2.262-291.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dihazi H, Kessler R, Eschrich K. High osmolarity glycerol (HOG) pathway-induced phosphorylation and activation of 6-phosphofructo-2-kinase are essential for glycerol accumulation and yeast cell proliferation under hyperosmotic stress. J Biol Chem. 2004;279:23961–8. doi: 10.1074/jbc.M312974200. [DOI] [PubMed] [Google Scholar]
- 26.Zhou Z, Gartner A, Cade R, Ammerer G, Errede B. Pheromone-induced signal transduction in Saccharomyces cerevisiae requires the sequential function of three protein kinases. Mol Cell Biol. 1993;13:2069–80. doi: 10.1128/mcb.13.4.2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grimsrud PA, den Os D, Wenger CD, Swaney DL, Schwartz D, Sussman MR, Ané JM, Coon JJ. Large-scale phosphoprotein analysis in Medicago truncatula roots provides insight into in vivo kinase activity in legumes. Plant Physiol. 2010;152:19–28. doi: 10.1104/pp.109.149625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Khan GA, Declerck M, Sorin C, Hartmann C, Crespi M, Lelandais-Brière C. MicroRNAs as regulators of root development and architecture. Plant Mol Biol. 2011;77:47–58. doi: 10.1007/s11103-011-9793-x. [DOI] [PubMed] [Google Scholar]
- 29.Schoenbeck MA, Samac DA, Fedorova M, Gregerson RG, Gantt JS, Vance CP. The alfalfa (Medicago sativa) TDY1 gene encodes a mitogen-activated protein kinase homolog. Mol Plant Microbe Interact. 1999;12:882–93. doi: 10.1094/MPMI.1999.12.10.882. [DOI] [PubMed] [Google Scholar]
- 30.Liu JZ, Horstman HD, Braun E, Graham MA, Zhang C, Navarre D, Qiu WL, Lee Y, Nettleton D, Hill JH, et al. Soybean homologs of MPK4 negatively regulate defense responses and positively regulate growth and development. Plant Physiol. 2011;157:1363–78. doi: 10.1104/pp.111.185686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fernandez-Pascual M, Lucas MM, de Felipe MR, Boscá L, Hirt H, Golvano MP. Involvement of mitogen-activated protein kinases in the symbiosis Bradyrhizobium-Lupinus. J Exp Bot. 2006;57:2735–42. doi: 10.1093/jxb/erl038. [DOI] [PubMed] [Google Scholar]
- 32.Cooper JE. Early interactions between legumes and rhizobia: disclosing complexity in a molecular dialogue. J Appl Microbiol. 2007;103:1355–65. doi: 10.1111/j.1365-2672.2007.03366.x. [DOI] [PubMed] [Google Scholar]
- 33.Parniske M. Intracellular accommodation of microbes by plants: a common developmental program for symbiosis and disease? Curr Opin Plant Biol. 2000;3:320–8. doi: 10.1016/S1369-5266(00)00088-1. [DOI] [PubMed] [Google Scholar]
- 34.Herouart D, Baudouin E, Frendo P, Harrison J, Santos R, Jamet A, Van de Sype G, Touati D, Puppo A. Reactive oxygen species, nitric oxide and glutathione: a key role in the establishment of the legume-Rhizobium symbiosis? Plant Physiol Biochem. 2002;40:619–24. doi: 10.1016/S0981-9428(02)01415-8. [DOI] [Google Scholar]
- 35.Schmutz J, Cannon SB, Schlueter J, Ma J, Mitros T, Nelson W, Hyten DL, Song Q, Thelen JJ, Cheng J, et al. Genome sequence of the palaeopolyploid soybean. Nature. 2010;463:178–83. doi: 10.1038/nature08670. [DOI] [PubMed] [Google Scholar]
- 36.Sato S, Nakamura Y, Kaneko T, Asamizu E, Kato T, Nakao M, Sasamoto S, Watanabe A, Ono A, Kawashima K, et al. Genome structure of the legume, Lotus japonicus. DNA Res. 2008;15:227–39. doi: 10.1093/dnares/dsn008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Young ND, Cannon SB, Sato S, Kim D, Cook DR, Town CD, Roe BA, Tabata S. Sequencing the genespaces of Medicago truncatula and Lotus japonicus. Plant Physiol. 2005;137:1174–81. doi: 10.1104/pp.104.057034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gepts P. Phaseolus vulgaris (Beans). In: Sydney B, Jeffrey HM, eds. Encyclopedia of Genetics. New York: Academic Press, 2001:1444-5. [Google Scholar]
- 39.Lynch M, Conery JS. The evolutionary fate and consequences of duplicate genes. Science. 2000;290:1151–5. doi: 10.1126/science.290.5494.1151. [DOI] [PubMed] [Google Scholar]
- 40.Zhang JZ. Evolution by gene duplication: an update. Trends Ecol Evol. 2003;18:292–8. doi: 10.1016/S0169-5347(03)00033-8. [DOI] [Google Scholar]
- 41.Dufresne A, Garczarek L, Partensky F. Accelerated evolution associated with genome reduction in a free-living prokaryote. Genome Biol. 2005;6:R14. doi: 10.1186/gb-2005-6-2-r14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Abada EA, Sung H, Dwivedi M, Park B-J, Lee S-K, Ahnn J. C. elegans behavior of preference choice on bacterial food. Mol Cells. 2009;28:209–13. doi: 10.1007/s10059-009-0124-x. [DOI] [PubMed] [Google Scholar]
- 43.Coolon JD, Jones KL, Todd TC, Carr BC, Herman MA. Caenorhabditis elegans genomic response to soil bacteria predicts environment-specific genetic effects on life history traits. PLoS Genet. 2009;5:e1000503. doi: 10.1371/journal.pgen.1000503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Horiuchi J, Prithiviraj B, Bais HP, Kimball BA, Vivanco JM. Soil nematodes mediate positive interactions between legume plants and rhizobium bacteria. Planta. 2005;222:848–57. doi: 10.1007/s00425-005-0025-y. [DOI] [PubMed] [Google Scholar]
- 45.Schranz ME, Mitchell-Olds T. Independent ancient polyploidy events in the sister families Brassicaceae and Cleomaceae. Plant Cell. 2006;18:1152–65. doi: 10.1105/tpc.106.041111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Force A, Lynch M, Pickett FB, Amores A, Yan YL, Postlethwait J. Preservation of duplicate genes by complementary, degenerative mutations. Genetics. 1999;151:1531–45. doi: 10.1093/genetics/151.4.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ohno S. Evolution by gene duplication. Berlin, New York: Springer-Verlag, 1970. [Google Scholar]
- 48.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. Clustal W and clustal X version 2.0. Bioinformatics. 2007;23:2947–8. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 49.Bailey TL, Elkan C. The value of prior knowledge in discovering motifs with MEME. Proceedings / International Conference on Intelligent Systems for Molecular Biology; ISMB International Conference on Intelligent Systems for Molecular Biology 1995; 3:21-9. [PubMed] [Google Scholar]
- 50.Petsko GA, Ringe D. Protein structure and function. Oxford: New Science Press, 2004. [Google Scholar]
- 51.Rost B. Twilight zone of protein sequence alignments. Protein Eng. 1999;12:85–94. doi: 10.1093/protein/12.2.85. [DOI] [PubMed] [Google Scholar]
- 52.Lesk AM. Introduction to protein architecture: the structural biology of proteins. Oxford, New York: Oxford University Press, 2001. [Google Scholar]
- 53.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–9. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–91. doi: 10.2307/2408678. [DOI] [PubMed] [Google Scholar]
- 55.Drummond AJ, Ashton B, Buxton S, Cheung M, Cooper A, Heled J, Kearse M, Moir R, Stones-Havas S, Sturrock S, et al. Geneious v5.5, available from http://www.geneious.com 2011.
- 56.Rambaldi D, Ciccarelli FD. FancyGene: dynamic visualization of gene structures and protein domain architectures on genomic loci. Bioinformatics. 2009;25:2281–2. doi: 10.1093/bioinformatics/btp381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Severin AJ, Woody JL, Bolon YT, Joseph B, Diers BW, Farmer AD, Muehlbauer GJ, Nelson RT, Grant D, Specht JE, et al. RNA-Seq Atlas of Glycine max: a guide to the soybean transcriptome. BMC Plant Biol. 2010;10:160. doi: 10.1186/1471-2229-10-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–10. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nakano M, Nobuta K, Vemaraju K, Tej SS, Skogen JW, Meyers BC. Plant MPSS databases: signature-based transcriptional resources for analyses of mRNA and small RNA. Nucleic Acids Res. 2006;34:D731–5. doi: 10.1093/nar/gkj077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Battke F, Symons S, Nieselt K. Mayday--integrative analytics for expression data. BMC Bioinformatics. 2010;11:121. doi: 10.1186/1471-2105-11-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.