Abstract
The RAB5 GTPase ARA6 of Arabidopsis thaliana is known to be involved in endosomal trafficking by targeting vesicles to the plasma membrane. During this process AtARA6 is working in close relationship with the SNARE protein VAMP727 (vesicle associated membrane protein 727). Recently, ARA6 of the characean green algae Chara australis (CaARA6) was shown to have properties similar to AtARA6, pointing to similar trafficking pathways. In order to gain further insight into the vesicle trafficking machinery of Characeae, C. australis was analyzed for homologous proteins of the VAMP72-family. A CaVAMP72 protein was detected and classified by protein sequence alignment and phylogenetic analyses.
Keywords: ARA6, RAB5 GTPase, Characeae, Chara australis, endosomal trafficking, plasma membrane, SNARE protein, vesicle associated membrane protein, VAMP
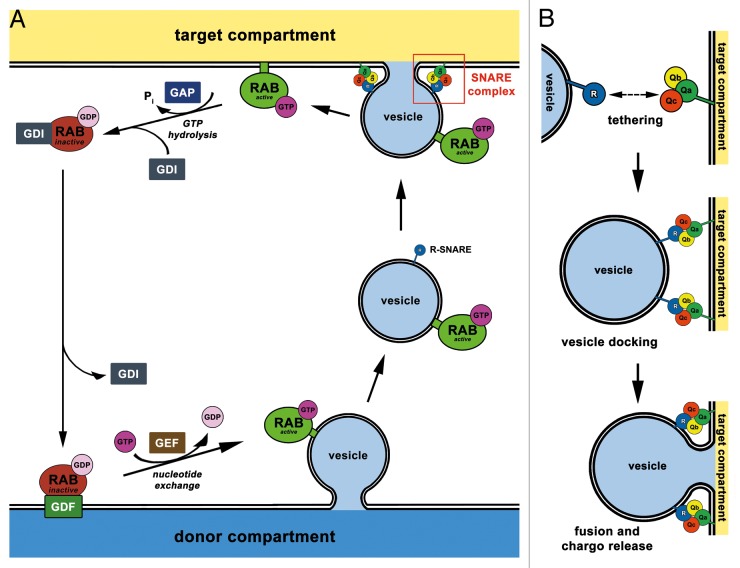
Intracellular vesicle trafficking to and from the plasma membrane as well as between different organelles is a characteristic attribute of eukaryotic cells. These membrane interactions are tightly controlled by a molecular machinery allowing fast response to a multitude of intracellular and extracellular conditions. Since two decades endocytosis is a well-established field of research in plant sciences and one of its highlights was the identification and characterization of RAB GTPases in plants. RAB GTPases are key regulators in membrane trafficking and had long been well known in animal and yeast cells.1 The RAB protein family consists of small molecules (molecular mass 20 – 25 kD) that are localized in their activated (GTP bound) state at membranes, where they recruit different effector molecules and promote downstream reactions (e.g., tethering of transport vesicles or other organelles to target membranes). The GTPase cycle of RAB proteins is conventionally dependent on different regulatory proteins like GDP dissociation inhibitors (GDI), GEFs (guanine nucleotide exchange factors), GDFs (GDI displacement factors), and GAPs (GTPase activating proteins; for a review see ref. 2.). A schematic overview of the RAB GTPase cycle is shown in Figure 1A.
Figure 1. Schematic overview of vesicular trafficking: RAB GTPase cycle and SNARE complex formation. (A) RAB proteins cycle between active, GTP-bound (green) and inactive, GDP-bound (dark red) state during vesicular trafficking. Several regulatory proteins are involved in this process: RAB GTPase activating protein (GAP, dark blue) increases GTP hydrolysis on active RAB proteins. GDP dissociation inhibitor (GDI, dark gray) solubilises RABs by masking the membrane association site until the next round of GTPase cycle. For signaling, RAB protein has to travel to a proper organelle membrane carrying the GDI displacement factor (GDF, dark green) which removes the GDI. The RAB guanine nucleotide exchange factor (GEF, dark brown) enables exchange of bound GDP for GTP in order to reactivate RAB protein. SNARE complexes (red box in A) are involved in the fusion of vesicle and target compartment membranes. GTP, magenta; GDP, light pink; (B) SNARE complex formation, vesicle docking, and membrane fusion. R-SNARE proteins reside on vesicular membranes and bind to Q-SNARES (Qa, Qb, and Qc) at the target compartment membrane. The created SNARE complex (Qa, Qb, Qc, and R-SNARE) enables vesicle docking and membrane fusion. SNARE proteins: R, R-SNARE (blue); Qa, Qa-SNARE (green); Qb, Qb-SNARE (yellow); Qc, Qc-SNARE (red);
A subgroup of RAB GTPases, the so called RAB5 family, is responsible for endosomal trafficking in animal, yeast, and plant cells.1 The RAB5 group of Arabidopsis thaliana consists of three proteins: RHA1 (AtRABF2a), ARA7 (AtRABF2b), and ARA6 (AtRABF1).3,4 ARA7 and RHA1 structurally resemble mammalian RAB5 GTPases and exhibit similar functions in endocytosis and transport toward the vacuole (for example see refs. 5–7. ARA6, however, is plant-specific and one of the most noticeable hallmarks of plant RAB proteins. ARA6 differs from conventional RAB5s in the protein sequence responsible for membrane anchoring and shows an interesting abundance throughout different plant species: until lately ARA6 was detected in land plants including bryophytes and lycophytes, whereas algae only showed conventional RAB5 members.4 Therefore, ARA6 was assumed to be land plant specific.8
This hypothesis was recently refuted, as a next generation sequencing of a close relative to land plants, the charophycean green alga Chara australis revealed an ortholog of ARA6 (CaARA6 or CaRABF1).9 CaARA6 was shown to bear high sequence similarities to well known land plant ARA6 proteins (compare Figure 1 in Hoepflinger et al.9) and functional studies revealed further resemblances, but also some differences between ARA6 proteins of C. australis and A. thaliana (AtARA6). One of these similarities was the presence of comparable amounts of intrinsic GTPase activity in recombinant AtARA6 and CaARA6, respectively.9 Direct comparison of subcellular ARA6 distribution using distinct fluorescent tags demonstrated that both CaARA6 and AtARA6 localized at multivesicular endosomes (MVEs) when transiently expressed in tobacco9 and needed N-terminal glycine and cysteine for correct membrane anchoring.4 Furthermore, the nucleotide-free mutant CaAra6N172I and the constitutively active GTP-locked form CaAra6Q118L localized at the plasma membrane, like their counterparts in A. thaliana.4,9 A notable difference between both constitutively active Ara6 mutants of Arabidopsis and Chara was the absence of GFP-tagged CaAra6Q118L at the tonoplast. This difference was likely due to a functional divergence mediated by the N-terminal region of CaARA6, which showed an extra stretch of about 20 amino acids compared with AtARA6.9 Immunolabeling of electron microscopical sections of Chara internodal cells confirmed localization of wildtype CaARA6 at MVEs (late endosomes; compare ref. 4, 9.) and revealed additional ARA6 epitopes at the trans-Golgi network (TGN; considered to be an early endosome) and at the plasma membrane including charasomes even under normal, unstressed conditions.9,10 Charasomes are structured plasma membrane elaborations involved in environmental acidification, which increase the efficiency of HCO3- utilization and photosynthesis (for references see ref. 11.). Summarizing, an involvement in an endosomal trafficking pathway to the plasma membrane including MVEs and TGN was stated for CaARA6.
New insights: Assigning CaARA6 to a concrete endosomal trafficking pathway?
ARA6 of A. thaliana, which is known to be involved in endosomal trafficking pathways by targeting vesicles to the plasma membrane, works in close relationship with another plant unique R-SNARE protein: the vesicle associated membrane protein 727 (AtVAMP727, At3g54300). Members of the SNARE (soluble N-ethyl-maleimide sensitive factor attachment protein receptor) superfamily are key components in mediating specific fusions of transport vesicles and target membranes in eukaryotic cells. A model of SNARE complex formation is shown in Figure 1B. SNAREs contain a helical region, the so called SNARE domain, which is used for protein classification: Qa-, Qb-, Qc-, and R-SNAREs; in which Q-SNAREs contain a glutamine (Q) residue and R-SNAREs an arginine (R) residue at a specific site – the zero layer – within the C-terminal synaptobrevin domain. Tightly clustered SNARE complexes are mostly formed by four SNAREs, one protein of each SNARE group (Qa/Qb/Qc/R; for a review see ref. 2.), where the zero ionic layer structurally forms the center of the complex. Until now, only a few complete SNARE complexes involved in different cellular processes are identified in A. thaliana: two SNARE complexes participating in cytokinesis comprising of the same proteins, either in form of a trimer in the plasma membrane (Qa/Qb,c/R), or as a tetramer (Qa/Qb/Qc/R) at the endomembrane system. Another SNARE complex is described as taking part in pathogen response at the plasma membrane. A fourth one containing VAMP727 is mediating membrane fusion of endosomes and the vacuole by forming a complex with the three Q-SNAREs: SYP22, SYP5 and VTI11. Furthermore, AtVAMP727 is binding to SYP121 at the plasma membrane, forming a complex of which Qb- and Qc-SNARES still remain to be elucidated.12-14 AtVAMP727 is a special R-SNARE protein, as it contains a unique, approximately 20 amino acid insertion in the N-terminal longin domain.15 So far, close homologs of AtVAMP727 have only been detected in seed plants. The question arises how trafficking pathways of endosomes had developed during the evolution of land plants. Since SNARE-proteins are key players in docking and fusion events involved in vesicle trafficking, we searched for homologous proteins of the VAMP72-family in the characean green alga Chara australis, a species closely related to land plants.
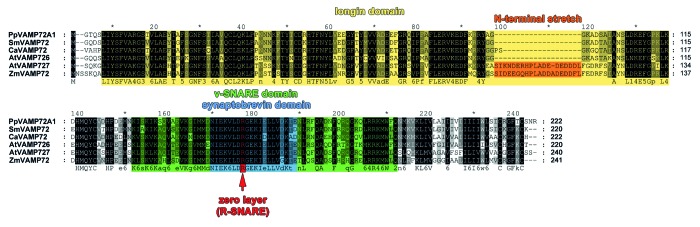
Therefore, AtVAMP727 was used as template sequence for a screening of our C. australis 454 database (as described in ref. 9.). An interesting contig was found and named CaVAMP72-family like. The coding sequence of this gene comprises 669 bp which encode a protein of 222 amino acids with a calculated molecular mass of 25.4 g mol−1 and an isoelectric point of 8.66. A protein sequence alignment (blastp16) revealed high sequence similarity to a VAMP72-family protein of Physcomitrella patens (NCBI reference sequence: XP_001777330), which is described in NCBI as similar to the VAMP72-family of R-SNARE proteins, a protein family specific to green plants involved in vesicle trafficking to the plasma membrane. Furthermore, this alignment displayed a clear classification of CaVAMP72 to R-SNARE proteins of the VAMP72 family. As shown in Figure 2, CaVAMP72-family like protein contains all domains described for VAMP72 proteins: the about 110 amino acid N-terminal longin domain that is characteristic for VAMPs (Prosite domain: PS50859); the v-SNARE domain (vesicle-SNARE, Prosite domain: PS50892) defining SNARES residing at vesicle membranes and binding to their counterparts on target membranes (t-SNAREs) in the process of vesicle docking; and the synaptobrevin domain (Prosite domain: PS00417) containing the characteristic arginine (R) residue in the zero ionic layer of R-SNAREs.
Figure 2. Protein sequence alignment of VAMP72-family members. Multiple sequence alignment of amino acids from CaVAMP72-family like protein with other species was performed using ClustalW.17 Identical residues are highlighted in black, conserved domains are shown in different shades of gray. Aligned sequences are: Physcomitrella patens VAMP72 (PpVAMP72A1, XP_001777330), Selaginella moellendorfii VAMP (SmVAMP, XP_002960391), Chara australis VAMP72-family like protein, Arabidopsis thaliana VAMP726 (AtVAMP726, NP_171968), Arabidopsis thaliana VAMP727 (AtVAMP727, NP_190998), and Zea mays VAMP727 (ZmVAMP727, NP_001136721).
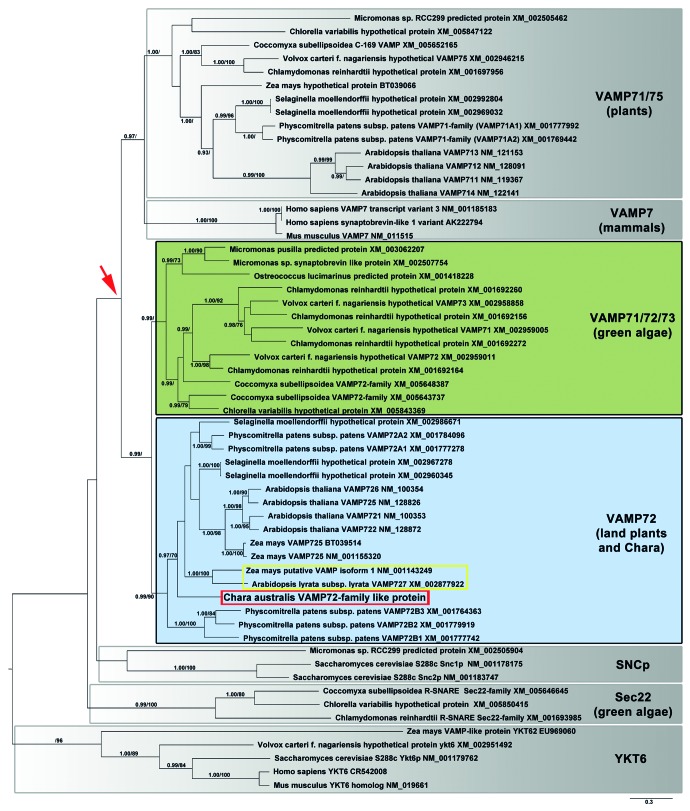
In order to further classify CaVAMP72 protein in the large family of SNAREs, phylogenetic analyses were performed. As shown in Figure 3, all aligned VAMPs divide into two major groups (red arrow in Figure 3), where the clade containing VAMP71 and 75 members of different plant species is more closely related to mammalian VAMP7s than to other plant proteins. Interestingly, all aligned plant VAMP71 and 75 proteins are grouped together into one clade independent of the species. Contrary, plant VAMP72 proteins divide into sister clades. One clade consisting of VAMP72 proteins of green algae (excluding C. australis) that are grouped together with VAMP71 and 73 (see green box in Figure 3), and a second clade of land plant VAMP72s including CaVAMP72 (see Figure 3: blue box and red frame within). Therefore, it can be assumed that CaVAMP72 is more closely related to VAMP72 proteins of land plants (blue box in Figure 3) than to VAMPs of other green algae. VAMP727 sequences of Arabidopsis thaliana (XM_002877922) and Zea mays (NM_001143249) cluster together and constitute an own group within land plant VAMP72s (see yellow frame in Figure 3), which is most probably due to their specific N-terminal stretch in the longin domain.
Figure 3. Unrooted maximum clade credibility tree of different vesicle-SNAREs calculated with BEAST. Branches with posterior probabilities (PP) ≥ 0.95%, which reflect the posterior median node heights for the clades, and ML bootstrap support (MLB) ≥ 70% were considered as strongly supported. The bar specifies the substitutions per site and indicates the branch lengths. The different clades are highlighted in color and termed after their protein denotation (right side).
To summarize our results in the context of current research, the following can be stated: In Arabidopsis thaliana the SNARE protein VAMP727 targets vesicular transport to the plasma membrane by working in close relationship with ARA6. In the charophycean green alga Chara australis, CaARA6 was also shown to localize at the plasma membrane. Therefore, it can be assumed that a VAMP-family protein can be involved in charophycean vesicular trafficking too. VAMP727 proteins are described to differ from other VAMP72-family members by an additional insertion of 20 amino acids in the longin domain. The SNARE protein CaVAMP72 described in this study does not possess the additional amino acids of VAMP727 proteins, but phylogenetic analyses reveal a clear relationship to land plant VAMP72s. Therefore, CaVAMP72 can be assumed to perform an intermediate function between VAMP727 and other VAMP72-family proteins of land plants. Further studies have to be performed in order to determine the exact role of the VAMP72-family like protein in charophycean green algae.
Methods
Sequence alignment and phylogenetic analyses
Nucleotide sequence of Chara australis was aligned with sequences of land plants, bryophytes, lycophytes, green algae, mammals, and yeast retrieved from NCBI GenBank using the program Geneious 6.1.2 created by Biomatters. The alignment in FASTA format was converted into a Nexus-file by the web-portal alignment.18 Afterwards, the data set was analyzed with the program jModelTest 2.1.1 using Akaiko Information Criterion (AIC) scores to achieve the optimal substitution model for phylogenetic analysis.19,20 The analysis was computed in BEAST 1.7.421 on the basis of the GTR+G+I model, but with fixed values for the gamma shape and the proportion of invariant sites calculated with jModelTest. Three parallel analyses were run for 20,000,000 chains, which were performed under a lognormal relaxed clock and every 1000th tree was sampled. The three obtained log-files were controlled with the program Tracer v1.5 if the analyses were suitable for the final phylogenetic tree configuration.22 The tree-files with branch lengths in units of substitutions of each run were combined to one common tree-file using LogCombiner1.7.4 in BEAST. Finally, this file was used to summarize the sampled trees with a burn-in of 50% (30,000 trees) to a maximum clade credibility tree by TreeAnnotator1.7.4 in BEAST. The data set was also analyzed by ML bootstrapping in RAxML with the graphical front-end raxmlGUI 1.3 using 1000 replicates.23,24 All trees were illustrated using the program FigTree v1.3.1.25
Glossary
Abbreviations:
- RAB
rat sarcoma-related protein in brain
- MVE
multivesicular endosome
- TGN
trans-Golgi network
- PP
posterior probability
- MLB
maximum likelihood bootstrap
- VAMP
vesicle associated membrane protein
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We are grateful to Dr. Frantisek Baluska for kindly inviting us to submit this communication and to Dr. Aniela Sommer for valuable discussion. This research was funded by the Austrian Science Fund (project no. P 22957-B20 to IF).
References
- 1.Schwartz SL, Cao C, Pylypenko O, Rak A, Wandinger-Ness A. . Rab GTPases at a glance. J Cell Sci 2007; 120:3905 - 10; http://dx.doi.org/ 10.1242/jcs.015909; PMID: 17989088 [DOI] [PubMed] [Google Scholar]
- 2.Saito C, Ueda T. Chapter 4: functions of RAB and SNARE proteins in plant life. Int Rev Cell Mol Biol 2009; 274:183-233. [DOI] [PubMed] [Google Scholar]
- 3.Ueda T, Uemura T, Sato MH, Nakano A. . Functional differentiation of endosomes in Arabidopsis cells. Plant J 2004; 40:783 - 9; http://dx.doi.org/ 10.1111/j.1365-313X.2004.02249.x; PMID: 15546360 [DOI] [PubMed] [Google Scholar]
- 4.Ueda T, Yamaguchi M, Uchimiya H, Nakano A. . Ara6, a plant-unique novel type Rab GTPase, functions in the endocytic pathway of Arabidopsis thaliana. EMBO J 2001; 20:4730 - 41; http://dx.doi.org/ 10.1093/emboj/20.17.4730; PMID: 11532937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sohn EJ, Kim ES, Zhao M, Kim SJ, Kim H, Kim YW, Lee YJ, Hillmer S, Sohn U, Jiang L, et al. . Rha1, an Arabidopsis Rab5 homolog, plays a critical role in the vacuolar trafficking of soluble cargo proteins. Plant Cell 2003; 15:1057 - 70; http://dx.doi.org/ 10.1105/tpc.009779; PMID: 12724533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kotzer AM, Brandizzi F, Neumann U, Paris N, Moore I, Hawes C. . AtRabF2b (Ara7) acts on the vacuolar trafficking pathway in tobacco leaf epidermal cells. J Cell Sci 2004; 117:6377 - 89; http://dx.doi.org/ 10.1242/jcs.01564; PMID: 15561767 [DOI] [PubMed] [Google Scholar]
- 7.Dhonukshe P, Baluska F, Schlicht M, Hlavacka A, Samaj J, Friml J, Gadella TW Jr.. . Endocytosis of cell surface material mediates cell plate formation during plant cytokinesis. Dev Cell 2006; 10:137 - 50; http://dx.doi.org/ 10.1016/j.devcel.2005.11.015; PMID: 16399085 [DOI] [PubMed] [Google Scholar]
- 8.Ebine K, Ueda T. . Unique mechanism of plant endocytic/vacuolar transport pathways. J Plant Res 2009; 122:21 - 30; http://dx.doi.org/ 10.1007/s10265-008-0200-x; PMID: 19082690 [DOI] [PubMed] [Google Scholar]
- 9.Hoepflinger MC, Geretschlaeger A, Sommer A, Hoeftberger M, Nishiyama T, Sakayama H, Hammerl P, Tenhaken R, Ueda T, Foissner I. . Molecular and biochemical analysis of the first ARA6 homologue, a RAB5 GTPase, from green algae. [Epub ahead of print] J Exp Bot 2013; 64:5553 - 68; http://dx.doi.org/ 10.1093/jxb/ert322; PMID: 24127512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Viotti C, Bubeck J, Stierhof YD, Krebs M, Langhans M, van den Berg W, van Dongen W, Richter S, Geldner N, Takano J, et al. . Endocytic and secretory traffic in Arabidopsis merge in the trans-Golgi network/early endosome, an independent and highly dynamic organelle. Plant Cell 2010; 22:1344 - 57; http://dx.doi.org/ 10.1105/tpc.109.072637; PMID: 20435907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schmölzer PM, Höftberger M, Foissner I. . Plasma membrane domains participate in pH banding of Chara internodal cells. Plant Cell Physiol 2011; 52:1274 - 88; http://dx.doi.org/ 10.1093/pcp/pcr074; PMID: 21659328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ebine K, Fujimoto M, Okatani Y, Nishiyama T, Goh T, Ito E, Dainobu T, Nishitani A, Uemura T, Sato MH, et al. . A membrane trafficking pathway regulated by the plant-specific RAB GTPase ARA6. Nat Cell Biol 2011; 13:853 - 9; http://dx.doi.org/ 10.1038/ncb2270; PMID: 21666683 [DOI] [PubMed] [Google Scholar]
- 13.Kwon C, Neu C, Pajonk S, Yun HS, Lipka U, Humphry M, Bau S, Straus M, Kwaaitaal M, Rampelt H, et al. . Co-option of a default secretory pathway for plant immune responses. Nature 2008; 451:835 - 40; http://dx.doi.org/ 10.1038/nature06545; PMID: 18273019 [DOI] [PubMed] [Google Scholar]
- 14.El Kasmi F, Krause C, Hiller U, Stierhof YD, Mayer U, Conner L, Kong L, Reichardt I, Sanderfoot AA, Jürgens G. . SNARE complexes of different composition jointly mediate membrane fusion in Arabidopsis cytokinesis. Mol Biol Cell 2013; 24:1593 - 601; http://dx.doi.org/ 10.1091/mbc.E13-02-0074; PMID: 23515225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Uemura T, Ueda T, Ohniwa RL, Nakano A, Takeyasu K, Sato MH. . Systematic analysis of SNARE molecules in Arabidopsis: dissection of the post-Golgi network in plant cells. Cell Struct Funct 2004; 29:49 - 65; http://dx.doi.org/ 10.1247/csf.29.49; PMID: 15342965 [DOI] [PubMed] [Google Scholar]
- 16.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. . Basic local alignment search tool. J Mol Biol 1990; 215:403 - 10; PMID: 2231712 [DOI] [PubMed] [Google Scholar]
- 17.Goujon M, McWilliam H, Li W, Valentin F, Squizzato S, Paern J, Lopez R. . A new bioinformatics analysis tools framework at EMBL-EBI. Nucleic Acids Res 2010; 38:W695-9; http://dx.doi.org/ 10.1093/nar/gkq313; PMID: 20439314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Glez-Peña D, Gómez-Blanco D, Reboiro-Jato M, Fdez-Riverola F, Posada D. . ALTER: program-oriented conversion of DNA and protein alignments. Nucleic Acids Res 2010; 38:W14-8; http://dx.doi.org/ 10.1093/nar/gkq321; PMID: 20439312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Darriba D, Taboada GL, Doallo R, Posada D. . jModelTest 2: more models, new heuristics and parallel computing. Nat Methods 2012; 9:772; http://dx.doi.org/ 10.1038/nmeth.2109; PMID: 22847109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guindon S, Gascuel O. . A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol 2003; 52:696 - 704; http://dx.doi.org/ 10.1080/10635150390235520; PMID: 14530136 [DOI] [PubMed] [Google Scholar]
- 21.Drummond AJ, Rambaut A. . BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol 2007; 7:214; http://dx.doi.org/ 10.1186/1471-2148-7-214; PMID: 17996036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rambaut A, Drummond AJ. Tracer v1.4, Available from http://beast.bio.ed.ac.uk/Tracer. 2007.
- 23.Stamatakis A. . RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 2006; 22:2688 - 90; http://dx.doi.org/ 10.1093/bioinformatics/btl446; PMID: 16928733 [DOI] [PubMed] [Google Scholar]
- 24.Silvestro D, Michalak I. . raxmlGUI: a graphical front-end for RAxML. Org Divers Evol 2012; 12:335 - 7; http://dx.doi.org/ 10.1007/s13127-011-0056-0 [DOI] [Google Scholar]
- 25.Rambaut A. (2006-2009). FigTree: tree figure drawing tool. Version 1.3.1. http://tree.bio.ed.ac.uk/software/figtree/