Abstract
Salicylic acid (SA) is a prominent signaling molecule during biotic and abiotic stresses in plants biosynthesized via cinnamate and isochorismate pathways. Cinnamate 4-hydroxylase (C4H) and isochorismate synthase (ICS) are the main enzymes in phenylpropanoid and isochorismate pathways, respectively. To investigate the actual roles of these genes in resistance mechanism to environmental stresses, here, the coding sequences of these enzymes in safflower (Carthamus tinctorius), as an oilseed industrial medicinal plant, were partially isolated and their expression profiles during salinity stress, wounding, and salicylic acid treatment were monitored. As a result, safflower ICS (CtICS) and C4H (CtC4H) were induced in early time points after wounding (3–6 h). Upon salinity stress, CtICS and CtC4H were highly expressed for the periods of 6–24 h and 3–6 h after treatment, respectively. It seems evident that ICS expression level is SA concentration dependent as if safflower treatment with 1 mM SA could induce ICS much stronger than that with 0.1 mM, while C4H is less likely to be so. Based on phylogenetic analysis, safflower ICS has maximum similarity to its ortholog in Vitis vinifera up to 69%, while C4H shows the highest similarity to its ortholog in Echinacea angustifolia up to 96%. Overall, the isolated genes of CtICS and CtC4H in safflower could be considered in plant breeding programs for salinity tolerance as well as for pathogen resistance.
Keywords: Cinnamate 4-hydroxylase, Isochorismate synthase, Safflower, Salicylic acid, Salinity, Wounding
Introduction
Safflower (Carthamus tinctorius) belonging to Asteraceae is mainly cultivated as an oilseed plant; however, its petals and kernels are rich in natural antioxidants.1 It is evident that safflower extracts of leaves, roots, kernels, and especially petals are able to scavenge the reactive oxygen species (ROS) and contain the active compounds such as carthamin and polyphenols.2 According to FAO statistics in 2012, the world production of safflower in 2010 reached to more that 6 × 105 tons out of about 8 × 105 hectares.
Plant immune responses are modulated by a fine-tuned crosstalk among the major signaling molecules, i.e., salicylic acid (SA), jasmonic acid (JA), and ethylene (Et).3 Shikimate pathway, also present in bacteria and fungi, is one of the most crucial pathways for primary and secondary metabolite biosynthesis in plants.4 This is a part of the biosynthetic pathway of volatile organic compounds and is recognized as the biosynthetic pathway of aromatic compounds.5 This metabolic pathway converts the carbohydrates, sequentially, to produce the ultimate product, chorismate. Chorismate per se is the last common precursor for phenylalanine, tyrosine, and tryptophan.4 Aromatic amino acids, consumed during protein synthesis, are precursors for a vast range of secondary metabolites pivotal for plant growth, development, and defense responses.6,7
Besides being involved in plant growth and development, photosynthesis, transpiration, etc., salicylic acid is a key signaling molecule in response to environmental stresses. SA biosynthesis in plants occurs via two pathways, i.e., cinnamate (phenylpropanoid) and isochorismate. The cinnamate pathway can be further split into the paths involving benzoic acid8 and o-coumarate as precursors (Fig. 1). Genomic studies in Arabidopsis have revealed that the major portion of SA synthesis befalls via isochorismate pathway.6 However, plant species capable of SA biosynthesis via isochorismate pathway are relatively minor compared with the species sharing the cinnamate pathway. Isochorismate synthase (ICS) catalyzes the conversion of isochorismate to finally yield SA.7 Phenylpropanoid pathway is, as well, one of the main plant pathways for secondary metabolite biosynthesis.6 It is estimated that 20 percent of carbon fixation by photosynthesis is directly contributing to this pathway.9 The products of this pathway include different classes of flavonoids, e.g., chalcones, flavonones, flavones, flavonoles, anthocyanins, and isoflavonones (Fig. 1). Flavonoids are generally biosynthesized in many plants and take diverse tasks as flower pigments, UV protectants, and antimicrobials.6,9,10 Phenolic compounds are synthesized in the cells in unstressed condition; however a/biotic stresses alter their synthesis rates. In reality, alterations in activities of biosynthesizing or degrading enzymes of these products affect their quantities in the cell.11 Activity of phenylpropanoid pathway is exceptionally crucial for plant protection against stress damages.12 Cinnamate 4-hydroxylase (C4H) is one of the leading enzymes in phenylpropanoid pathway, which catalyzes the second step of the pathway in converting the trans-cinnamate to coumarate. C4H is the member of P450 protein family and catalyzes the monooxygenation of vast variety of substrates.13 The flavonoid pathway is a part of phenylpropanoid pathway, synthesizing also other secondary metabolites like alkaloids, lignin, suberin, and monolignols. It is evident that flavonoids act as antimicrobials and antioxidants during microbial challenges in some plants.14,15
Figure 1. Salicylic acid biosynthesis pathways. Isochorismate synthase (ICS) in isochorismate pathway. The 1–7 in shikimate pathway refers to 7 steps to produce the final product chorismate. Isochorismate synthase (ICS) and cinnamate 4-hydroxylase (C4H) enzymes in respective isochorismate and phenylpropanoid pathways are shown in red. The scheme was adapted after refs 61-64.
Mechanical wounding in plants might be resulted from many environmental stress sources, e.g., wind, sand, rainfall, herbivores, etc. As a rule, plants react to wounding through induction of diverse genes given that an open wound is well prone to microbial infections. Therefore, local expression of defense genes in wound periphery prevents the entry of assaulting microorganisms.16 Wound signaling in plants is a complex process including many regulatory molecules and other inducible defense responses.17 Among the molecular components of wound signal perception and transduction, jasmonate and MAP kinases are of high significance. Plants activate the phenylpropanoid metabolism as a critical defense response against wounding and pathogenic attacks. As a matter of fact, augmentation of carbon flow via this pathway leads to the accumulation of defense compounds with structural and/or antimicrobial properties.18,19
Abiotic stresses are within the main causes of yield loss, worldwide, and salinity, in particular, is becoming a global agricultural crisis.20 Salinity imposes a double-loss on agriculture by limiting both the yield and the arable lands, thereby necessitating a deep understanding of plant responses to this abiotic stress.21 Tolerance to salinity is a complicated trait and is governed by several quantitative trait loci (QTLs). These QTLs are involved in many pathways, concurrently, including salinity signal perception and transduction culminating in the overall response of plant.20 High soil salinity results in ion toxicity as well as hyperosmosis, which by itself leads to ROS production and, ultimately, programmed cell death.22 Different abiotic stresses pathways share common components in their signaling cascades with a sophisticated interplay23 for maintenance of the homeostasis and lowering the fitness cost.24 In Arabidopsis, for instance, salinity and drought stress signaling cascades involve the phenylpropanoid pathway.25 The cinnamate-benzoate path is more common than cinnamate-coumarate one for SA biosynthesis in stress condition in different plant species, e.g., water deficit as well as UV irradiation in barley,26 ozone in tobacco,27 thermal stimuli in pea,28 and salt stress in rice.29 In this study, regardless of possible involvement of cinnamate-benzoate pathway, we report the molecular cloning and functional analyses of 2 important genes, isochorismate synthase and cinnamate 4-hydroxylase, in safflower, involved in responses to stresses, i.e., salinity stress, wounding, and salicylic acid treatment.
Results and Discussion
Isolation of CtICS from safflower
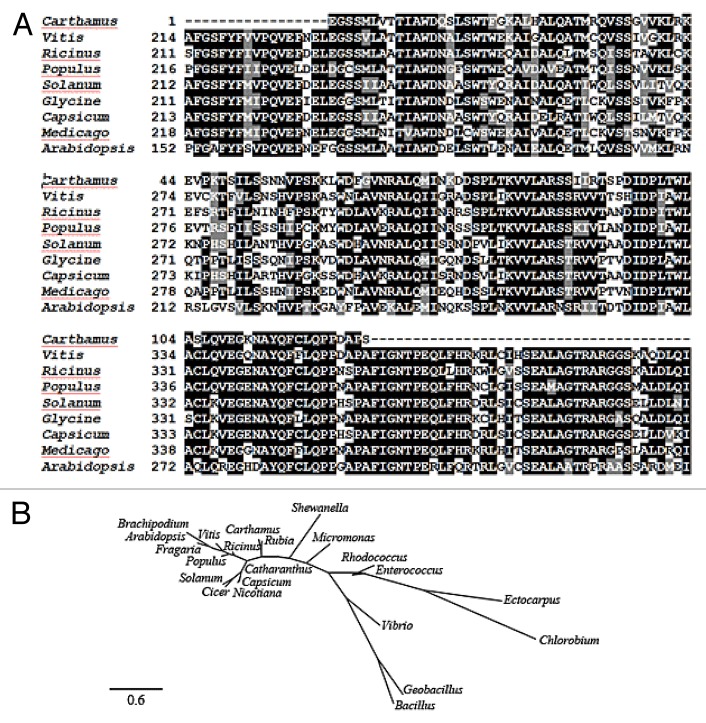
Several ICS cDNAs have been isolated from prokaryotes as well as eukaryotes. Isochorismate synthase (EC 5.4.4.2) belonging to the family of isomerases is known to catalyze the conversion of chorismate to isochorismate. It is implicated that ICSs mediate production of salicylate from chorismate,30 synthesis of phenolate siderophore amonabactin,31 biosynthesis of menaquinone,32 etc. in prokaryotes. In plants, very few peer-reviewed reports on ICS isolation and characterization are available. In Arabidopsis, it is demonstrated that ICS is involved in the synthesis of salicylic acid required for both local and systemic acquired resistance.33 As well, it is involved in phylloquinone (vitamin K1) synthesis.34 To look for ICS gene in C. tinctorius as an alternative source for salicylic acid biosynthesis, the available ICS coding sequences in GenBank were considered to design isolating primers (Table 1). The validated partial sequence of CtICS was deposited in GenBank under accession number of JQ772500. The inferred amino acid sequence of CtICS (Accession: AFI57884) was blasted against the non-redundant protein sequences database, revealing that ICS in safflower (Asteraceae) shows considerable identities to orthologs in Vitis vinifera (Vitaceae), Ricinus communis (Euphorbiaceae), Populus tremuloides (Salicaceae), Solanum lycopersicum (Solanaceae), Glycine max (Fabaceae), Capsicum annuum (Solanaceae), Medicago truncatula (Fabaceae), and Arabidopsis thaliana (Brassicaceae) as shown in Figure 2A. The maximum identity, however, of CtICS is to chloroplastic isochorismate synthase 2 (Accession: XP_002267681) of grape (Vitis vinifera) up to 69%. Phylogenetic analysis of deduced amino acid sequence of CtICS in parallel with its eukaryotic and prokaryotic orthologs demonstrated that ICS gene is rather conserved in prokaryotes and eukaryotes (Fig. 2B). However, two main clusters are constructed each devoted to either plants or bacteria. As briefly mentioned earlier, isochorismate synthase orthologs are extensively characterized in bacteria, believed to be involved in siderophore and salicylic acid biosyntheses. As well, isochorismate synthases in plants are involved in salicylic acid biosynthesis, required for both local and systemic acquired resistance, while SA synthesized through phenylpropanoid pathway seems to potentiate the plant cell death.33 As expected, the plant ICSs cluster together aside from bacterial ICSs indicating that although being rather conserved in terms of involving in SA biosynthesis in bacteria and plants, plant ICSs are separate from their bacterial orthologs from the evolutionary point of view.
Table 1. Sequences of primes used in this study. Primers for each gene were designed according to the most conserved domains of orthologs in close species. The cDNA sequences of orthologs in GenBank were used for primer design.
| Gene | Primer | Sequence (5`- 3`) | Amplicon size |
|---|---|---|---|
| Cinnamate 4-hydroxylase | Ct-C4H-fwd | ATGGGGCAGC GCAACCT | 343 |
| Ct-C4H-rev | ACATGTTGTT GTACATCATC AGCTG | ||
| Isochorismate synthase | Ct-ICS-fwd | CTCAGGTTGA GTTTGATGAG CT | 419 |
| Ct-ICS-rev | CTCTGGAGTG TTTCCAATGA A | ||
| Isochorismate synthase (internal) | Ct-ICS2-fwd | ATGGACATTT GGGAAAGCAC | 195 |
| Ct-ICS2-rev | TTGGTAAGGG GTGAGTCGTC | ||
| 18S rRNA | 18S-fwd | ACTCACCTCA AGACT | 205 |
| 18S-rev | CTTTGGCACA TCC |
Figure 2. Amino acid sequence alignment (A) and phylogenetic analysis (B) of CtICS orthologs. The known ICS sequences fall into 2 main clusters each devoted to either plants or bacteria. The evolutionary history was inferred using the Neighbor-Joining method. The evolutionary distances were computed using the Poisson correction method and are in the units of the number of amino acid substitutions per site. All positions containing gaps and missing data were eliminated from the data set. Phylogenetic tree was constructed at Phylogeny.fr. Accession numbers of sequences are: Carthamus tinctorius (AFI57884), Ricinus communis (XP_002511526), Vitis vinifera (XP_002267681), Populus tremuloides (ACX46384), Fragaria vesca subsp. vesca (XP_004301164), Solanum lycopersicum (NP_001234794), Glycine max (XP_003522193), Capsicum annuum (AAW66457), Arabidopsis thaliana (AAC97926), Cicer arietinum (XP_004514127), Nicotiana tabacum (AAW67000), Catharanthus roseus (Q9ZPC0), Brachypodium distachyon (XP_003578021), Rubia cordifolia (ABK79678), Rhodococcus rhodnii (WP_010837681), Ectocarpus siliculosus (CBN77562), Bacillus cereus (WP_001003908), Micromonas pusilla (XP_003064233), Chlorobium limicola (YP_001944055), Geobacillus thermoglucosidasius (YP_004586325), Enterococcus gallinarum (WP_003127529), Shewanella frigidimarina (WP_011639322), and Vibrio anguillarum (NP_943589). The scale bar indicates 0.6 substitutions per site.
Isolation of safflower C4H (CtC4H)
Using conserved sequences of C4Hs, CtC4H partial sequence (343 bp) was cloned from safflower. The validated sequence of CtC4H was deposited in GenBank (Accession: JN998608). The inferred amino acid sequence of CtC4H (Accession: AFK25795) was blasted against non-redundant protein sequences database, revealing that the CtC4H shows the highest identity to its ortholog in Echinacea angustifolia up to 96% (Fig. 3A). The phylogenetic analyses based on amino acid sequences of C4H orthologs in different plant species disclosed that C4H gene is much conserved in Asterales, i.e., Carthamus tinctorius, Cynara cardunculus, Echinacea angustifolia, Gynura bicolor, Zinnia violacea, Helianthus tuberosus, and Artemisia annua constructing a separate cluster in the phylogenetic tree (Fig. 3B). However, as depicted in Figure 3B, cinnamate hydroxylases genes are considerably conserved in plants. The multi-enzymatic pathway of phenylpropanoid is in charge of biosynthesis of many secondary metabolites essential for many developmental processes in plants including disease resistance,3 and cinnamate 4-hydroxylase is the second enzyme in this pathway catalyzing the hydroxylation of trans-cinnamic acid to p-coumaric acid. In plant physiology, cinnamate hydroxylases control carbon flux to: 1) pigments influential on pollination and UV protection, 2) numerous phytoalexins synthesized by plants when challenged by pathogens, and 3) lignins,35 presenting it as one of the most critical enzymes in plants growth and development.
Figure 3. Amino acid sequence alignment (A) and phylogenetic analysis (B) of CtC4H with other orthologs in GenBank. The evolutionary history was inferred using the Neighbor-Joining method. Evolutionary distances were computed using the Poisson correction method and are in the units of the number of amino acid substitutions per site. All positions containing gaps and missing data were eliminated from the data set (Complete deletion option). Phylogenetic analyses were conducted at Phylogeny.fr. The sequences for C4H genes: Carthamus tinctorius (AFK25795), Cynara cardunculus var scolymus (CAM84301), Echinacea angustifolia (ACF74449), Helianthus tuberosus (CAA78982), Gynura bicolor (BAJ17666), Agastache foeniculum (BAJ22964), Coffea arabica (CAJ41419), Populus kitakamiensis (BAA11578), Artemisia annua (ADO16181), Zinnia violacea (AAB42024), Salvia miltiorrhiza (ABC75596), Solanum lycopersicum (XP_004242478), Glycine max (ACR44227), Fragaria vesca subsp. vesca (XP_004294725), Scutellaria baicalensis (ADN32769), Astragalus chrysochlorus (ACY06865), Vitis vinifera (CAN77208), Capsicum annuum (ACF19421), Cucumis sativus (CAK95273), Pisum sativum (AAG09205), and Arabidopsis thaliana (CAP08837). The scale bar indicates 0.04 substitutions per site.
CtICS and CtC4H responses to wounding
Induction of CtC4H in response to wounding stress takes place at 3 h after treatment (hat) and levels off thereafter, while CtICS highly induces a bit later at 6 hat with a decrease afterward (Fig. 4A). In Arabidopsis, C4H expression is clearly higher in roots than in leaves.36 Several lines of evidence substantiate the participation of phenylpropanoid pathway in wound signaling.18,37-39 Wounding affects the phenylpropanoid pathway by inducing PAL40,41 and C4H.42 PAL as the just upstream gene of C4H in this cascade is proved to be upregulated by wounding via different mediatory signals including, but not limited to, ethylene, methyl jasmonate, and abscisic acid.38 According to Mizutani et al. (1997),36 a C4H promoter region of 907 bp contains all of the 3 cis-acting elements (boxes P, A, and L) conserved among the PAL and 4CL genes, which per se substantiates that C4H expression is coordinated in Arabidopsis with PAL1 and 4CL in response to light and wounding. In this context, while wounding induces the PAL promoter, which is the starting gene in phenylpropanoid pathway and results in PAL mRNA accumulation and elevated expression, inhibitors of PAL transcription can reduce the responses to wounding.38 It is documented in lettuce that the PAL activity level correlates with the wound severity and contributes to wounding signal transduction and accumulation of phenolic compounds involved in secondary processes like tissue browning.38 As well, in parsley PAL and 4CL expression levels are locally boosted in response to wounding. Likewise, in tobacco plants transgenic for parsley 4CL promoter fragment, wounding results in the reporter gene induction.18 PAL transcript accumulation pattern in lettuce in response to wounding looks like a slight induction during the first 6 h with a climax at 12 h and leveling off at 24 h accompanied by ethylene production. Methyl jasmonate is one of the wound signal transducers, which increases the PAL mRNA transcript in cell culture. In addition, ABA can enhance the PAL activity after wounding in potato cell culture during suberization of cell wall.38 Mechanical wounding in leaf periphery of pine leads to abundant expression of phenylpropanoid and flavonoid genes. In this study, accordingly, CtC4H, induces significantly at 3 h after wounding and levels off later in a steady-state manner (Fig. 4A). Batard et al. (1997)43 reported that in Jerusalem Artichoke tubers wounding caused a biphasic induction of C4H at 10 and 20 h after wounding followed by steady-state content of C4H messenger decreasing until 50 h and continuing rather constantly for the next 2 days. C4H protein and enzymatic activity clearly showed the comparable pattern of transcript induction; however the peaks were shifted 10–20 h later than that observed for mRNA. Likewise, activity was to some extent delayed compared with protein accumulation. Comparably, in tea, the expression levels of C4H and PAL in response to wounding was induced at 12 h after treatment up to 45% relatively to 0 h and decreased thereafter.44 Here, the highest induction of CtICS, in contrast, is observed at 6 h after wounding (Fig. 4A). Flavonoid pathway produces, via phenylpropanoid pathway, certain secondary metabolites, which contribute to cell wall reinforcement as a defense response. According to present study, phenylpropanoid pathway in safflower is activated rapidly after wounding followed by activation, with delay, of isochorismate pathway, showing that both pathways get activated soon after wounding stress to eventually contribute to SA biosynthesis, which is generally approved as a fundamental signal transducer in plant defense.3 On the other hand, phenylpropanoid pathway contributes to production of a broad range of secondary metabolites crucial for plant protection against environmental threats. After 24 h, the gene transcriptions of both CtC4H and CtICS decrease (Fig. 4A) to prevent the unnecessary loss of resources after activation of downstream pathways, which could as such be the consequence of a negative feedback from downstream signals.
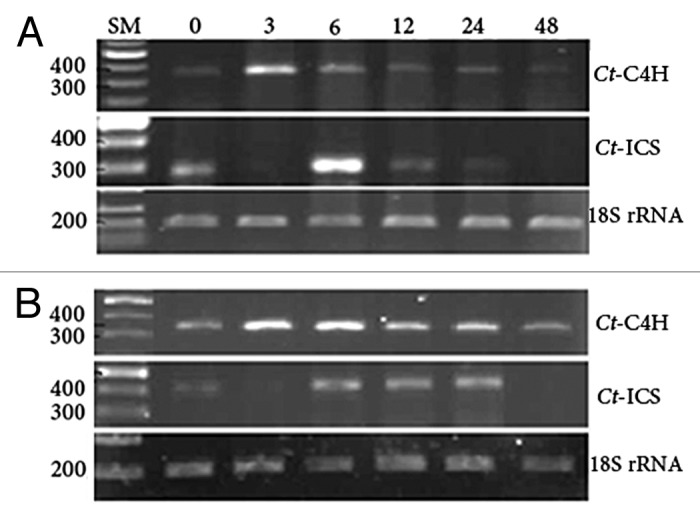
Figure 4. Expression profiles of CtC4H and CtICS genes after wounding (A) and during salinity stress (B). Samplings were performed at 0, 3, 6, 12, 24, and 48 h after treatment. The RNA was extracted from all treated plants and after DNaseI treatment, total RNA was reverse transcribed to cDNA. The cDNA amounts were first normalized by 18S rRNA PCR product intensity. For CtICS, the internal primers based on isolated sequence were designed and used for RT-PCR. Intensity of PCR products at different time points refers to as temporal expression level of the genes. 18S rRNA transcription levels serve as internal control.
Expression of CtICS and CtC4H in response to salinity stress
During salinity stress, CtC4H induces at 3 h after treatment (hat) and slowly settles down after 12 h. CtICS, however, is induced by salinity stress from 6 to 24 hat and calms down thereafter (Fig. 4B). It is proven that the salinity stress causes the induction of aromatic amino acids biosynthesis. Concurrent induction of both genes implicates that the lignin biosynthesis should also be boosted up by salinity stress, which by itself contributes to cell wall fortification.45 Among aromatic amino acids, tryptophan is the activator of positive feedback for phenylalanine (forerunner of phenylpropanoid pathway) and tyrosine biosyntheses. It is documented that expression of tryptophan biosynthesizing enzymes is induced in response to biotic as well as abiotic stresses including salinity.45 Since shikimate pathway triggers the tryptophan biosynthesis in plants and chorismate is the last common precursor of tryptophan, phenylalanine and tyrosine, when challenging with salinity, the higher activation of shikimate pathway and consequent higher chorismate production can lead to a higher activity of chorismate-driven pathways, e.g., phenylpropanoid and isochorismate. It has been recorded that phenylpropanoid pathway induces in salt-sensitive genotypes of rice, IR29, in response to salinity stress. On the other hand, Arabidopsis mutant uvs66, deficient in one of the genes in phenylpropanoid pathway and highly sensitive to UV, is highly sensitive to salinity.46 Association of phenylpropanoid pathway with salinity, ozone, and UV stresses concerns the oxidative stress.46 It is acknowledged that the long-term salt accumulation in plants leads to diminishing photosynthetic activity, increasing oxidative stress, and finally imposing an anaerobic condition as well as metabolic damages.45 This pathway retains a dual function in: 1) secondary metabolites production, e.g., lignin for cell wall fortification, and 2) scavenging the oxidative stress factors in consequence of salinity stress. Moreover, this pathway contributes to SA production. In wheat, the salinity-derived growth impairments could be evaded by exogenous application of SA. It results in augmented activation of antioxidant enzymes catalase, peroxidase, glutathione reductase, ascorbate peroxidase, and superoxide dismutase.47 Lee et al. (2010)48 also reported that although SA is not essential for Arabidopsis seed germination under normal growth condition, it does play a promotional role in seed germination under high salinity circumstances by decreasing the oxidative damage. Hence, induction of the corresponding components in those pathways namely C4H and ICS during salinity stress looks inevitable. Accordingly, results of this study revealed that in salinity-stressed plants, CtC4H gene is induced early after stress in 3–6 h and settles down subsequently (Fig. 4B). Salinity, on the other hand, induces the CtICS fairly later at 6–24 hat (Fig. 4B). Thus, after salinity stress, the phenylpropanoid pathway is quickly activated followed by activation of isochorismate pathway; altogether giving rise to SA and other metabolites biosyntheses and culminating in the overall protection of plant.
Responses of CtC4H and CtICS to SA treatment
In contrast to CtC4H, induction of CtICS gene upon SA treatment seems to be concentration-dependent. In 0.1 mM SA condition, CtC4H is rather highly induced in a biphasic manner at 3 hat and to some extent at 24 and 48 hat (Fig. 5A), which gets more intensified upon treatment with 1 mM SA (Fig. 5B). In contrast, the induction patterns for CtICS are different upon treatment with different SA concentrations. Treatment of plants with 1 mM SA results in a comparatively stronger induction of CtICS, even at 0 h, however in the following time points until 18 hours after treatment (hat) a decline in gene expression is observed. Again, at 24 h, an intense induction is visible, which repeats at 48 hat. This pattern is not evident when 0.1 mM SA is applied on plants. In plants treated with 1 mM SA, CtC4H gene was more transcribed at 3 hat followed by a drop at 6 hat, a second phase of high transcription from 12 to 24 hat, and the final drop at 48 hat (Fig. 5B). In return, the CtICS gene with a very slow ramp during the first 12 h gets induced at 24–48 hat (Fig. 5B), indicating that in safflower SA biosynthesis is controlled probably by phenylpropanoid rather isochorismate pathway. In Nicotiana benthamiana, Catinot et al. (2008)49 showed that ICS is, like PAL, crucial for SA biosynthesis as if ICS-silenced plants do not accumulate SA following UV exposure or pathogenic attack. Interestingly, the lower concentration of SA (0.1 mM) induces both genes weakly at only 3 hat (Fig. 5A), suggesting the SA responsiveness of both genes. In real life, pathogenic attacks elicit the de novo SA biosynthesis, which per se triggers the immune responses, particularly expression of pathogenesis-related (PR) genes.50 Accordingly, exogenous SA can amply induce the CtC4H and CtICS genes in safflower leading to higher supply of pivotal precursors for defense compounds, i.e., propanoids, flavonoids, and lignins. Salicylic acid has a crucial role in defense responses against pathogens and abiotic stimuli as such higher SA content can induce more strongly and much durably the defense arsenals. Evidently, Arabidopsis ICS1 expression is influenced by a sort of positive feedback from SA, which is for itself the ultimate product of ICS1 gene.51 It is as well documented that transgenic Brassica rapa plants expressing bacterial isochorismate synthase accumulate SA significantly higher than the corresponding control plants.52 In contrast, plants deficient in responsiveness to SA are super susceptible to environmental stresses. For instance, Arabidopsis SA mutant, i.e., eds1, eds2, eds5, pad4, and sid2 are highly susceptible to fungal and bacterial infections.53 Mutant plants of sid2 line are impaired in SA synthesis due to a mutation in the gene of isochorismate synthase isozyme, ICS1; a crucial factor in induction of SAR in Arabidopsis. It has been recently shown that Arabidopsis ICS1 is required for SA biosynthesis.51 Moreover, ICS1 gene expression analysis using transgenic plants expressing ICS1 promoter:reporter gene constructs reveled that SA positively regulates ICS1, however ICS2 is unaffected by SA. Auto-regulation of Arabidopsis ICS1 by SA is inferred by sustained elevated level of ICS1 during 6–48 h after SA treatment. Interestingly, supplying exogenous SA to ics1-mutant plants does not induce the downstream defense genes to the same extent as seen in wild type counterparts.51 In this study, treatment of safflower plants with SA elevated the ICS gene expression rather soon and maintained it high for approximately 48 h. This is confirmed when higher concentration of SA is applied (Fig. 5B). In another study,54 ICS-silenced tomato plants were recruited to investigate the role of SA in delaying nonhost hypersensitive response (HR) cell death. As a result, it was shown that likely due to SA pathway suppression in ICS-RNAi plants, a late (48 h) and mild HR was observed in response to infiltration of Pseudomonas syringae pv tabaci alone or P. syringae pv tabaci+phytotoxin coronatine.54 SA concentration-dependency is evident not only in defense responses but also in other developmental phenomena. SA treatment of seeds before germination improves the germination rate, seedling growth, and the net assimilation, while high SA concentration has inhibitory effects.55 Priming of rice seeds with low SA concentration before planting decreases the ROS production level rooted in cadmium. As well, it increases the content of different antioxidant enzymes namely catalase, guaiacol peroxidase, glutathione reductase, and superoxide dismutase resulting in a protection against oxidative damages. Additionally, SA induces the H2O2 accumulation in tissues infected with microbial pathogens leading to onset of hypersensitive response. Furthermore, SA affects the lipid peroxidation, which per se plays a role in defense response and systemic acquired resistance (SAR) in plants during pathogenic attacks.55 SAR immunizes the plants to a wide range of pathogens, even less similar to the primary infecting pathogen. As a matter of fact, SA is a significant signal for HR and SAR responses.24 However, this might not be excluded that differences in sensitivities of CtC4H and CtICS to SA might be attributed to their differences in affinity to SA, given that generally high affinity mechanism is likely inhibited at relatively higher concentrations of signaling molecules.
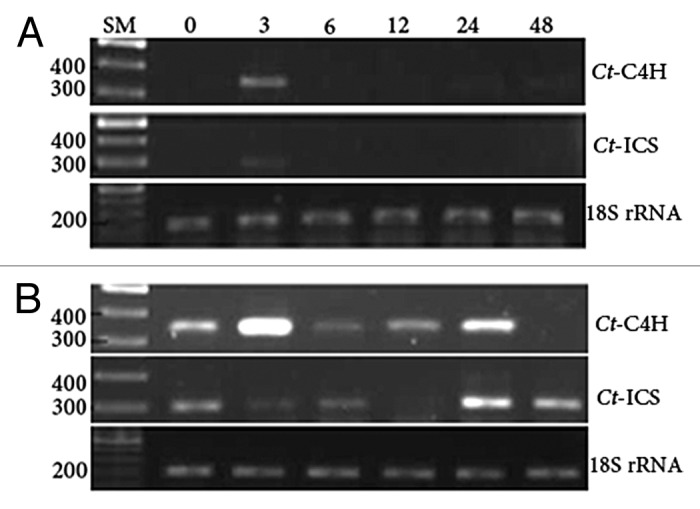
Figure 5. Expression profiles of CtC4H and CtICS genes after SA treatment of 0.1 mM (A) and 1 mM (B). Samplings were performed at 0, 3, 6, 12, 24, and 48 h after treatment. Total RNA was prepared from all plants followed by DNaseI treatment. Then, RNA was reverse transcribed to cDNA. For CtICS, the internal primers were used for RT-PCR. Intensity of PCR products at different time points refers to as temporal expression level of the genes. 18S rRNA transcription levels serve as internal control.
To conclude, we determined in safflower the partial coding sequences of 2 important genes in phenylpropanoid and isochorismate pathways, i.e., cinnamate 4-hydroxylase and isochorismate synthase, which are very critical in plant resistance and adaptation to biotic and abiotic stress circumstances. Both genes get highly induced after wounding, salinity stress, and salicylic acid treatment to contribute to safflower fitness. The findings of this study regarding the molecular cloning and functional characterization of these genes may be applicable in breeding programs for salinity tolerance as well as for pathogen resistance in safflower and other plants.
Materials and Methods
Plant materials and growth condition
Seeds of safflower var. 22–191 (kindly provided by Dr Mahammadinejad, Shahid Bahonar University of Kerman) were sterilized with 70% ethanol and sodium hypochlorite (5% chlorine) for respective 2 and 15 min. Prior to germination, seed were vernalized for 2 h and then seeded in petri dishes on sterile, water-soaked filter papers. The germinated seeds were transplanted into pots filled with sand and grown in glasshouse at 26 ± 2 °C and photoperiod of 16 h with irrigation regime of every 3 days. The plants were fertilized by Hoagland solution once a week.
Isolation of partial sequences of CtICS and CtC4H genes
Genomic DNA isolation was performed using leaf samples according to Saghai-Maroof et al. (1984).56 The isolating primers were designed using the highly conserved domains, in taxonomically close species, of coding sequences of C4H (Cynara scolymus, Zinnia violacea, and Gynura bicolor) and ICS (Solanum lycopersicum, Capsicum annuum, Ricinus communis, Catharanthus roseus, and Rubia cordifolia). The primers were synthesized by Eurofin MWG Operon (Germany) with the given sequences (Table 1). Amplification of CtICS and CtC4H partial coding sequences was performed using genomic DNA. Polymerase chain reactions (PCR) were performed in final volume of 25 µL including 10 pmol of specific primers for each gene. The annealing temperature for CtICS and CtC4H were 50 and 54 °C, respectively.
Cloning of CtICS and CtC4H amplicons for sequencing
The InsTAclone™ PCR Cloning Kit (Thermo SCIENTIFIC, # K1213) was used to clone the PCR products of CtICS and CtC4H into sequencing plasmid according to manufacturer instruction. The E. coli strains for cloning of CtICS and CtC4H were HD5α and JM107, respectively. Briefly, bacterial strains were transformed with pTZ57R/T cloning vector harboring the isolated fragments. Next, the recombinant colonies were picked based on blue/white screening and plasmid DNA was extracted using GF-1 Plasmid DNA Extraction Kit (Vivantis). Sequencing of isolated coding region of genes was performed by Faza Pajooh Biotech (Tehran, Iran) using M13 universal primers. The sequencing results were checked using Chromas Lite 2.01 (Technelysium) after clipping the flanking sequences of cloning vector.
Homology and phylogenetic analyses
Homology analyses of amino acid sequences of isolated genes were performed using ClustalW.57 The evolutionary history was inferred using the Neighbor-Joining method.58 The evolutionary distances were computed using the Poisson correction method.59 All positions containing gaps and missing data were eliminated from the data set (Complete deletion option). Phylogenetic analyses were conducted at Phylogeny.fr.60
Gene expression experiments
Mechanical wounding of leaves was done on 14-d-old seedlings. The wounding was performed by sterile blunt-nosed thumb forceps punched rather equally onto the leaves. Salinity stress was applied, as well, on 14-d-old seedlings. The plants were irrigated with 150 mM NaCl solution. Similar seedlings were used for salicylic acid treatments in 2 groups of 0.1 mM, and 1 mM SA solutions. The solutions were sprayed onto the leaves. Samplings for all 3 assays were performed at 0, 3, 6, 12, 24, and 48 hat. All experiments were initiated at 8 AM to consider the possible diurnal rhythm in the genes expression profiling.
RNA extraction and cDNA synthesis
Total RNA was extracted using RNX™ Plus Kit (Cinnagen) from all treated plants following the manufacturer instruction. After DNaseI treatment of all RNA samples, 1 µg of total RNA was reverse transcribed to cDNA by RevertAid First Strand cDNA Synthesis Kit (Thermo SCIENTIFIC, # K1691).
Semi-quantitative RT-PCR
The cDNA amounts were first normalized by 18S rRNA PCR product intensity. For CtICS, the internal primers based on isolated sequence were designed and used for RT-PCR. PCR thermal profile was as follows: 98 °C (5 min) followed by 35 cycles of 98 °C (20 s.), 52 °C (20 s.), and 72 °C (1 min), and finally 10 min incubation at 72 °C for final extension. A separate experiment was performed to confirm the linear amplification in the condition used. More and less intensity of PCR products at different time points were judged for respective more and less transcription level of each gene.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
The authors thank the anonymous reviewers for their valuable comments on the manuscript. Rahnamaeian M acknowledges the research grant from Shahid Bahonar University of Kerman (Iran). Rahnamaeian M and Vilcinskas A acknowledge the Ministry for Science and Art of the State of Hesse (Germany) for funding both the LOEWE research focus “Insect Biotechnology” and the Fraunhofer project group “Bioresources.”
Footnotes
These authors contributed equally to this work.
References
- 1.Martinez N, Sosa M, Higa R, Fornes D, Capobianco E, Jawerbaum A. Dietary treatments enriched in olive and safflower oils regulate seric and placental matrix metalloproteinases in maternal diabetes. Placenta. 2012;33:8–16. doi: 10.1016/j.placenta.2011.10.015. [DOI] [PubMed] [Google Scholar]
- 2.Hiramatsu M, Takahashi T, Komatsu M, Kido T, Kasahara Y. Antioxidant and neuroprotective activities of Mogami-benibana (safflower, Carthamus tinctorius Linne) Neurochem Res. 2009;34:795–805. doi: 10.1007/s11064-008-9884-5. [DOI] [PubMed] [Google Scholar]
- 3.Rahnamaeian M, Vilcinskas A. Defense gene expression is potentiated in transgenic barley expressing antifungal peptide Metchnikowin throughout powdery mildew challenge. J Plant Res. 2012;125:115–24. doi: 10.1007/s10265-011-0420-3. [DOI] [PubMed] [Google Scholar]
- 4.Herrmann KM. The shikimate pathway as an entry to aromatic secondary metabolism. Plant Physiol. 1995;107:7–12. doi: 10.1104/pp.107.1.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Maffei ME, Gertsch J, Appendino G. Plant volatiles: production, function and pharmacology. Nat Prod Rep. 2011;28:1359–80. doi: 10.1039/c1np00021g. [DOI] [PubMed] [Google Scholar]
- 6.Chen Z, Zheng Z, Huang J, Lai Z, Fan B. Biosynthesis of salicylic acid in plants. Plant Signal Behav. 2009;4:493–6. doi: 10.4161/psb.4.6.8392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van Tegelen LJP, Moreno PRH, Croes AF, Verpoorte R, Wullems GJ. Purification and cDNA cloning of isochorismate synthase from elicited cell cultures of Catharanthus roseus. Plant Physiol. 1999;119:705–12. doi: 10.1104/pp.119.2.705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.León J, Shulaev V, Yalpani N, Lawton MA, Raskin I. Benzoic acid 2-hydroxylase, a soluble oxygenase from tobacco, catalyzes salicylic acid biosynthesis. Proc Natl Acad Sci U S A. 1995;92:10413–7. doi: 10.1073/pnas.92.22.10413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yu ZZ, Fu CX, Han YS, Li YX, Zhao DX. Salicylic acid enhances Jaceosidin and Syringin production in cell cultures of Saussurea medusa. Biotechnol Lett. 2006;28:1027–31. doi: 10.1007/s10529-006-9035-5. [DOI] [PubMed] [Google Scholar]
- 10.Havsteen BH. The biochemistry and medical significance of the flavonoids. Pharmacol Ther. 2002;96:67–202. doi: 10.1016/S0163-7258(02)00298-X. [DOI] [PubMed] [Google Scholar]
- 11.Pourcel L, Irani NG, Koo AJ, Bohorquez-Restrepo A, Howe GA, Grotewold E. A chemical complementation approach reveals genes and interactions of flavonoids with other pathways. Plant J. 2013;74:383–97. doi: 10.1111/tpj.12129. [DOI] [PubMed] [Google Scholar]
- 12.Sanchez-Ballesta MT, Romero I, Jiménez JB, Orea JM, González-Ureña Á, Escribano MI, Merodio C. Involvement of the phenylpropanoid pathway in the response of table grapes to low temperature and high CO2 levels. Postharvest Biol Technol. 2007;46:29–35. doi: 10.1016/j.postharvbio.2007.04.001. [DOI] [Google Scholar]
- 13.Schoch GA, Nikov GN, Alworth WL, Werck-Reichhart D. Chemical inactivation of the cinnamate 4-hydroxylase allows for the accumulation of salicylic acid in elicited cells. Plant Physiol. 2002;130:1022–31. doi: 10.1104/pp.004309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Morkunas I, Narożna D, Nowak W, Samardakiewicz S, Remlein-Starosta D. Cross-talk interactions of sucrose and Fusarium oxysporum in the phenylpropanoid pathway and the accumulation and localization of flavonoids in embryo axes of yellow lupine. J Plant Physiol. 2011;168:424–33. doi: 10.1016/j.jplph.2010.08.017. [DOI] [PubMed] [Google Scholar]
- 15.Khastkhodaei S, Sharifi G, Salahi R, Rahnamaeian M, Moattar F. Clinical efficacy of Stragol herbal heart drop in ischemic heart failure of stable chest angina. Eur J Intern Med. 2011;3:e201–7. doi: 10.1016/j.eujim.2011.07.004. [DOI] [Google Scholar]
- 16.Corpas FJ, Leterrier M, Valderrama R, Airaki M, Chaki M, Palma JM, Barroso JB. Nitric oxide imbalance provokes a nitrosative response in plants under abiotic stress. Plant Sci. 2011;181:604–11. doi: 10.1016/j.plantsci.2011.04.005. [DOI] [PubMed] [Google Scholar]
- 17.Rakwal R, Agrawal GK. Wound signaling-coordination of the octadecanoid and MAPK pathways. Plant Physiol Biochem. 2003;41:855–61. doi: 10.1016/S0981-9428(03)00142-6. [DOI] [Google Scholar]
- 18.Ellard-Ivey M, Douglas CJ. Role of jasmonates in the elicitor-and wound-inducible expression of defense genes in parsley and transgenic tobacco. Plant Physiol. 1996;112:183–92. doi: 10.1104/pp.112.1.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rahnamaeian M. Antimicrobial peptides: modes of mechanism, modulation of defense responses. Plant Signal Behav. 2011;6:1325–32. doi: 10.4161/psb.6.9.16319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ouyang B, Yang T, Li H, Zhang L, Zhang Y, Zhang J, Fei Z, Ye Z. Identification of early salt stress response genes in tomato root by suppression subtractive hybridization and microarray analysis. J Exp Bot. 2007;58:507–20. doi: 10.1093/jxb/erl258. [DOI] [PubMed] [Google Scholar]
- 21.Jamil M, Iqbal W, Bangash A, Rehman SUR, Imran QM, Rha EUIS. Constitutive expression of OSC3H33, OSC3H50 and OSC3H37 genes in rice under salt stress. Pak J Bot. 2010;42:4003–9. [Google Scholar]
- 22.Gong Z, Koiwa H, Cushman MA, Ray A, Bufford D, Kore-eda S, Matsumoto TK, Zhu J, Cushman JC, Bressan RA, et al. Genes that are uniquely stress regulated in salt overly sensitive (sos) mutants. Plant Physiol. 2001;126:363–75. doi: 10.1104/pp.126.1.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Knight H, Knight MR. Abiotic stress signalling pathways: specificity and cross-talk. Trends Plant Sci. 2001;6:262–7. doi: 10.1016/S1360-1385(01)01946-X. [DOI] [PubMed] [Google Scholar]
- 24.Rahnamaeian M, Langen G, Imani J, Khalifa W, Altincicek B, von Wettstein D, Kogel KH, Vilcinskas A. Insect peptide metchnikowin confers on barley a selective capacity for resistance to fungal ascomycetes pathogens. J Exp Bot. 2009;60:4105–14. doi: 10.1093/jxb/erp240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li ZY, Xu ZS, He GY, Yang GX, Chen M, Li LC, Ma YZ. A mutation in Arabidopsis BSK5 encoding a brassinosteroid-signaling kinase protein affects responses to salinity and abscisic acid. Biochem Biophys Res Commun. 2012;426:522–7. doi: 10.1016/j.bbrc.2012.08.118. [DOI] [PubMed] [Google Scholar]
- 26.Bandurska H, Cieślak M. The interactive effect of water deficit and UV-B radiation on salicylic acid accumulation in barley roots and leaves. Environ Exp Bot. 2013;94:9–18. doi: 10.1016/j.envexpbot.2012.03.001. [DOI] [Google Scholar]
- 27.Drzewiecka K, Borowiak K, Bandurska H, Golinski P. Salicylic acid - a potential biomarker of tobacco Bel-W3 cell death developed as a response to ground level ozone under ambient conditions. Acta Biol Hung. 2012;63:231–49. doi: 10.1556/ABiol.63.2012.2.6. [DOI] [PubMed] [Google Scholar]
- 28.Pan Q, Zhan J, Liu H, Zhang J, Chen J, Wen P, Huang W. Salicylic acid synthesized by benzoic acid 2-hydroxylase participates in the development of thermotolerance in pea plants. Plant Sci. 2006;171:226–33. doi: 10.1016/j.plantsci.2006.03.012. [DOI] [Google Scholar]
- 29.Sawada H, Shim I-S, Usui K. Induction of benzoic acid 2-hydroxylase and salicylic acid biosynthesis-Modulation by salt stress in rice seedlings. Plant Sci. 2006;171:263–70. doi: 10.1016/j.plantsci.2006.03.020. [DOI] [Google Scholar]
- 30.Harrison AJ, Yu M, Gårdenborg T, Middleditch M, Ramsay RJ, Baker EN, Lott JS. The structure of MbtI from Mycobacterium tuberculosis, the first enzyme in the biosynthesis of the siderophore mycobactin, reveals it to be a salicylate synthase. J Bacteriol. 2006;188:6081–91. doi: 10.1128/JB.00338-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Barghouthi S, Payne SM, Arceneaux JE, Byers BR. Cloning, mutagenesis, and nucleotide sequence of a siderophore biosynthetic gene (amoA) from Aeromonas hydrophila. J Bacteriol. 1991;173:5121–8. doi: 10.1128/jb.173.16.5121-5128.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cole ST, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, Gordon SV, Eiglmeier K, Gas S, Barry CE, 3rd, et al. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393:537–44. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- 33.Garcion C, Lohmann A, Lamodière E, Catinot J, Buchala A, Doermann P, Métraux JP. Characterization and biological function of the ISOCHORISMATE SYNTHASE2 gene of Arabidopsis. Plant Physiol. 2008;147:1279–87. doi: 10.1104/pp.108.119420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gross J, Cho WK, Lezhneva L, Falk J, Krupinska K, Shinozaki K, Seki M, Herrmann RG, Meurer J. A plant locus essential for phylloquinone (vitamin K1) biosynthesis originated from a fusion of four eubacterial genes. J Biol Chem. 2006;281:17189–96. doi: 10.1074/jbc.M601754200. [DOI] [PubMed] [Google Scholar]
- 35.Bell-Lelong DA, Cusumano JC, Meyer K, Chapple C. Cinnamate-4-hydroxylase expression in Arabidopsis. Regulation in response to development and the environment. Plant Physiol. 1997;113:729–38. doi: 10.1104/pp.113.3.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mizutani M, Ohta D, Sato R. Isolation of a cDNA and a genomic clone encoding cinnamate 4-hydroxylase from Arabidopsis and its expression manner in planta. Plant Physiol. 1997;113:755–63. doi: 10.1104/pp.113.3.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Richard S, Lapointe G, Rutledge RG, Séguin A. Induction of chalcone synthase expression in white spruce by wounding and jasmonate. Plant Cell Physiol. 2000;41:982–7. doi: 10.1093/pcp/pcd017. [DOI] [PubMed] [Google Scholar]
- 38.Campos R, Nonogaki H, Suslow T, Saltveit ME. Isolation and characterization of a wound inducible phenylalanine ammonia-lyase gene (LsPAL1) from Romaine lettuce leaves. Physiol Plant. 2004;121:429–38. doi: 10.1111/j.1399-3054.2004.00336.x. [DOI] [Google Scholar]
- 39.Mellway RD, Tran LT, Prouse MB, Campbell MM, Constabel CP. The wound-, pathogen-, and ultraviolet B-responsive MYB134 gene encodes an R2R3 MYB transcription factor that regulates proanthocyanidin synthesis in poplar. Plant Physiol. 2009;150:924–41. doi: 10.1104/pp.109.139071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schulz W, Eiben HG, Hahlbrock K. Expression in Escherichia coli of catalytically active phenylalanine ammonia-lyase from parsley. FEBS Lett. 1989;258:335–8. doi: 10.1016/0014-5793(89)81687-4. [DOI] [PubMed] [Google Scholar]
- 41.Logemann E, Parniske M, Hahlbrock K. Modes of expression and common structural features of the complete phenylalanine ammonia-lyase gene family in parsley. Proc Natl Acad Sci U S A. 1995;92:5905–9. doi: 10.1073/pnas.92.13.5905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chapple C. Molecular-genetic analysis on plant cytochrome P450-dependent monooxygenases. Annu Rev Plant Physiol Plant Mol Biol. 1998;49:311–43. doi: 10.1146/annurev.arplant.49.1.311. [DOI] [PubMed] [Google Scholar]
- 43.Batard Y, Schalk M, Pierrel MA, Zimmerlin A, Durst F, Werck-Reichhart D. Regulation of the Cinnamate 4-Hydroxylase (CYP73A1) in Jerusalem Artichoke Tubers in Response to Wounding and Chemical Treatments. Plant Physiol. 1997;113:951–9. doi: 10.1104/pp.113.3.951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Singh K, Kumar S, Rani A, Gulati A, Ahuja PS. Phenylalanine ammonia-lyase (PAL) and cinnamate 4-hydroxylase (C4H) and catechins (flavan-3-ols) accumulation in tea. Funct Integr Genomics. 2009;9:125–34. doi: 10.1007/s10142-008-0092-9. [DOI] [PubMed] [Google Scholar]
- 45.Kim JK, Bamba T, Harada K, Fukusaki E, Kobayashi A. Time-course metabolic profiling in Arabidopsis thaliana cell cultures after salt stress treatment. J Exp Bot. 2007;58:415–24. doi: 10.1093/jxb/erl216. [DOI] [PubMed] [Google Scholar]
- 46.Walia H, Wilson C, Condamine P, Liu X, Ismail AM, Zeng L, Wanamaker SI, Mandal J, Xu J, Cui X, et al. Comparative transcriptional profiling of two contrasting rice genotypes under salinity stress during the vegetative growth stage. Plant Physiol. 2005;139:822–35. doi: 10.1104/pp.105.065961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Al-Whaibi MH, Siddiqui MH, Basalah MO. Salicylic acid and calcium-induced protection of wheat against salinity. Protoplasma. 2012;249:769–78. doi: 10.1007/s00709-011-0322-1. [DOI] [PubMed] [Google Scholar]
- 48.Lee S, Kim SG, Park CM. Salicylic acid promotes seed germination under high salinity by modulating antioxidant activity in Arabidopsis. New Phytol. 2010;188:626–37. doi: 10.1111/j.1469-8137.2010.03378.x. [DOI] [PubMed] [Google Scholar]
- 49.Catinot J, Buchala A, Abou-Mansour E, Métraux JP. Salicylic acid production in response to biotic and abiotic stress depends on isochorismate in Nicotiana benthamiana. FEBS Lett. 2008;582:473–8. doi: 10.1016/j.febslet.2007.12.039. [DOI] [PubMed] [Google Scholar]
- 50.Belchí-Navarro S, Almagro L, Sabater-Jara AB, Fernández-Pérez F, Bru R, Pedreño MA. Induction of trans-resveratrol and extracellular pathogenesis-related proteins in elicited suspension cultured cells of Vitis vinifera cv Monastrell. J Plant Physiol. 2013;170:258–64. doi: 10.1016/j.jplph.2012.10.003. [DOI] [PubMed] [Google Scholar]
- 51.Hunter LJR, Westwood JH, Heath G, Macaulay K, Smith AG, Macfarlane SA, Palukaitis P, Carr JP. Regulation of RNA-dependent RNA polymerase 1 and isochorismate synthase gene expression in Arabidopsis. PLoS One. 2013;8:e66530. doi: 10.1371/journal.pone.0066530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Simoh S, Linthorst HJ, Lefeber AW, Erkelens C, Kim HK, Choi YH, Verpoorte R. Metabolic changes of Brassica rapa transformed with a bacterial isochorismate synthase gene. J Plant Physiol. 2010;167:1525–32. doi: 10.1016/j.jplph.2010.06.022. [DOI] [PubMed] [Google Scholar]
- 53.Kunkel BN, Brooks DM. Cross talk between signaling pathways in pathogen defense. Curr Opin Plant Biol. 2002;5:325–31. doi: 10.1016/S1369-5266(02)00275-3. [DOI] [PubMed] [Google Scholar]
- 54.Lee S, Ishiga Y, Clermont K, Mysore KS. Coronatine inhibits stomatal closure and delays hypersensitive response cell death induced by nonhost bacterial pathogens. PeerJ. 2013;1:e34. doi: 10.7717/peerj.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hayat Q, Hayat S, Irfan M, Ahmad A. Effect of exogenous salicylic acid under changing environment: a review. Environ Exp Bot. 2012;68:14–25. doi: 10.1016/j.envexpbot.2009.08.005. [DOI] [Google Scholar]
- 56.Saghai-Maroof MA, Soliman KM, Jorgensen RA, Allard RW. Ribosomal DNA spacer-length polymorphisms in barley: mendelian inheritance, chromosomal location, and population dynamics. Proc Natl Acad Sci U S A. 1984;81:8014–8. doi: 10.1073/pnas.81.24.8014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. ClustalW and ClustalX version 2. Bioinformatics. 2007;23:2947–8. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 58.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–25. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 59.Zuckerkandl E, Pauling L. Evolutionary divergence and convergence in proteins. In: Evolving Genes and Proteins. Bryson V and Vogel HJ, eds. New York: Academic Press, 1965: 97-166 [Google Scholar]
- 60.Dereeper A, Guignon V, Blanc G, Audic S, Buffet S, Chevenet F, Dufayard JF, Guindon S, Lefort V, Lescot M, et al. Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 2008;36:W465-9. doi: 10.1093/nar/gkn180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Maeda H, Dudareva N. The shikimate pathway and aromatic amino Acid biosynthesis in plants. Annu Rev Plant Biol. 2012;63:73–105. doi: 10.1146/annurev-arplant-042811-105439. [DOI] [PubMed] [Google Scholar]
- 62.Naoumkina MA, Zhao Q, Gallego-Giraldo L, Dai X, Zhao PX, Dixon RA. Genome-wide analysis of phenylpropanoid defence pathways. Mol Plant Pathol. 2010;11:829–46. doi: 10.1111/j.1364-3703.2010.00648.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Vogt T. Phenylpropanoid biosynthesis. Mol Plant. 2010;3:2–20. doi: 10.1093/mp/ssp106. [DOI] [PubMed] [Google Scholar]
- 64.Wang X. Structure, function, and engineering of enzymes in isoflavonoid biosynthesis. Funct Integr Genomics. 2011;11:13–22. doi: 10.1007/s10142-010-0197-9. [DOI] [PubMed] [Google Scholar]