Abstract
Transcription factors are DNA binding proteins that regulate gene expression. The nitrogen fixing symbiosis established between legume plants and soil bacteria is a complex interaction, in which plants need to integrate signals derived from the symbiont and the surrounding environment to initiate the developmental program of nodule organogenesis and the infection process. Several transcription factors that play critical roles in these processes have been reported in the past decade, including proteins of the GRAS and NF-Y families. Recently, we reported the characterization of a new GRAS domain containing-protein that interacts with a member of the C subunit of the NF-Y family, which plays an important role in nodule development and the progression of bacterial infection during the symbiotic interaction. The connection between transcription factors of these families highlights the significance of multimeric complexes in the fabulous capacity of plants to integrate and respond to multiple environmental stimuli.
Keywords: Nuclear Factor Y, legume, nitrogen fixation, nodulation, root development, transcription factors
The interaction between legumes and rhizobia has been the focus of intensive research over the last years. This symbiotic association is responsible for the majority of the nitrogen incorporation into biological systems and has the potential to reduce the chemical fertilization used in agricultural systems. In addition to the economical and social importance of the biological nitrogen fixation, this interaction between a eukaryotic and a prokaryotic organism is a fascinating biological system, in which mutual recognition leads to developmental changes in both symbionts. Low nitrogen availability in the soil, combined with the presence of compatible bacteria, triggers the development of a new root organ, the nodule, where nitrogen fixation takes place. Rhizobia reach the nodule through a tubular structure called the infection thread, where bacteria progress toward the cortical cells. These two genetic programs -nodule organogenesis and infection- are independent but strongly coordinated.1 Attempts to decipher the mechanisms underlying the mutualistic symbiosis between legume plants and rhizobia have resulted in the identification of a significant number of transcription factors that are required for nodulation, helping to understand the exquisite complexity of the plant response to nitrogen availability. In this work, we will briefly review the transcription factors already described in the genetic programs that are involved in the infection and nodule organogenesis, with emphasis in the recent discovery of a new GRAS regulator that participates in both, nitrogen-fixing symbiosis and lateral root growth.2
Transcriptional Regulators Involved in Symbiosis
Signal molecules secreted by rhizobia (like an oligo-polysaccharide called Nod Factor) can activate a signal transduction pathway that ultimately results in the activation of transcription factors in the nuclei of epidermal and cortical cells. These regulatory proteins recognize and bind to specific sequences of DNA located in the promoter of certain genes, controlling gene activity in response to environmental cues. Based on sequence similarity and the characteristics of their DNA-binding motifs, transcription factors have been classified into different families, which generally are conserved among eukaryotic organisms. Even though several transcription factors were identified by their capacity to bind to nodulin promoters, the most significant advances have aroused from genetic approaches by positional cloning of genes whose mutation altered infection or nodule development. The first transcription factor cloned in this way was NODULE INCEPTION (NIN).3 Other genes, such as Astray,4 Ethylene Response Factor Required for Nodulation 1 (ERN1) and ERN2,5,6 RR1,7 CYCLOPS,8 Nodulation Signaling Pathway 1 (NSP1) and NSP2,9,10 Nuclear Factor Y (NF-Y) A1 (formerly MtHap2–1)11 and NF-YC112 were later added to the list (Fig. 1). The last ones belong to the GRAS (NSP1 and NSP2) and NF-Y (NF-YA and NF-YC) families of transcriptional regulators.
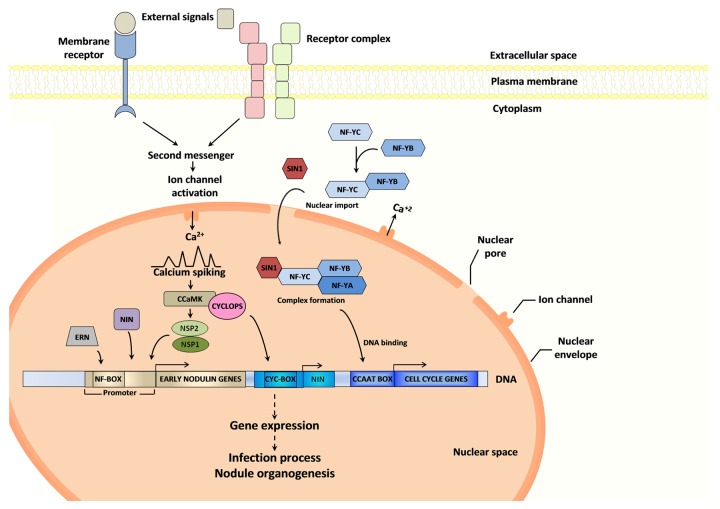
Figure 1. Transcription factors involved in nodulation. The nodulation signaling pathway is triggered when receptors located in the plasma membrane sense rhizobia derived signals. Upon receptor activation, oscillations of calcium concentration (calcium spiking) are produced in and around the nucleus. A nuclear located calcium and calmodulin dependent protein kinase (CCaMK) has been proposed as the protein that deciphers the calcium signal. Downstream from CCaMK, several transcriptional regulators are required for symbiosis establishment. CYCLOPS interacts and is phosphorylated by CCaMK, activating its capacity to recognize the NIN promoter. Nodulation signaling 1 (NSP1) and NSP2 both encode GRAS transcription factors that are essential for nodulation and activate the expression of early nodulation genes (nodulins). NIN and ERN are also involved in the transcriptional regulation of nodulins. Members of the NF-Y gene family are also required for nodulation. NF-YB would interact with NF-YC subunit in the cytoplasm prior to the nuclear import of the heterodimer. Once inside the nucleus, they interact with NF-YA to form the functional heterotrimer that binds with high affinity to CCAAT elements. SIN1 (a member of the GRAS family) physically interacts with the C subunit of the NF-Y transcription factor in the nucleus. This putative multimeric complex participates, directly or indirectly, in the transcriptional control of cell cycle genes.
NF-Y Transcriptional Regulators Involved in Symbiosis
NF-Ys are heterotrimeric complexes that bind to CCAAT boxes. They are composed by the NF-YA, NF-YB and NF-YC subunits. Whereas each subunit of NF-Y is encoded by one or two members in yeasts and mammals, these gene families have several members in plants. It is an open question whether this expansion has resulted in a functional diversification to cope with plant-specific responses. Individual NF-Y subunits have been shown to be involved in multiple developmental events and/or responses to environmental cues. In legumes, members of each subunit of NF-Y genes families have been implicated in the development of indeterminate (MtNF-YA1 in Medicago truncatula) and determinate nodules (LjNF-YA1 in Lotus japonicus and PvNF-YC1 in Phaseolus vulgaris). MtNF-YA1 was characterized as a symbiosis-specific transcription factor that plays a key role during nodule development by controlling meristem persistence.11 MtNF-YA1 gene expression is restricted to the meristematic zone of indeterminate nodules by a regulatory mechanism of mRNA decay involving miRNA169 at early stages of symbiosis. More recently, it was shown that expression of MtNF-YA1 is strongly upregulated during early stages of the symbiotic interaction, particularly in the root hairs of the infection zone. Moreover, mutant plants carrying a premature stop codon in the MtNF-YA1 gene form aberrant (i.e., thick and branched) infection threads that fail to progress to the cortical cells.13 On the other hand, Soyano et al. described that knockdown of LjNF-YA1 inhibited root nodule organogenesis, but not the infection process in L. japonicus.14 This report also showed that NIN is a direct regulator of the LjNF-YA1 and LjNF-YB1 genes. Interestingly, a recent report has shown that CYCLOPS is phosphorylated by the protein kinase CCaMK, exposing its DNA binding domain. In this conformation, CYCLOPS can recognize and bind to a specific region in the NIN promoter, initiating a transcriptional cascade that involves NF-YA1 and leads to nodule organogenesis.8
Reverse genetic studies performed in common bean (P. vulgaris) revealed that the NF-YC1 subunit is required for nodule organogenesis and rhizobial infection.12,15 Overexpression of NF-YC1 was sufficient to improve nodulation efficiency of a relatively poor competitive rhizobium strain, possibly trough activation of the G2/M transition cell cycle genes.12
The number of genes that encodes for each subunit of the NF-Y complex in plants results in a wide number of potential trimeric complexes (1690 in Arabidopsis).16 In addition, NF-Y complexes or individual subunits have been shown to associate with other transcriptional regulators, like SIN1 or transcription factors of the bZIP17 or MAD box families,18 further expanding the number of putative multimeric complexes that can be formed. This modular system provides an amazing versatility for the plant to integrate developmental programs with different environmental stimuli.
GRAS-Domain Transcription Factors and Their Roles in Symbiosis
The GRAS-type transcription factors constitute an important family of plant-specific proteins, whose initials come from the three members initially identified: Gibberellic-ACID INSENSITIVE (GAI), repressor of GAI (RGA) and SCARECROW (SCR).19 They play different roles in the development of stem and root, in the gibberellic acid signaling and in the signal transduction pathways of phytochromes A and B.20 In M. truncatula and L. japonicus, two proteins of this family, NSP1 and NSP2, were shown to be required for nodulation. They mediate different Nod Factor induced responses, such as root hair deformation, infection thread formation, cortical cell divisions and expression of nodulation genes known as nodulins.9,10,21 Interestingly, NSP1 and NSP2 can form homo- and hetero- dimers and associate to promoters of early induced nodulins.22 The complex formed by NSP1 and NSP2 can enhance the action of ERN on the transcriptional activity of the early nodulin 11 (ENOD11) during the progression of rhizobia infection.6 Both proteins are also involved in mycorrhization, a symbiotic interaction with fungi that is present in the majority of land plants.23 More recently, another GRAS protein, called RAM1 (Required for Arbuscular Mycorrhization 1), was shown to play a specific role in the formation of arbuscular mycorrhiza through its interaction with NSP2.24
A New Connection Between GRAS and NF-Ys
In a recent work, we identified a new GRAS-domain containing protein, named Scarecrow like 13 (SCL13) Involved in Nodulation (SIN1), which physically interacts with the C subunit of the NF-Y complex from common bean.2 The GRAS family has been divided into eight subfamilies. Unlike NSP1 and NSP2, which belongs to the SHORT ROOT (SHR) and HAIRY MERISTEM (HAM) subfamilies, respectively, SIN1 is part of the PAT1 subfamily and highly similar to Arabidopsis SCL13. Both in yeast and in planta experiments have established that the NF-YC1 subunit physically interacts with SIN1. SIN1 is expressed in aerial and root tissues, reaching higher levels in roots and mature nodules. Post-transcriptional gene silencing of SIN1 using RNA interference (RNAi) showed that the product of this gene plays a critical role in lateral root elongation and the establishment of successful symbiosis between P. vulgaris and Rhizobium etli.2 The symbiotic phenotype was similar to that observed in NF-YC1 silenced root, but the effect on nodule development and infection thread progression was comparatively milder.12,15 Interestingly, SIN1 silenced roots failed to upregulate mRNA levels of G2/M cell cycle genes and NF-YA1 in response to rhizobia infection. This suggests that SIN1 can act through its interaction with the NF-Y complex, but also might have a positive feedback on NF-Y gene regulation during nodule formation.
In addition to its role in nodulation, SIN1 showed to play a role during the elongation of lateral roots. This transcription factor with a dual role is an interesting connection between the developmental programs of two root organs, nodules and lateral roots. When LjNF-YA1 was overexpressed, the tips of lateral roots showed malformations, an effect that was enhanced by the co-expression with LjNF- YB1.14 This phenotype was accompanied by an increment of cell division in non-meristematic tissue, suggesting that the A1 and B1 subunits of LjNF-Y can modulate the activity of cell cycle genes. It would be interesting to explore the possible connections between NF-Y and GRAS transcription factors with the auxin/cytokinin balance, which are critical to initiate both lateral roots and nodules.25,26,27
Overexpression of SIN1 Affects Nodulation in Common Bean
As a complementary strategy to the post-transcriptional silencing, a fusion of SIN1 with the FLAG and HIS tags was ectopically expressed under the control of the CaMV 35S promoter. The nodulation phenotype of transformed roots was compared with control plants transformed with the plasmid p35S:GFPGUS (Fig. 2A). Unexpectedly, plants overexpressing SIN1 showed a reduced number of nodules per root as compared with control plants (Fig. 2B). Overexpression of SIN1 also affected the diameter of the nodules (Fig. 2C), producing a growth impair similar to that observed in RNAi roots. These results support the role of SIN1 in the nodulation process, specifically in the nodule organogenesis, and suggest that a fine-tuned balance between individual components of the same transcriptional complex is critical to maintain its function.
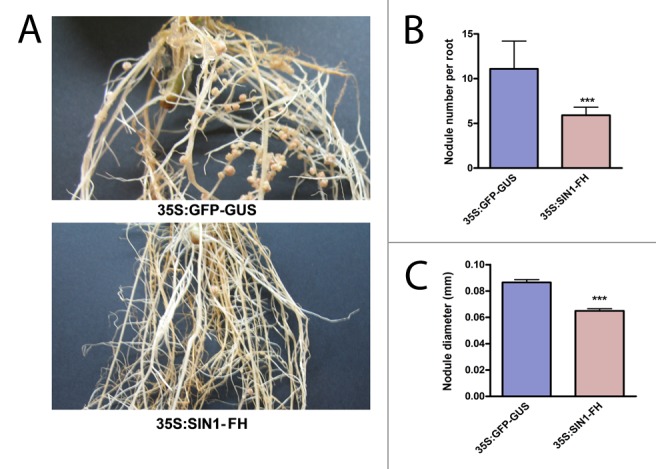
Figure 2. Overexpression of SIN1 resulted in the reduction of the number and size of root nodules. (A) Representative pictures illustrating the reduced nodule density and size found in 35S:SIN1-FH (FLAG-HIS) roots (upper panel) as compared with the control roots 35S:GFP-GUS (lower panel) at 17 dpi with R. etli. The complete open reading frame of SIN1 was cloned in the p35S:FH vector.28 Nodule number per root (B) and nodule size (C) developed in 35S:GFP-GUS or 35S:SIN1-FH roots at 17 dpi with R. etli. Error bars represent the SEM. Asterisks indicate significant differences with the control in an unpaired two-tailed t test with P < 0.001.
Concluding remarks and perspectives
The fact that plants of P. vulgaris with reduced or ectopic expression of SIN1 exhibited a significant reduction in the number and size of nodules formed by rhizobia raises the hypothesis that fine-tuned levels of SIN1 are required for an efficient performance of NF-YC1. NF-YC1 controls, directly or indirectly, the expression of G2/M transition cell cycle genes.12 SIN1 is also required for upregulation of these cell cycle genes in response to rhizobia.2 A key question is whether the complex formed by SIN1 and NF-YC1 can bind to promoters of these genes to regulate cell proliferation during nodule formation. Chromatin immunoprecipitation (ChIP) experiments followed by high throughput sequencing of isolated DNA fragments would certainly help to reveal downstream genes controlled by this complex during the interaction of legume roots with nitrogen fixing bacteria.
In addition to the role described in the plant response during symbiosis, SIN1 is also involved in lateral root elongation, suggesting that this is a shared component of two different root developmental programs. On the other hand, as previously mentioned, NF-Y subunits also have been implicated in these two developmental processes. It will be interesting to characterize at global scale the overlapping transcriptional networks governed by GRAS and NF-Y proteins during lateral root and nodule formation.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
This work was financially supported by grants from ANPCyT, Argentina (PICT 2008/0443 and 2010/2431) and by the ANR-09-BLAN-0033-01 HAPIHUB project. AN and MBaudin are part of ‘Laboratoire d’Excellence’ (LABEX) entitled TULIP (ANR-10-LABX-41). CR, JC, OMA, MEZ and FAB are funded by CONICET. MBaudin is funded by an INRA CJS (Contrat Jeune Scientifique) contract.
References
- 1.Oldroyd GE, Downie JA. Coordinating nodule morphogenesis with rhizobial infection in legumes. Annu Rev Plant Biol. 2008;59:519–46. doi: 10.1146/annurev.arplant.59.032607.092839. [DOI] [PubMed] [Google Scholar]
- 2.Battaglia M, et al. The NF-YC1 interacting protein SIN1, a member of the GRAS family, is required for nodule organogenesis, infection thread progression and lateral root growth. Plant Physiol. 2014;164:1430–42. doi: 10.1104/pp.113.230896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schauser L, Roussis A, Stiller J, Stougaard J. A plant regulator controlling development of symbiotic root nodules. Nature. 1999;402:191–5. doi: 10.1038/46058. [DOI] [PubMed] [Google Scholar]
- 4.Nishimura R, Ohmori M, Kawaguchi M. The novel symbiotic phenotype of enhanced-nodulating mutant of Lotus japonicus: astray mutant is an early nodulating mutant with wider nodulation zone. Plant Cell Physiol. 2002;43:853–9. doi: 10.1093/pcp/pcf098. [DOI] [PubMed] [Google Scholar]
- 5.Middleton PH, Jakab J, Penmetsa RV, Starker CG, Doll J, Kaló P, Prabhu R, Marsh JF, Mitra RM, Kereszt A, et al. An ERF transcription factor in Medicago truncatula that is essential for Nod factor signal transduction. Plant Cell. 2007;19:1221–34. doi: 10.1105/tpc.106.048264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cerri MR, Frances L, Laloum T, Auriac MC, Niebel A, Oldroyd GE, Barker DG, Fournier J, de Carvalho-Niebel F. Medicago truncatula ERN transcription factors: regulatory interplay with NSP1/NSP2 GRAS factors and expression dynamics throughout rhizobial infection. Plant Physiol. 2012;160:2155–72. doi: 10.1104/pp.112.203190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Plet J, Wasson A, Ariel F, Le Signor C, Baker D, Mathesius U, Crespi M, Frugier F. MtCRE1-dependent cytokinin signaling integrates bacterial and plant cues to coordinate symbiotic nodule organogenesis in Medicago truncatula. Plant J. 2011;65:622–33. doi: 10.1111/j.1365-313X.2010.04447.x. [DOI] [PubMed] [Google Scholar]
- 8.Singh S, Katzer K, Lambert J, Cerri M, Parniske M. CYCLOPS, a DNA-binding transcriptional activator, orchestrates symbiotic root nodule development. Cell Host Microbe. 2014;15:139–52. doi: 10.1016/j.chom.2014.01.011. [DOI] [PubMed] [Google Scholar]
- 9.Kaló P, Gleason C, Edwards A, Marsh J, Mitra RM, Hirsch S, Jakab J, Sims S, Long SR, Rogers J, et al. Nodulation signaling in legumes requires NSP2, a member of the GRAS family of transcriptional regulators. Science. 2005;308:1786–9. doi: 10.1126/science.1110951. [DOI] [PubMed] [Google Scholar]
- 10.Smit P, Raedts J, Portyanko V, Debellé F, Gough C, Bisseling T, Geurts R. NSP1 of the GRAS protein family is essential for rhizobial Nod factor-induced transcription. Science. 2005;308:1789–91. doi: 10.1126/science.1111025. [DOI] [PubMed] [Google Scholar]
- 11.Combier JP, Frugier F, de Billy F, Boualem A, El-Yahyaoui F, Moreau S, Vernié T, Ott T, Gamas P, Crespi M, et al. MtHAP2-1 is a key transcriptional regulator of symbiotic nodule development regulated by microRNA169 in Medicago truncatula. Genes Dev. 2006;20:3084–8. doi: 10.1101/gad.402806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zanetti ME, Blanco FA, Beker MP, Battaglia M, Aguilar OM. A C subunit of the plant nuclear factor NF-Y required for rhizobial infection and nodule development affects partner selection in the common bean-Rhizobium etli symbiosis. Plant Cell. 2010;22:4142–57. doi: 10.1105/tpc.110.079137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Laporte P, Lepage A, Fournier J, Catrice O, Moreau S, Jardinaud MF, Mun JH, Larrainzar E, Cook DR, Gamas P, et al. The CCAAT box-binding transcription factor NF-YA1 controls rhizobial infection. J Exp Bot. 2014;65:481–94. doi: 10.1093/jxb/ert392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Soyano T, Kouchi H, Hirota A, Hayashi M. Nodule inception directly targets NF-Y subunit genes to regulate essential processes of root nodule development in Lotus japonicus. PLoS Genet. 2013;9:e1003352. doi: 10.1371/journal.pgen.1003352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mazziotta L, Reynoso MA, Aguilar OM, Blanco FA, Zanetti ME. Transcriptional and functional variation of NF-YC1 in genetically diverse accessions of Phaseolus vulgaris during the symbiotic association with Rhizobium etli. Plant Biol (Stuttg) 2013;15:808–18. doi: 10.1111/j.1438-8677.2012.00683.x. [DOI] [PubMed] [Google Scholar]
- 16.Laloum T, De Mita S, Gamas P, Baudin M, Niebel A. CCAAT-box binding transcription factors in plants: Y so many? Trends Plant Sci. 2013;18:157–66. doi: 10.1016/j.tplants.2012.07.004. [DOI] [PubMed] [Google Scholar]
- 17.Liu JX, Howell SH. bZIP28 and NF-Y transcription factors are activated by ER stress and assemble into a transcriptional complex to regulate stress response genes in Arabidopsis. Plant Cell. 2010;22:782–96. doi: 10.1105/tpc.109.072173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Masiero S, Imbriano C, Ravasio F, Favaro R, Pelucchi N, Gorla MS, Mantovani R, Colombo L, Kater MM. Ternary complex formation between MADS-box transcription factors and the histone fold protein NF-YB. J Biol Chem. 2002;277:26429–35. doi: 10.1074/jbc.M202546200. [DOI] [PubMed] [Google Scholar]
- 19.Pysh LD, Wysocka-Diller JW, Camilleri C, Bouchez D, Benfey PN. The GRAS gene family in Arabidopsis: sequence characterization and basic expression analysis of the SCARECROW-LIKE genes. Plant J. 1999;18:111–9. doi: 10.1046/j.1365-313X.1999.00431.x. [DOI] [PubMed] [Google Scholar]
- 20.Bolle C. The role of GRAS proteins in plant signal transduction and development. Planta. 2004;218:683–92. doi: 10.1007/s00425-004-1203-z. [DOI] [PubMed] [Google Scholar]
- 21.Heckmann AB, Lombardo F, Miwa H, Perry JA, Bunnewell S, Parniske M, Wang TL, Downie JA. Lotus japonicus nodulation requires two GRAS domain regulators, one of which is functionally conserved in a non-legume. Plant Physiol. 2006;142:1739–50. doi: 10.1104/pp.106.089508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hirsch S, Kim J, Muñoz A, Heckmann AB, Downie JA, Oldroyd GE. GRAS proteins form a DNA binding complex to induce gene expression during nodulation signaling in Medicago truncatula. Plant Cell. 2009;21:545–57. doi: 10.1105/tpc.108.064501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu W, Kohlen W, Lillo A, Op den Camp R, Ivanov S, Hartog M, Limpens E, Jamil M, Smaczniak C, Kaufmann K, et al. Strigolactone biosynthesis in Medicago truncatula and rice requires the symbiotic GRAS-type transcription factors NSP1 and NSP2. Plant Cell. 2011;23:3853–65. doi: 10.1105/tpc.111.089771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gobbato E, Marsh JF, Vernié T, Wang E, Maillet F, Kim J, Miller JB, Sun J, Bano SA, Ratet P, et al. A GRAS-type transcription factor with a specific function in mycorrhizal signaling. Curr Biol. 2012;22:2236–41. doi: 10.1016/j.cub.2012.09.044. [DOI] [PubMed] [Google Scholar]
- 25.Mathesius U. Goldacre paper: Auxin: at the root of nodule development? Funct Plant Biol. 2008;35:651–68. doi: 10.1071/FP08177. [DOI] [PubMed] [Google Scholar]
- 26.Lavenus J, Goh T, Roberts I, Guyomarc’h S, Lucas M, De Smet I, Fukaki H, Beeckman T, Bennett M, Laplaze L. Lateral root development in Arabidopsis: fifty shades of auxin. Trends Plant Sci. 2013;18:450–8. doi: 10.1016/j.tplants.2013.04.006. [DOI] [PubMed] [Google Scholar]
- 27.Oldroyd GE, Murray JD, Poole PS, Downie JA. The rules of engagement in the legume-rhizobial symbiosis. Annu Rev Genet. 2011;45:119–44. doi: 10.1146/annurev-genet-110410-132549. [DOI] [PubMed] [Google Scholar]
- 28.Zanetti ME, Chang IF, Gong F, Galbraith DW, Bailey-Serres J. Immunopurification of polyribosomal complexes of Arabidopsis for global analysis of gene expression. Plant Physiol. 2005;138:624–35. doi: 10.1104/pp.105.059477. [DOI] [PMC free article] [PubMed] [Google Scholar]